Effectiveness of Bioinks and the Clinical Value of 3D Bioprinted Glioblastoma Models: A Systematic Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Study Selection and Eligibility Criteria
2.3. Data Extraction
2.4. Quality Assessment
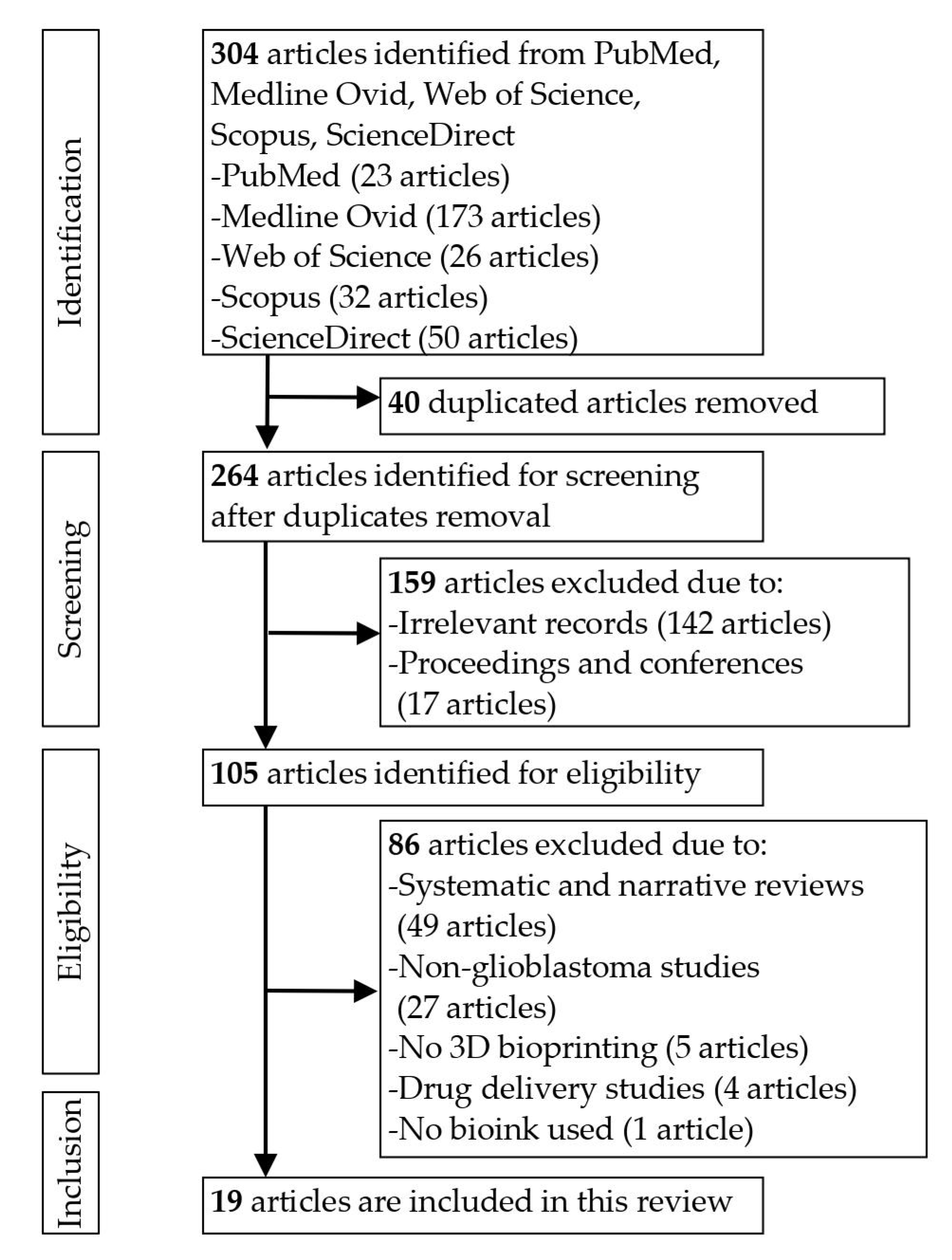
3. Results
3.1. Quality Evaluation
3.2. Characteristics of the Included Studies
3.2.1. Cell and Animal Models
3.2.2. Bioinks, 3D Bioprinting and Crosslinking Methods
3.3. Physical Properties and Biocompatibility Measures
3.4. Drug Response
4. Discussion
4.1. Overview of the Included Studies
4.2. Bioink Materials and Combination
4.3. Physical Properties of the Bioink Scaffolds
4.4. Biocompatibility and Cellular Response
4.5. Drug Response and Clinical Value
4.6. Study Limitations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stanković, T.; Ranđelović, T.; Dragoj, M.; Stojković Burić, S.; Fernández, L.; Ochoa, I.; Pérez-García, V.M.; Pešić, M. In vitro biomimetic models for glioblastoma-a promising tool for drug response studies. Drug Resist. Updat. 2021, 55, 100753. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Oliva, R.; Dominguez-Garcia, S.; Carrascal, L.; Abalos-Martinez, J.; Pardillo-Diaz, R.; Verastegui, C.; Castro, C.; Nunez-Abades, P.; Geribaldi-Doldan, N. Evolution of Experimental Models in the Study of Glioblastoma: Toward Finding Efficient Treatments. Front. Oncol. 2021, 10, 614295. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.L.; Ma, C.; Lan, Q.; Xu, T. 3D bioprinted glioma stem cells for brain tumor model and applications of drug susceptibility. Biofabrication 2016, 8, 045005. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro-Oncology 2019, 21, v1–v100. [Google Scholar] [CrossRef] [PubMed]
- Allahdini, F.; Amirjamshidi, A.; Reza-Zarei, M.; Abdollahi, M. Evaluating the prognostic factors effective on the outcome of patients with glioblastoma multiformis: Does maximal resection of the tumor lengthen the median survival? World Neurosurg. 2010, 73, 128–134. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Taylor, O.G.; Brzozowski, J.S.; Skelding, K.A. Glioblastoma Multiforme: An Overview of Emerging Therapeutic Targets. Front. Oncol. 2019, 9, 963. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Abelseth, E.; de la Vega, L.; Willerth, S.M. Bioprinting a novel glioblastoma tumor model using a fibrin-based bioink for drug screening. Mater. Today Chem. 2019, 12, 78–84. [Google Scholar] [CrossRef]
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of clinical trial success rates and related parameters. Biostatistics 2019, 20, 273–286. [Google Scholar] [CrossRef]
- Melchels, F.P.W.; Domingos, M.A.N.; Klein, T.J.; Malda, J.; Bartolo, P.J.; Hutmacher, D.W. Additive manufacturing of tissues and organs. Prog. Polym. Sci. 2012, 37, 1079–1104. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.Y.; Lee, H.; Kim, Y.; Yoo, S.Y.; Chung, W.J.; Kim, G. Phage as versatile nanoink for printing 3-D cell-laden scaffolds. Acta Biomater. 2016, 29, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.F.; Zayats, M.; Guerrero-Cazares, H.; Quiñones-Hinojosa, A.; Searson, P.C. Influence of basement membrane proteins and endothelial cell-derived factors on the morphology of human fetal-derived astrocytes in 2D. PLoS ONE 2014, 9, e92165. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef] [PubMed]
- Tufanaru, C.; Munn, Z.; Aromataris, E.; Campbell, J.; Hopp, L. Chapter 3: Systematic Reviews of Effectiveness. In JBI Manual for Evidence Synthesis; Aromataris, E., Munn, Z., Eds.; JBI: Adelaide, SA, Australia, 2020. [Google Scholar]
- Wang, X.; Dai, X.; Zhang, X.; Ma, C.; Li, X.; Xu, T.; Lan, Q. 3D bioprinted glioma cell-laden scaffolds enriching glioma stem cells via epithelial–mesenchymal transition. J. Biomed. Mater. Res.-Part A 2019, 107, 383–391. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Ding, J.; Long, X.; Zhang, H.; Zhang, X.; Jiang, X.; Xu, T. 3D bioprinted glioma microenvironment for glioma vascularization. J. Biomed. Mater. Res.-Part A 2021, 109, 915–925. [Google Scholar] [CrossRef]
- Han, S.; Kim, S.; Chen, Z.; Shin, H.K.; Lee, S.Y.; Moon, H.E.; Paek, S.H.; Park, S. 3D bioprinted vascularized tumour for drug testing. Int. J. Mol. Sci. 2020, 21, 2993. [Google Scholar] [CrossRef] [Green Version]
- Haring, A.P.; Thompson, E.G.; Hernandez, R.D.; Laheri, S.; Harrigan, M.E.; Lear, T.; Sontheimer, H.; Johnson, B.N. 3D Printed Multiplexed Competitive Migration Assays with Spatially Programmable Release Sources. Adv. Biosyst. 2020, 4, e1900225. [Google Scholar] [CrossRef]
- Heinrich, M.A.; Bansal, R.; Lammers, T.; Zhang, Y.S.; Michel Schiffelers, R.; Prakash, J. 3D-Bioprinted Mini-Brain: A Glioblastoma Model to Study Cellular Interactions and Therapeutics. Adv. Mater. 2019, 31, e1806590. [Google Scholar] [CrossRef]
- Tricinci, O.; De Pasquale, D.; Marino, A.; Battaglini, M.; Pucci, C.; Ciofani, G. A 3D Biohybrid Real-Scale Model of the Brain Cancer Microenvironment for Advanced In Vitro Testing. Adv. Mater. Technol. 2020, 5, 2000540. [Google Scholar] [CrossRef]
- Utama, R.H.; Atapattu, L.; O’Mahony, A.P.; Fife, C.M.; Baek, J.; Allard, T.; O’Mahony, K.J.; Ribeiro, J.C.C.; Gaus, K.; Kavallaris, M.; et al. A 3D Bioprinter Specifically Designed for the High-Throughput Production of Matrix-Embedded Multicellular Spheroids. iScience 2020, 23, 101621. [Google Scholar] [CrossRef]
- Yi, H.G.; Jeong, Y.H.; Kim, Y.; Choi, Y.J.; Moon, H.E.; Park, S.H.; Kang, K.S.; Bae, M.; Jang, J.; Youn, H.; et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, X.; Dai, X.; Zhang, X.; Zhang, J.; Xu, T.; Lan, Q. Bioprinting of glioma stem cells improves their endotheliogenic potential. Colloids Surf. B Biointerfaces 2018, 171, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, X.; Dai, X.; Zhang, X.; Zhang, J.; Xu, T.; Lan, Q. Coaxial extrusion bioprinted shell-core hydrogel microfibers mimic glioma microenvironment and enhance the drug resistance of cancer cells. Colloids Surf. B Biointerfaces 2018, 171, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Promjantuek, W.; Heebkaew, N.; Rujanapun, N.; Noisa, P. Fabrication of 3D calcium-alginate scaffolds for human glioblastoma modeling and anticancer drug response evaluation. J. Cell. Physiol. 2019, 234, 20085–20097. [Google Scholar] [CrossRef]
- Bakirci, E.; Schaefer, N.; Dahri, O.; Hrynevich, A.; Strissel, P.; Strick, R.; Dalton, P.D.; Villmann, C. Melt Electrowritten In Vitro Radial Device to Study Cell Growth and Migration. Adv. Biosyst. 2020, 4, e2000077. [Google Scholar] [CrossRef]
- Smits, I.P.M.M.; Blaschuk, O.W.; Willerth, S.M. Novel N-cadherin antagonist causes glioblastoma cell death in a 3D bioprinted co-culture model. Biochem. Biophys. Res. Commun. 2020, 529, 162–168. [Google Scholar] [CrossRef]
- Tang, M.; Tiwari, S.K.; Agrawal, K.; Tan, M.; Dang, J.; Tam, T.; Tian, J.; Wan, X.; Schimelman, J.; You, S.; et al. Rapid 3D Bioprinting of Glioblastoma Model Mimicking Native Biophysical Heterogeneity. Small 2021, 17, e2006050. [Google Scholar] [CrossRef]
- Chadwick, M.; Yang, C.; Liu, L.; Gamboa, C.M.; Jara, K.; Lee, H.; Sabaawy, H.E. Rapid Processing and Drug Evaluation in Glioblastoma Patient-Derived Organoid Models with 4D Bioprinted Arrays. iScience 2020, 23, 101365. [Google Scholar] [CrossRef]
- Hermida, M.A.; Kumar, J.D.; Schwarz, D.; Laverty, K.G.; Di Bartolo, A.; Ardron, M.; Bogomolnijs, M.; Clavreul, A.; Brennan, P.M.; Wiegand, U.K.; et al. Three dimensional in vitro models of cancer: Bioprinting multilineage glioblastoma models. Adv. Biol. Regul. 2020, 75, 100658. [Google Scholar] [CrossRef]
- Tang, M.; Xie, Q.; Gimple, R.C.; Zhong, Z.; Tam, T.; Tian, J.; Kidwell, R.L.; Wu, Q.; Prager, B.C.; Qiu, Z.; et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 2020, 30, 833–853. [Google Scholar] [CrossRef]
- Lee, S.Y.; Ma, J.; Khoo, T.S.; Abdullah, N.; Nik Md Noordin Kahar, N.N.F.; Abdul Hamid, Z.A.; Mustapha, M. Polysaccharide-Based Hydrogels for Microencapsulation of Stem Cells in Regenerative Medicine. Front. Bioeng. Biotechnol. 2021, 9, 735090. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.A.; Madlambayan, G.; Mooney, D.J. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials 1999, 20, 45–53. [Google Scholar] [CrossRef]
- Andersen, T.; Auk-Emblem, P.; Dornish, M. 3D Cell Culture in Alginate Hydrogels. Microarrays 2015, 4, 133–161. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.A.; Mooney, D.J. Alginate type and RGD density control myoblast phenotype. J. Biomed. Mater. Res. 2002, 60, 217–223. [Google Scholar] [CrossRef]
- Kuen, Y.L.; Alsberg, E.; Hsiong, S.; Comisar, W.; Linderman, J.; Ziff, R.; Mooney, D. Nanoscale Adhesion Ligand Organization Regulates Osteoblast Proliferation and Differentiation. Nano Lett. 2004, 4, 1501–1506. [Google Scholar] [CrossRef] [Green Version]
- Edgar, J.M.; Robinson, M.; Willerth, S.M. Fibrin hydrogels induce mixed dorsal/ventral spinal neuron identities during differentiation of human induced pluripotent stem cells. Acta Biomater. 2017, 51, 237–245. [Google Scholar] [CrossRef]
- Liu, J.; Tan, Y.; Zhang, H.; Zhang, Y.; Xu, P.; Chen, J.; Poh, Y.C.; Tang, K.; Wang, N.; Huang, B. Soft fibrin gels promote selection and growth of tumorigenic cells. Nat. Mater. 2012, 11, 734–741. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.; Douglas, S.; Willerth, S.M. Mechanically stable fibrin scaffolds promote viability and induce neurite outgrowth in neural aggregates derived from human induced pluripotent stem cells. Sci. Rep. 2017, 7, 6250. [Google Scholar] [CrossRef]
- Weisel, J.W.; Litvinov, R.I. Mechanisms of fibrin polymerization and clinical implications. Blood 2013, 121, 1712–1719. [Google Scholar] [CrossRef] [Green Version]
- Butler, M.F.; Ng, Y.F.; Pudney, P.D.A. Mechanism and kinetics of the crosslinking reaction between biopolymers containing primary amine groups and genipin. J. Polym. Sci. Part A Polym. Chem. 2003, 41, 3941–3953. [Google Scholar] [CrossRef]
- Chen, H.J.; Wei, Z.; Sun, J.; Bhattacharya, A.; Savage, D.J.; Serda, R.; Mackeyev, Y.; Curley, S.A.; Bu, P.; Wang, L.; et al. A recellularized human colon model identifies cancer driver genes. Nat. Biotechnol. 2016, 34, 845–851. [Google Scholar] [CrossRef] [Green Version]
- Dunne, L.W.; Huang, Z.; Meng, W.; Fan, X.; Zhang, N.; Zhang, Q.; An, Z. Human decellularized adipose tissue scaffold as a model for breast cancer cell growth and drug treatments. Biomaterials 2014, 35, 4940–4949. [Google Scholar] [CrossRef]
- Wolf, K.J.; Chen, J.; Coombes, J.D.; Aghi, M.K.; Kumar, S. Dissecting and rebuilding the glioblastoma microenvironment with engineered materials. Nat. Rev. Mater. 2019, 4, 651–668. [Google Scholar] [CrossRef]
- Pedron, S.; Hanselman, J.S.; Schroeder, M.A.; Sarkaria, J.N.; Harley, B.A.C. Extracellular Hyaluronic Acid Influences the Efficacy of EGFR Tyrosine Kinase Inhibitors in a Biomaterial Model of Glioblastoma. Adv. Healthc. Mater. 2017, 6, 1700529. [Google Scholar] [CrossRef]
- Chen, L.; Xiao, Z.; Meng, Y.; Zhao, Y.; Han, J.; Su, G.; Chen, B.; Dai, J. The enhancement of cancer stem cell properties of MCF-7 cells in 3D collagen scaffolds for modeling of cancer and anti-cancer drugs. Biomaterials 2012, 33, 1437–1444. [Google Scholar] [CrossRef]
- Druecke, D.; Langer, S.; Lamme, E.; Pieper, J.; Ugarkovic, M.; Steinau, H.U.; Homann, H.H. Neovascularization of poly(ether ester) block-copolymer scaffolds in vivo: Long-term investigations using intravital fluorescent microscopy. J. Biomed. Mater. Res. Part A 2004, 68A, 10–18. [Google Scholar] [CrossRef]
- Heidenreich, A.C.; Pérez-Recalde, M.; González Wusener, A.; Hermida, É.B. Collagen and chitosan blends for 3D bioprinting: A rheological and printability approach. Polym. Test. 2020, 82, 106297. [Google Scholar] [CrossRef]
- Liu, P.; Shen, H.; Zhi, Y.; Si, J.; Shi, J.; Guo, L.; Shen, S.G. 3D bioprinting and in vitro study of bilayered membranous construct with human cells-laden alginate/gelatin composite hydrogels. Colloids Surf. B Biointerfaces 2019, 181, 1026–1034. [Google Scholar] [CrossRef]
- Xu, C.; Zhang Molino, B.; Wang, X.; Cheng, F.; Xu, W.; Molino, P.; Bacher, M.; Su, D.; Rosenau, T.; Willför, S.; et al. 3D printing of nanocellulose hydrogel scaffolds with tunable mechanical strength towards wound healing application. J. Mater. Chem. B 2018, 6, 7066–7075. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef]
- Jhala, D.; Vasita, R. A Review on Extracellular Matrix Mimicking Strategies for an Artificial Stem Cell Niche. Polym. Rev. 2015, 55, 561–595. [Google Scholar] [CrossRef]
- Banerjee, A.; Arha, M.; Choudhary, S.; Ashton, R.S.; Bhatia, S.R.; Schaffer, D.V.; Kane, R.S. The influence of hydrogel modulus on the proliferation and differentiation of encapsulated neural stem cells. Biomaterials 2009, 30, 4695–4699. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Mouw, J.K.; Weaver, V.M. Forcing form and function: Biomechanical regulation of tumor evolution. Trends Cell Biol. 2011, 21, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Florczyk, S.J.; Kievit, F.M.; Wang, K.; Erickson, A.E.; Ellenbogen, R.G.; Zhang, M. 3D porous chitosan-alginate scaffolds promote proliferation and enrichment of cancer stem-like cells. J. Mater. Chem. B 2016, 4, 6326–6334. [Google Scholar] [CrossRef]
- Wang, C.; Tong, X.; Yang, F. Bioengineered 3D brain tumor model to elucidate the effects of matrix stiffness on glioblastoma cell behavior using peg-based hydrogels. Mol. Pharm. 2014, 11, 2115–2125. [Google Scholar] [CrossRef]
- Polacheck, W.J.; Zervantonakis, I.K.; Kamm, R.D. Tumor cell migration in complex microenvironments. Cell. Mol. Life Sci. 2013, 70, 1335–1356. [Google Scholar] [CrossRef] [Green Version]
- Chauvet, D.; Imbault, M.; Capelle, L.; Demene, C.; Mossad, M.; Karachi, C.; Boch, A.L.; Gennisson, J.L.; Tanter, M. In Vivo Measurement of Brain Tumor Elasticity Using Intraoperative Shear Wave Elastography. Ultraschall Med. 2016, 37, 584–590. [Google Scholar] [CrossRef]
- Netti, P.A.; Baxter, L.T.; Boucher, Y.; Skalak, R.; Jain, R.K. Time-dependent behavior of interstitial fluid pressure in solid tumors: Implications for drug delivery. Cancer Res. 1995, 55, 5451–5458. [Google Scholar]
- Awad, O.; Yustein, J.T.; Shah, P.; Gul, N.; Katuri, V.; O’Neill, A.; Kong, Y.; Brown, M.L.; Toretsky, J.A.; Loeb, D.M. High ALDH activity identifies chemotherapy-resistant Ewing’s sarcoma stem cells that retain sensitivity to EWS-Fli1 inhibition. PLoS ONE 2010, 5, e13943. [Google Scholar] [CrossRef] [Green Version]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [Green Version]
- Hegi, M.E.; Diserens, A.-C.; Godard, S.; Dietrich, P.-Y.; Regli, L.; Ostermann, S.; Otten, P.; Van Melle, G.; De Tribolet, N.; Stupp, R. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 2004, 10, 1871–1874. [Google Scholar] [CrossRef] [Green Version]
- Vecchio, D.; Daga, A.; Carra, E.; Marubbi, D.; Baio, G.; Neumaier, C.E.; Vagge, S.; Corv, R.; Brisigotti, M.P.; Ravetti, J.L.; et al. Predictability, efficacy and safety of radiosensitization of glioblastoma-initiating cells by the ATM inhibitor KU-60019. Int. J. Cancer 2014, 135, 479–491. [Google Scholar] [CrossRef]
| References | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A: Low Risk of Bias B: High Risk of Bias C: Not Clear D: Not Applicable | Wang et al. [15] | Wang et al. [16] | Dai et al. [3] | Han et al. [17] | Haring et al. [18] | Heinrich et al. [19] | Tricinci et al. [20] | Utama et al. [21] | Yi et al. [22] | Lee et al. [8] | Wang et al. [23] | Wang et al. [24] | Chaicharoenau-domrung et al. [25] | Ba-kirci et al. [26] | Smits et al. [27] | Tang et al. [28] | Chadwick et al. [29] | Hermida et al. [30] | Tang et al. [31] |
| Checklist | |||||||||||||||||||
| No confusion about which variable comes first | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| The subjects involved in any of the comparisons were comparable | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| Other than the exposure or intervention of interest, the subjects involved in any comparisons received similar treatment/care | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| There was a control group | A | A | A | A | A | A | A | A | A | A | C | A | A | C | A | A | A | A | A |
| Multiple outcome assessments taken before and after the intervention/exposure | A | A | A | A | A | A | A | A | A | A | A | A | A | A | B | A | A | A | A |
| Completed follow-up | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D | D |
| Participants’ results measured in the same way in any comparisons | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| Reliable outcomes measured | A | A | A | C | A | A | C | A | A | C | A | A | A | A | A | A | A | A | A |
| Appropriate statistical analysis | A | A | A | A | A | A | C | A | A | A | A | A | A | A | A | A | A | A | A |
| Bioinks | Cells | Study Design | Printing Method | Crosslinking Methods | Drugs | Ref. |
|---|---|---|---|---|---|---|
| Gelatin, alginate, fibrinogen (GAF), transglutaminase | Human glioma cell line U118 | In vitro and in vivo | Extrusion | Scaffolds were immersed in calcium chloride (CaCl2) solution for 3 min and then thrombin for 15 min after printing | TMZ | [15] |
| Sodium alginate and gelatin | Human glioma cell line U118 and human glioma stem cell GSC23 | In vitro and in vivo | Extrusion | Scaffolds were immersed in CaCl2 solution for 3 min after printing | N/A | [16] |
| GAF | Glioma stem cell line SU3 and human glioblastoma cell line U87 | In vitro | Extrusion | Transglutaminase was added to a hydrogel system. Scaffolds were first immersed in CaCl2 and then thrombin after printing | TMZ | [3] |
| GAF | Human glioblastoma cell line U87, human vascular endothelial cells (HUVECs) and lung fibroblasts (LFs) | In vitro | Not mentioned | Scaffolds were immersed in CaCl2 solution for 3 min and then thrombin for 15 min after printing | TMZ, sunitinib (SU) | [17] |
| Alginate solution | Human glioblastoma cells D54-MG | In vitro | Extrusion | Calcium carbonate was added to a hydrogel system | N/A | [18] |
| GelMA | Mouse glioblastoma cells GL261 | In vitro | Extrusion | Photocrosslink | Carmustine (BCNU), AS1517499, BLZ945 | [19] |
| Magnetically-responsive cage-like scaffolds (MRCSs) | Human glioblastoma cell line U87, GFP-expressing U87 and human cerebral microvascular endothelial cell line (hCMEC/D3) | In vitro | Two-photon lithography | Photocrosslink | Antibody-functionalised nutlin-loaded nanostructured lipid carriers (Ab-Nut-NLCs) | [20] |
| Sodium alginate | Neuroblastoma SK-N-BE (2) and human glioblastoma cell line U87vIII | In vitro | Droplet-based bioprinting | CaCl2 | Doxorubicin | [21] |
| Brain decellularised ECM or collagen | Human glioblastoma cell line U87, patient-derived glioblastoma and HUVECs | In vitro | Glioblastoma-on-a-chip | No crosslinking applied | TMZ, cisplatin (CIS), KU60019 (KU), O6-benzylguanine (O6BG), methoxyamine (MX) | [22] |
| Fibrin, alginate, genipin | Human glioblastoma cell line U87 | In vitro | Microfluidic extrusion | CaCl2, chitosan, thrombin were mixed and connected to the bioprinter through the ‘cross-linker’ pneumatic channel | N/A | [8] |
| GAF, transglutaminase | Human glioma stem cell GSC23 | In vitro | Extrusion | Scaffolds were immersed in CaCl2 solution for 3 min and then thrombin for 15 min after printing | N/A | [23] |
| Sodium alginate | Human glioma cell line U118 and human glioma stem cells GSC23 | In vitro | Extrusion | CaCl2 was used as printing receiving platform | TMZ | [24] |
| Sodium alginate | Human glioblastoma cells U-251 | In vitro | Not mentioned | The scaffolds were crosslinked with 2% CaCl2 solution for 1 hr | Cordycepin, doxorubicin | [25] |
| Matrigel | Human glioblastoma cell line U87 | In vitro | Melt electrowriting | No crosslinking applied | N/A | [26] |
| Fibrinogen, alginate, genipin | Human glioblastoma cell line U87 and human astrocytes | In vitro | Extrusion | CaCl2, chitosan, thrombin | Compound 15 (N-cadherin antagonist) | [27] |
| GMHA and GelMA | Human patient-derived GSCs (TS576) and HUVECs | In vitro | Digital light processing | Photocrosslink with rapid polymerisation of each region with 20–30 s of light exposure | TMZ | [28] |
| PEGDA and BPADMA | Patient-derived glioblastoma cells | In vitro | Projection micro-stereolithography | Photocrosslink by using phenylbis (2,4,6-trimethylbenzoyl) phosphine as a photoinitiator to initiate polymerisation and Sudan I as a photo absorber to control UV light penetration | TMZ plus BEZ235 or niraparib plus BEZ235 | [29] |
| RGD-alginate, HA and collagen-1 | Human glioblastoma cell line U87, monocytic (MM6), glioblastoma stem cell line (G7, G144 and G166) | In vitro | Extrusion | CaCl2 was used as a crosslinking agent for 3 min | CIS and TMZ | [30] |
| GelMA and GMHA | Patient-derived GSCs, macrophages, astrocytes, and neural stem cells (NSCs) | In vitro and in vivo | Digital light processing | Photocrosslink with exposure time of 20 s for the core and 15 s for the periphery | Abiraterone, vemurafenib, ifosfamide, erlotinib, gefitinib, TMZ | [31] |
| Bioinks | Physical Characteristic | Cell Morphology | Biological Characteristic | Drug Response | Ref. |
|---|---|---|---|---|---|
| Gelatin, alginate, fibrinogen (GAF), transglutaminase | N/A | In vivo: Tumours formed by 3D cultured cells were larger than those formed by 2D cells on day 42 | Cell viability: Day 0: 89.06 ± 3.58% Day 15: 84.30 ± 2.67% | After treatment with TMZ, the viability of cells in 2D and 3D culture began to decrease; 3D cultured cells showed higher viability | [15] |
| Sodium alginate and gelatin | N/A | Diameters of tumour cell spheroids formed: GSC23: 27.13 ± 2.59 μm U118: 21.71 ± 1.43 μm In vivo: 3D-U118 and 3D-GSC23 had an outer capsule on their surface with numerous blood vessels. | Cell viability: U118: 2h: 84.28 ± 2.15% Day 15: 85.36 ± 1.82% GSC23: 2h: 83.79 ± 3.08% Day 15: 87.85 ± 2.32% | N/A | [16] |
| GAF | Scaffold swelling ratio: Crosslinked by TG: 518.18 ± 60.58% Crosslinked by CaCl2: 501.85 ± 62.31% Pore diameter: 2–4 µm | Cells developed into spheroids after three weeks and pushed the surrounding hydrogels aside to take up more space within the scaffolds. | Live/dead cell ratio: 86.92% | Growth rates (1600 µg/mL TMZ for 48h): SU3: 3D: 107.20 ± 4.94% 2D: 72.73 ± 3.38% U87: 3D: 87.85 ± 4.57% 2D: 39.07 ± 3.57% | [3] |
| GAF | Storage moduli: 0.7–9 kPa Loss moduli: 0.06–1.7 kPa | Average diameter of multicellular tumour spheroids after 3 days: ~250 µm. | The viability of the HUVECs and LFs: >90% after bioprinting, >80% on day seven. | TMZ or SU significantly reduced the size of MCTS. Both TMZ and SU together further reduced the tumour size. | [17] |
| Alginate solution | Penetration time: 25–100 ng/mL EGF: 2.8–2.5 h 50–400 μM BK: 0.5–0.2 h | N/A | 46% more glioblastoma cells migrate toward EGF. EGF: 20.7 ± 3.2% BK: 14.2 ± 2.9% | N/A | [18] |
| GelMA | G′: 1000 Pa; storage modulus: 10–20 Pa. G′ and G″ remained relatively stable with increasing shear rate. Average pore size: |17.08 ± 6.7 µm | Compared to empty wells, tumour cells showed a significantly higher migration toward RAW264.7 macrophages | Both RAW264.7 and GL261 cells remained viable for days 10 post-printing, and the cell-laden constructs displayed high metabolic activity. | IC50 of BCNU: 2D cell culture: 139 µM 3D mono-cultured GL261 cells: 581 µM 3D co-cultured RAW264.7: 887 µM. Tumours isolated from co-cultured treated with BLZ945, but not with AS1517499, showed slow growth. | [19] |
| Magnetically-responsive cage-like scaffolds (MRCSs) | N/A | One spheroid developed per MRCS after 5 days of growing | Immunofluorescence analysis against Ki-67 marker: the external layers of cells were in the interphase of the cell cycle, while the inner part cells were quiescent | About 70% of the GB cells inside the microcage were positive for ethidium homodimer-1. | [20] |
| Sodium alginate | N/A | Cells formed in a dense ball by using an individual alginate droplet | Cell viability: immediately pre-print and post-print: >98% after 72 h: >95% Percentage of cleaved caspase-3-positive cells: 3D bioprinted: 0.4% manual spheroid:1.7% | IC50 of doxorubicin: 1.06 to 1.48 mM | [21] |
| Brain decellularised ECM or collagen | N/A | GBM-28 cells showed increased invasion and a more spindle-like morphology in the BdECM gel than the collagen gel. | Both hydrogels demonstrated >90% cell viability, but proliferation was higher in the BdECM gel than the collagen gel after 10 days. | Survival percentage: GBM-28: CIS < TMZ GBM-37: slight decrease after CIS treatment Responsive to drug: CIS + KU, O6BG + MX, CIS + KU + O6BG with radiation: GBM-28-on-a-chip > GBM-37-on-a-chip O6BG was the most effective in suppressing the GBMs-on-chips. | [22] |
| Fibrin, alginate, genipin | N/A | Cells tended to form spheroids within the scaffolds and tended to grow in size and density with increased time within the scaffold. | Live/dead imaging: 88.78% ± 2.92% (post-printing) Cell viability: Day 1: 98.09% ± 0.89% Day 6: 91.78% ± 5.96% Day 9: 83.93% ± 5.75% Day 12: 86.12% ± 5.09% | N/A | [8] |
| GAF, transglutaminase | Pore size: 338.41 ± 23.18 μm Filament diameter: 324.27 ± 30.98 μm | Cells in 3D scaffolds gradually proliferated to form spheroids with full, uniform shapes and pushed the surrounding hydrogels away | Cell viability: after bioprinting: 86.27 ± 2.41% Day 15: 89.39 ± 1.86% | N/A | [23] |
| Sodium alginate | N/A | Core-U118 cells gradually proliferated into spheroids connected with each other until the formation of fiber-like cell aggregates. | Shell-GSC23/core-U118 (G/U) hydrogel Cell survival rate: 2 h: 93.72 ± 2.51% 15 days: 90.63 ± 1.54% | As the concentration of TMZ increased, the cell viability decreased gradually. G/U cultured cells showed greater viability than U microfiber-cultured cells. | [24] |
| Sodium alginate | Young’s modulus (kPa): Day 0: 131.0 ± 16.2 Day 7: 100.6 ± 9.6 Day 14: 73.2 ± 2.1 Day 21: 27.8 ± 7.4 Pore diameter: 100–400 μm Porosity: 89.5% | Spheroid diameter: Day 7: over 50% spheroids < 50 μm in diameter Day 14: up to 85% spheroids > 50 μm Day 21: 100% spheroids > 50 μm | Live/dead cells percentage: Day 7: 90.37 ± 1.76%, Day 14: 83.45 ± 3.79%, Day 21: 78.25 ± 5.11% | IC50 of doxorubicin: 2D: 1.98 ± 0.01 µg/mL 3D: 10.00 ± 1.0 µg/mL IC50 of cordycepin: 2D: 103.66 ± 10.26 µg/mL 3D: 207.33 ± 16.62 µg/mL | [25] |
| Matrigel | Elastic modulus: 4.5 mg/mL: 31 ± 5.6 Pa 6 mg/mL: 48 ± 9.2 Pa 8 mg/mL: 66 ± 4.4 Pa | Cell migration: 2 mg/mL: 1.9 ± 0.2 mm 4 mg/mL: 2.4 ± 0.5 mm 6 mg/mL: 3.2 ± 0.4 mm 8 mg/mL: 2.6 ± 0.4 mm | Cell percentage: 2 mg/mL: 14 ± 2.8% 6 mg/mL: 33 ± 6.3% 8 mg/mL: 31.2 ± 8.4% | N/A | [26] |
| Fibrinogen, alginate, genipin | N/A | Human glioblastoma cell U87 formed spheroids within the scaffolds after 6 days in culture. | N/A | 3D-printed glioblastoma cell viability: 1mM: 86.5 ± 6.9% 5mM: 54.1 ± 8.9% 10mM: 50.6 ± 2.8% 25mM: 50.1 ± 3.6% 50mM: 46.7 ± 9.4% Cell viability of co-cultures: 5 mM: day 16: 82.6 ± 14.8% day 22: 68.5 ± 2.4% day 30: 16.8 ± 1.0% 10 mM: day 16: 83.1 ± 3.9% day 22: 25.0 ± 2.4% day 30: 11.1 ± 1.0% | [27] |
| GMHA and GelMA | Mean stiffness: Stiff model: 21 kPa Soft model: 2 kPa Pore size: stiff ECM < soft ECM | HUVEC morphology: Stiff model: sprouted blood vessels and close contact with glioblastoma cells. Soft model: expansive growth without visible sprouting. | Hypoxia-related genes and hypoxia-associated angiogenesis markers upregulated in the stiff condition. No significant difference in proliferation marker MKI67 of cells between both models. More KI67-positive cells were present in the soft model. | IC50 of TMZ: sphere cultured TS576 cells: 30 × 10−6 M TMZ treatment: No significant difference in the cell viability between both models. TMZ treatment in co-culture condition: Cell viability significantly increased in stiff models but not in the soft model | [28] |
| PEGDA and BPADMA | Good in shape programming and recovery | PDSs: 100–300 mm PDOs: 400–600 mm | NESTIN-expressing cells in the outer rim coexpressed other GSC markers widely detected in PDSs | Combination therapy increased apoptosis in GBM#46 PDOs and could significantly reduce migration and invasion of GBM-PDO cells | [29] |
| RGD-alginate, HA and collagen-1 | Mean stiffness (kPa): 10 mM CaCl2: 11.9 50 mM CaCl2: 25.7 | Cell spreading and apparent adhesion in <24 h within RGD-alginate | Cell viability: glioblastoma cells: >90%G144, G166, G7: >90% | IC50 of TMZ: U87: 1994 ± 1.0 μM| G7: 748.8 ± 1.1 μM (2-fold higher than 2D cell culture) IC50 of cisplatin: U87: 69.8 ± 1.1 μM G7: 241 ± 1.1 μM | [30] |
| GelMA and GMHA | Stiffness: tumour cell core: 2.8 ± 0.6 kPa NPCs and astrocytes peripheral: 0.9 ± 0.2 kPa Porosity: 53% Pore size: 85 μm | N/A | Cells showed increased levels of the proliferative marker Ki67 and increased protein expression of the stemness markers OLIG2 and SOX2. | GSC23 showed increased resistance to erlotinib, gefitinib, and TMZ in any 3D model than in sphere cultures. Tetra-culture GSCs showed increased sensitivity to abiraterone and ifosfamide than GSCs triculture, while vemurafenib sensitivity remained unchanged. In vivo: Ifosfamide therapy reduced tumour growth | [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leong, S.W.; Tan, S.C.; Norhayati, M.N.; Monif, M.; Lee, S.-Y. Effectiveness of Bioinks and the Clinical Value of 3D Bioprinted Glioblastoma Models: A Systematic Review. Cancers 2022, 14, 2149. https://doi.org/10.3390/cancers14092149
Leong SW, Tan SC, Norhayati MN, Monif M, Lee S-Y. Effectiveness of Bioinks and the Clinical Value of 3D Bioprinted Glioblastoma Models: A Systematic Review. Cancers. 2022; 14(9):2149. https://doi.org/10.3390/cancers14092149
Chicago/Turabian StyleLeong, Shye Wei, Shing Cheng Tan, Mohd Noor Norhayati, Mastura Monif, and Si-Yuen Lee. 2022. "Effectiveness of Bioinks and the Clinical Value of 3D Bioprinted Glioblastoma Models: A Systematic Review" Cancers 14, no. 9: 2149. https://doi.org/10.3390/cancers14092149
APA StyleLeong, S. W., Tan, S. C., Norhayati, M. N., Monif, M., & Lee, S.-Y. (2022). Effectiveness of Bioinks and the Clinical Value of 3D Bioprinted Glioblastoma Models: A Systematic Review. Cancers, 14(9), 2149. https://doi.org/10.3390/cancers14092149






