Spatial Characterization of Tumor-Infiltrating Lymphocytes and Breast Cancer Progression
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets and Algorithms
2.1.1. The TCGA BRCA Dataset
2.1.2. The UNC CBCS Dataset
2.1.3. Selection of Validation (“TIL-Sensitive”) Cohort
2.2. Machine Learning Computational Pathology Algorithms
2.2.1. Generation of Composite Tumor/TIL Maps
2.2.2. Calculation of Percent Infiltration
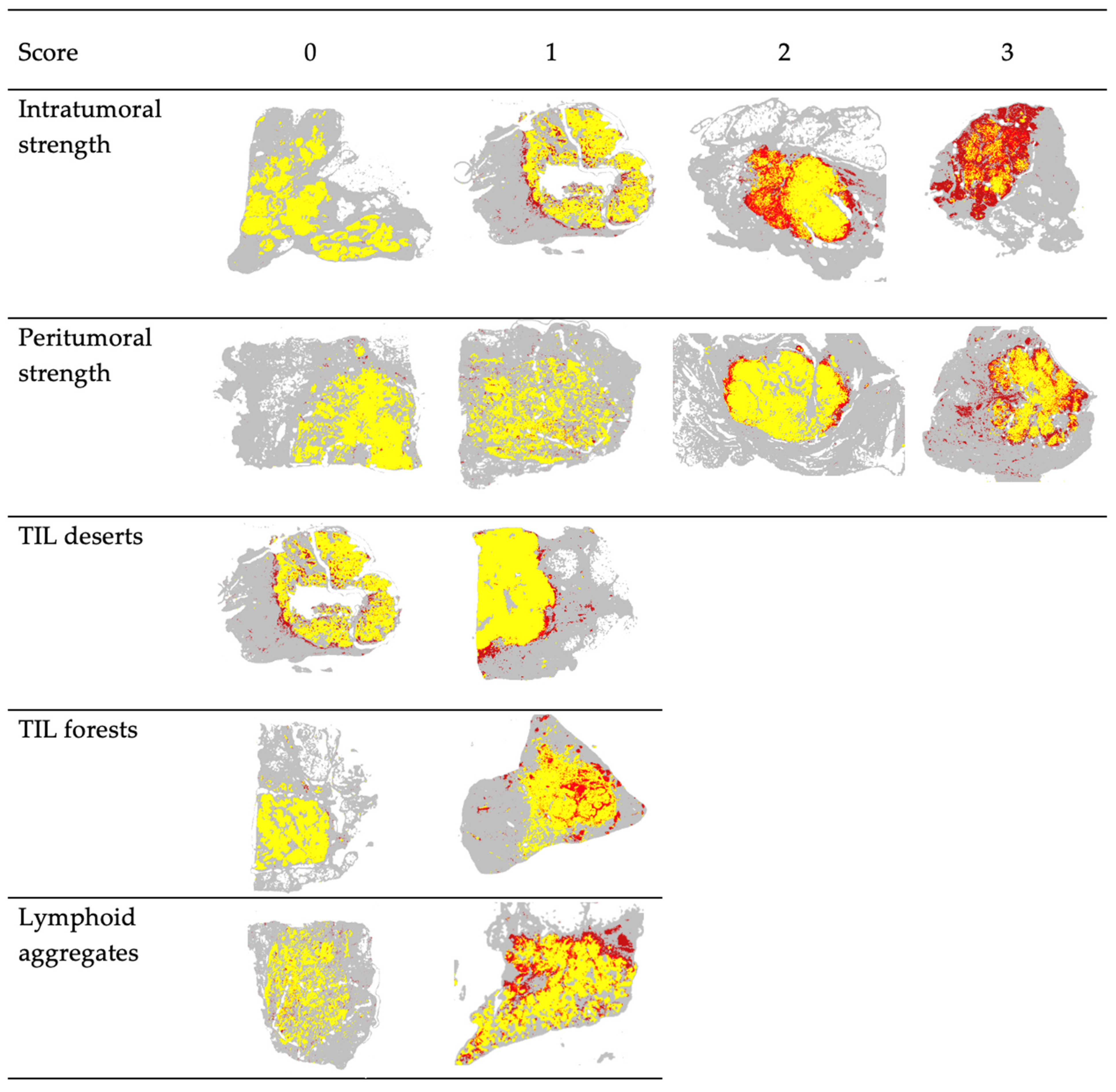
2.2.3. Spatial Feature Scoring
2.3. Assessment of Outcomes in Patient Studies with Computed TIL Infiltration
2.4. Evaluating Risk with Spatial Feature Inclusion
2.5. Statistical Tests
3. Results
3.1. Cohort and Lymphocyte Infiltration Characteristics
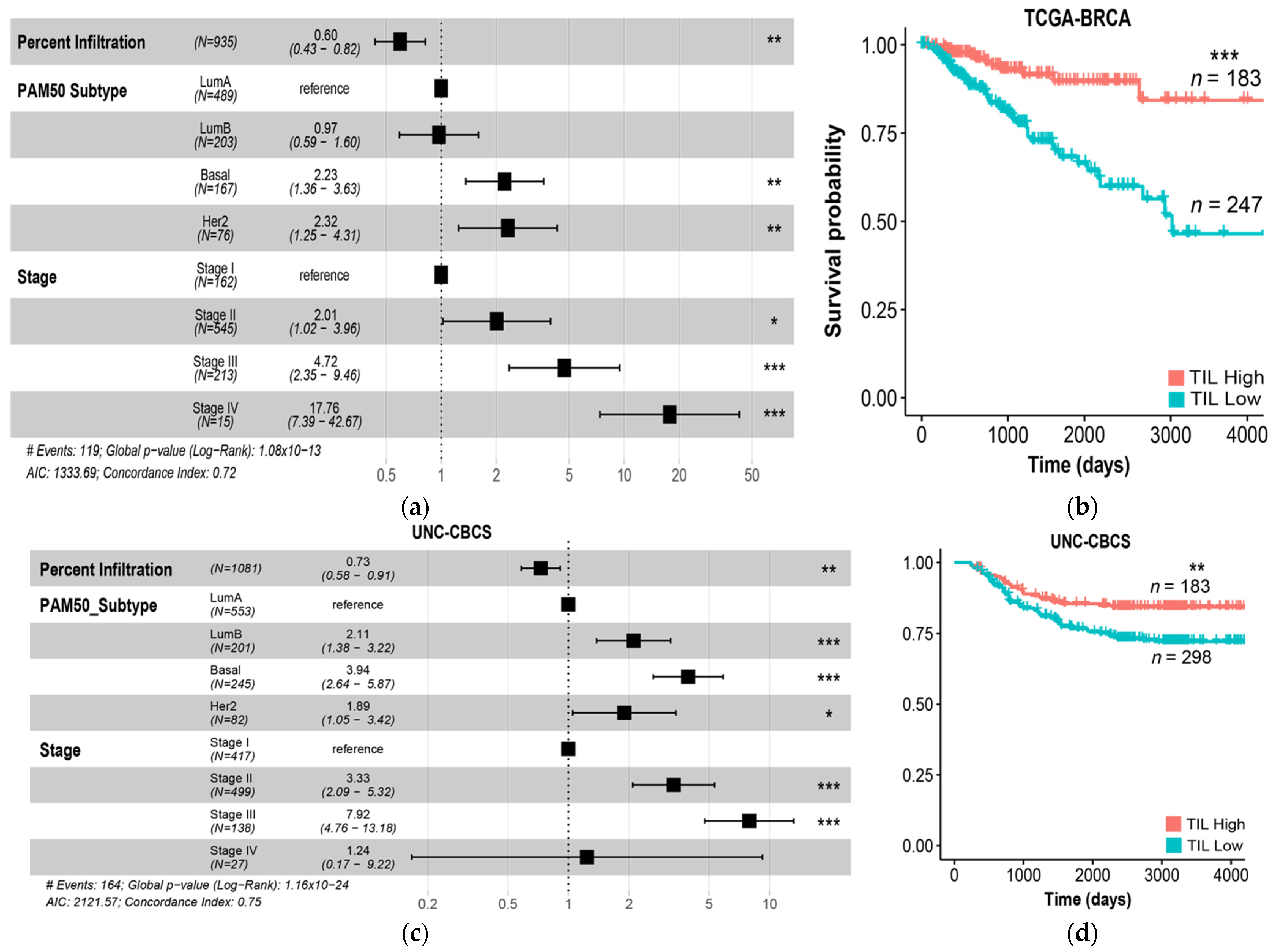
3.2. Computed Lymphocyte Infiltration from Tumor-TIL Maps Strongly Predicts PFI and Survival
3.3. Spatial Feature Interrater Agreement and Feature Correlation
3.4. Relationship between Percent TIL Infiltration and Spatial Features
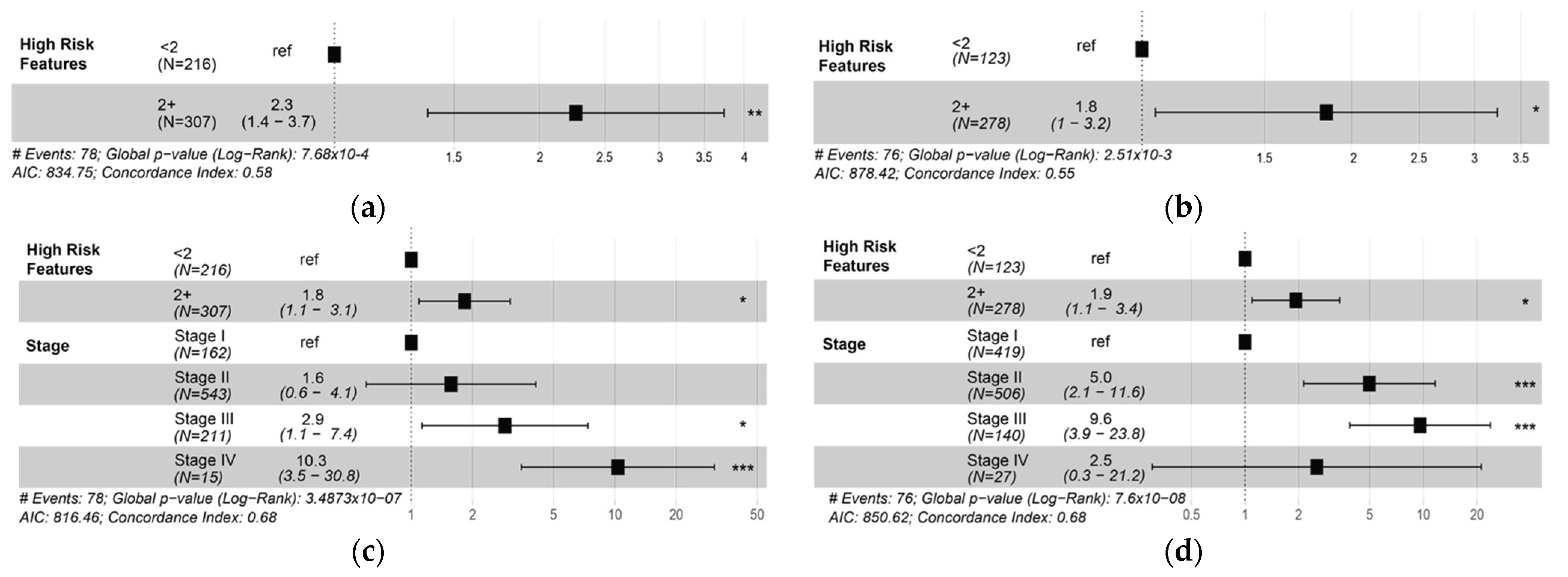
3.5. Presence of Two or More High-Risk Spatial Features Is Associated with Poor Prognosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Virchow, R. Die krankhaften Geschwülste; Dreissig Vorlesungen: Gehalten Während des Wintersemesters; Universität zu Berlin: Berlin, Germany, 1862; Volume 2, p. 1865. [Google Scholar]
- Ehrlich, P. Experimentelle Studien an Mausetumoren. Z Krebsforsch 1907, 5, 59–81. [Google Scholar] [CrossRef]
- Coley, W.B. The treatment of malignant tumors by repeated inoculations of erysipelas: With a report of ten original cases. Am. J. Med. Sci. 1893, 105, 487. [Google Scholar] [CrossRef]
- Coley, W.B. The classic: The treatment of malignant tumors by repeated inoculations of erysipelas: With a report of ten original cases. Clin. Orthop. Relat. Res. 1991, 262, 3–11. [Google Scholar] [CrossRef]
- Mihm, M.C., Jr.; Mulé, J.J. Reflections on the Histopathology of Tumor-Infiltrating Lymphocytes in Melanoma and the Host Immune Response. Cancer Immunol. Res. 2015, 3, 827–835. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity 2020, 52, 55–81. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Biswas, A.; De, S. Drivers of dynamic intratumor heterogeneity and phenotypic plasticity. Am. J. Physiol. Cell Physiol. 2021, 320, C750–C760. [Google Scholar] [CrossRef]
- MacCarty, W.; Mahle, A. Relation of differentiation and lympho-cytic infiltration to postoperative longevity in gastric carcinoma. Collected Papers 1921, 12, 32. [Google Scholar]
- Burnet, M. Cancer; a biological approach. I. The processes of control. Br. Med. J. 1957, 1, 779–786. [Google Scholar] [CrossRef]
- Thomas, L. Discussion in Cellular and Humoral Aspects of the Hypersensitive States; Hoeber-Harper: New York, NY, USA, 1959; p. 529. [Google Scholar]
- Hendry, S.; Salgado, R.; Gevaert, T.; Russell, P.A.; John, T.; Thapa, B.; Christie, M.; van de Vijver, K.; Estrada, M.V.; Gonzalez-Ericsson, P.I.; et al. Assessing Tumor-Infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method from the International Immuno-Oncology Biomarkers Working Group: Part 2: TILs in Melanoma, Gastrointestinal Tract Carcinomas, Non-Small Cell Lung Carcinoma and Mesothelioma, Endometrial and Ovarian Carcinomas, Squamous Cell Carcinoma of the Head and Neck, Genitourinary Carcinomas, and Primary Brain Tumors. Adv. Anat. Pathol. 2017, 24, 311–335. [Google Scholar] [CrossRef]
- Jayasurya, A.; Bay, B.-H.; Yap, W.-M.; Tan, N.-G. Lymphocytic Infiltration in Undifferentiated Nasopharyngeal Cancer. Arch. Otolaryngol. Head Neck Surg. 2000, 126, 1329–1332. [Google Scholar] [CrossRef][Green Version]
- Abousamra, S.; Gupta, R.; Hou, L.; Batiste, R.; Zhao, T.; Shankar, A.; Rao, A.; Chen, C.; Samaras, D.; Kurc, T.; et al. Deep Learning-Based Mapping of Tumor Infiltrating Lymphocytes in Whole Slide Images of 23 Types of Cancer. Front. Oncol. 2022, 11, 806603. [Google Scholar] [CrossRef] [PubMed]
- Abousamra, S.; Hou, L.; Gupta, R.; Chen, C.; Samaras, D.; Kurc, T.; Batiste, R.; Zhao, T.; Kenneth, S.; Saltz, J. Learning from Thresholds: Fully Automated Classification of Tumor Infiltrating Lymphocytes for Multiple Cancer Types. arXiv 2019, arXiv:1907.03960v1. Available online: https://arxiv.org/abs/1907.03960 (accessed on 11 April 2022).
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018, 23, 181. [Google Scholar] [CrossRef] [PubMed]
- Le, H.; Gupta, R.; Hou, L.; Abousamra, S.; Fassler, D.; Torre-Healy, L.; Moffitt, R.A.; Kurc, T.; Samaras, D.; Batiste, R.; et al. Utilizing Automated Breast Cancer Detection to Identify Spatial Distributions of Tumor-Infiltrating Lymphocytes in Invasive Breast Cancer. Am. J. Pathol. 2020, 190, 1491–1504. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Angell, H.K.; Bedognetti, D.; Marincola, F.M. The continuum of cancer immunosurveillance: Prognostic, predictive, and mechanistic signatures. Immunity 2013, 39, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Melichar, B.; ŠTudentovÁ, H.; KalÁBovÁ, H.; VitÁSkovÁ, D.; ČErmÁKovÁ, P.; HornychovÁ, H.; RyŠKa, A. Predictive and Prognostic Significance of Tumor-infiltrating Lymphocytes in Patients with Breast Cancer Treated with Neoadjuvant Systemic Therapy. Anticancer. Res. 2014, 34, 1115. [Google Scholar]
- Moore Jr, O.S.; Foote Jr, F.W. The relatively favorable prognosis of medullary carcinoma of the breast. Cancer 1949, 2, 635–642. [Google Scholar] [CrossRef]
- Angell, H.; Galon, J. From the immune contexture to the Immunoscore: The role of prognostic and predictive immune markers in cancer. Curr. Opin. Immunol. 2013, 25, 261–267. [Google Scholar] [CrossRef]
- Demaria, S.; Pikarsky, E.; Karin, M.; Coussens, L.M.; Chen, Y.C.; El-Omar, E.M.; Trinchieri, G.; Dubinett, S.M.; Mao, J.T.; Szabo, E.; et al. Cancer and inflammation: Promise for biologic therapy. J. Immunother. 2010, 33, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pagès, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- García-Aranda, M.; Redondo, M. Immunotherapy: A Challenge of Breast Cancer Treatment. Cancers 2019, 11, 1822. [Google Scholar] [CrossRef]
- Mittal, D.; Gubin, M.M.; Schreiber, R.D.; Smyth, M.J. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr. Opin. Immunol. 2014, 27, 16–25. [Google Scholar] [CrossRef]
- Mlecnik, B.; Bindea, G.; Pagès, F.; Galon, J. Tumor immunosurveillance in human cancers. Cancer Metastasis Rev. 2011, 30, 5–12. [Google Scholar] [CrossRef]
- Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Berger, A.; Bindea, G.; Meatchi, T.; Bruneval, P.; Trajanoski, Z.; Fridman, W.-H.; Pagès, F. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J. Clin. Oncol. 2011, 29, 610–618. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Taube, J.M.; Klein, A.; Brahmer, J.R.; Xu, H.; Pan, X.; Kim, J.H.; Chen, L.; Pardoll, D.M.; Topalian, S.L.; Anders, R.A. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 2014, 20, 5064–5074. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Uryvaev, A.; Passhak, M.; Hershkovits, D.; Sabo, E.; Bar-Sela, G. The role of tumor-infiltrating lymphocytes (TILs) as a predictive biomarker of response to anti-PD1 therapy in patients with metastatic non-small cell lung cancer or metastatic melanoma. Med. Oncol. 2018, 35, 25. [Google Scholar] [CrossRef]
- Angell, H.K.; Gray, N.; Womack, C.; Pritchard, D.I.; Wilkinson, R.W.; Cumberbatch, M. Digital pattern recognition-based image analysis quantifies immune infiltrates in distinct tissue regions of colorectal cancer and identifies a metastatic phenotype. Br. J. Cancer 2013, 109, 1618–1624. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Fridman, W.H.; Pagès, F. The adaptive immunologic microenvironment in colorectal cancer: A novel perspective. Cancer Res. 2007, 67, 1883–1886. [Google Scholar] [CrossRef]
- Clemente, C.G.; Mihm, M.C., Jr.; Bufalino, R.; Zurrida, S.; Collini, P.; Cascinelli, N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer 1996, 77, 1303–1310. [Google Scholar] [CrossRef]
- Naito, Y.; Saito, K.; Shiiba, K.; Ohuchi, A.; Saigenji, K.; Nagura, H.; Ohtani, H. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998, 58, 3491–3494. [Google Scholar]
- Pagès, F.; Berger, A.; Camus, M.; Sanchez-Cabo, F.; Costes, A.; Molidor, R.; Mlecnik, B.; Kirilovsky, A.; Nilsson, M.; Damotte, D.; et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N. Engl. J. Med. 2005, 353, 2654–2666. [Google Scholar] [CrossRef]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef]
- Amgad, M.; Stovgaard, E.S.; Balslev, E.; Thagaard, J.; Chen, W.; Dudgeon, S.; Sharma, A.; Kerner, J.K.; Denkert, C.; Yuan, Y.; et al. Report on computational assessment of Tumor Infiltrating Lymphocytes from the International Immuno-Oncology Biomarker Working Group. NPJ Breast Cancer 2020, 6, 16. [Google Scholar] [CrossRef]
- Burugu, S.; Asleh-Aburaya, K.; Nielsen, T.O. Immune infiltrates in the breast cancer microenvironment: Detection, characterization and clinical implication. Breast Cancer 2017, 24, 3–15. [Google Scholar] [CrossRef]
- Loi, S.; Drubay, D.; Adams, S.; Pruneri, G.; Francis, P.A.; Lacroix-Triki, M.; Joensuu, H.; Dieci, M.V.; Badve, S.; Demaria, S.; et al. Tumor-Infiltrating Lymphocytes and Prognosis: A Pooled Individual Patient Analysis of Early-Stage Triple-Negative Breast Cancers. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 559–569. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef]
- Wein, L.; Savas, P.; Luen, S.J.; Virassamy, B.; Salgado, R.; Loi, S. Clinical Validity and Utility of Tumor-Infiltrating Lymphocytes in Routine Clinical Practice for Breast Cancer Patients: Current and Future Directions. Front. Oncol. 2017, 7, 156. [Google Scholar] [CrossRef]
- Luen, S.J.; Griguolo, G.; Nuciforo, P.; Campbell, C.; Fasani, R.; Cortes, J.; Untch, M.; Lin, S.J.; Savas, P.; Fox, S.B.; et al. On-treatment changes in tumor-infiltrating lymphocytes (TIL) during neoadjuvant HER2 therapy (NAT) and clinical outcome. J. Clin. Oncol. 2019, 37, 574. [Google Scholar] [CrossRef]
- Luen, S.J.; Salgado, R.; Dieci, M.V.; Vingiani, A.; Curigliano, G.; Gould, R.E.; Castaneda, C.; D’Alfonso, T.; Sanchez, J.; Cheng, E.; et al. Prognostic implications of residual disease tumor-infiltrating lymphocytes and residual cancer burden in triple-negative breast cancer patients after neoadjuvant chemotherapy. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 236–242. [Google Scholar] [CrossRef]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl. Cancer Inst. 2005, 97, 1180–1184. [Google Scholar] [CrossRef]
- Simon, R.M.; Paik, S.; Hayes, D.F. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl. Cancer Inst. 2009, 101, 1446–1452. [Google Scholar] [CrossRef]
- Gao, Z.-h.; Li, C.-x.; Liu, M.; Jiang, J.-y. Predictive and prognostic role of tumour-infiltrating lymphocytes in breast cancer patients with different molecular subtypes: A meta-analysis. BMC Cancer 2020, 20, 1150. [Google Scholar] [CrossRef]
- Savas, P.; Salgado, R.; Denkert, C.; Sotiriou, C.; Darcy, P.K.; Smyth, M.J.; Loi, S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016, 13, 228–241. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, Z.; Wang, Z.; Wu, P.; Qiu, F.; Huang, J. Prognostic and predictive value of tumor-infiltrating lymphocytes in breast cancer: A systematic review and meta-analysis. Clin. Transl. Oncol. 2016, 18, 497–506. [Google Scholar] [CrossRef]
- Balic, M.; Thomssen, C.; Würstlein, R.; Gnant, M.; Harbeck, N. St. Gallen/Vienna 2019: A Brief Summary of the Consensus Discussion on the Optimal Primary Breast Cancer Treatment. Breast Care 2019, 14, 103–110. [Google Scholar] [CrossRef]
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1194–1220. [Google Scholar] [CrossRef]
- Morigi, C. Highlights of the 16th St Gallen International Breast Cancer Conference, Vienna, Austria, 20–23 March 2019: Personalised treatments for patients with early breast cancer. Ecancermedicalscience 2019, 13, 924. [Google Scholar] [CrossRef]
- Tan, P.H.; Ellis, I.; Allison, K.; Brogi, E.; Fox, S.B.; Lakhani, S.; Lazar, A.J.; Morris, E.A.; Sahin, A.; Salgado, R.; et al. The 2019 World Health Organization classification of tumours of the breast. Histopathology 2020, 77, 181–185. [Google Scholar] [CrossRef]
- De Angelis, C.; Nagi, C.; Hoyt, C.C.; Liu, L.; Roman, K.; Wang, C.; Zheng, Y.; Veeraraghavan, J.; Sethunath, V.; Nuciforo, P.; et al. Evaluation of the Predictive Role of Tumor Immune Infiltrate in Patients with HER2-Positive Breast Cancer Treated with Neoadjuvant Anti-HER2 Therapy without Chemotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 738–745. [Google Scholar] [CrossRef]
- Kos, Z.; Roblin, E.; Kim, R.S.; Michiels, S.; Gallas, B.D.; Chen, W.; van de Vijver, K.K.; Goel, S.; Adams, S.; Demaria, S.; et al. Pitfalls in assessing stromal tumor infiltrating lymphocytes (sTILs) in breast cancer. NPJ Breast Cancer 2020, 6, 17. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Allott, E.H.; Shan, Y.; Chen, M.; Sun, X.; Garcia-Recio, S.; Kirk, E.L.; Olshan, A.F.; Geradts, J.; Earp, H.S.; Carey, L.A.; et al. Bimodal age distribution at diagnosis in breast cancer persists across molecular and genomic classifications. Breast Cancer Res. Treat. 2020, 179, 185–195. [Google Scholar] [CrossRef]
- Benefield, H.C.; Zabor, E.C.; Shan, Y.; Allott, E.H.; Begg, C.B.; Troester, M.A. Evidence for Etiologic Subtypes of Breast Cancer in the Carolina Breast Cancer Study. Cancer Epidemiol Biomarkers Prev 2019, 28, 1784–1791. [Google Scholar] [CrossRef]
- Carey, L.A.; Perou, C.M.; Livasy, C.A.; Dressler, L.G.; Cowan, D.; Conway, K.; Karaca, G.; Troester, M.A.; Tse, C.K.; Edmiston, S.; et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006, 295, 2492–2502. [Google Scholar] [CrossRef]
- Haddad, S.A.; Lunetta, K.L.; Ruiz-Narváez, E.A.; Bensen, J.T.; Hong, C.C.; Sucheston-Campbell, L.E.; Yao, S.; Bandera, E.V.; Rosenberg, L.; Haiman, C.A.; et al. Hormone-related pathways and risk of breast cancer subtypes in African American women. Breast Cancer Res. Treat. 2015, 154, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Huo, D.; Hu, H.; Rhie, S.K.; Gamazon, E.R.; Cherniack, A.D.; Liu, J.; Yoshimatsu, T.F.; Pitt, J.J.; Hoadley, K.A.; Troester, M.; et al. Comparison of Breast Cancer Molecular Features and Survival by African and European Ancestry in The Cancer Genome Atlas. JAMA Oncol. 2017, 3, 1654–1662. [Google Scholar] [CrossRef] [PubMed]
- Parada, H., Jr.; Sun, X.; Fleming, J.M.; Williams-DeVane, C.R.; Kirk, E.L.; Olsson, L.T.; Perou, C.M.; Olshan, A.F.; Troester, M.A. Race-associated biological differences among luminal A and basal-like breast cancers in the Carolina Breast Cancer Study. Breast Cancer Res. 2017, 19, 131. [Google Scholar] [CrossRef] [PubMed]
- Troester, M.A.; Sun, X.; Allott, E.H.; Geradts, J.; Cohen, S.M.; Tse, C.K.; Kirk, E.L.; Thorne, L.B.; Mathews, M.; Li, Y.; et al. Racial Differences in PAM50 Subtypes in the Carolina Breast Cancer Study. J. Natl. Cancer Inst. 2018, 110, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; Gulsuner, S.; Lee, M.K.; Troester, M.A.; Olshan, A.F.; Earp, H.S.; Perou, C.M.; King, M.-C. Inherited predisposition to breast cancer in the Carolina Breast Cancer Study. NPJ Breast Cancer 2021, 7, 6. [Google Scholar] [CrossRef]
- Williams, L.A.; Casbas-Hernandez, P.; Nichols, H.B.; Tse, C.K.; Allott, E.H.; Carey, L.A.; Olshan, A.F.; Troester, M.A. Risk factors for Luminal A ductal carcinoma in situ (DCIS) and invasive breast cancer in the Carolina Breast Cancer Study. PLoS ONE 2019, 14, e0211488. [Google Scholar] [CrossRef]
- Heng, Y.J.; Lester, S.C.; Tse, G.M.; Factor, R.E.; Allison, K.H.; Collins, L.C.; Chen, Y.Y.; Jensen, K.C.; Johnson, N.B.; Jeong, J.C.; et al. The molecular basis of breast cancer pathological phenotypes. J. Pathol. 2017, 241, 375–391. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e411. [Google Scholar] [CrossRef]
- Koch, A.D.; Nicolai, J.P.; de Vries, J. Breast cancer and the role of breast size as a contributory factor. Breast 2004, 13, 272–275. [Google Scholar] [CrossRef]
- Gupta, R.; Le, H.; Van Arnam, J.; Belinsky, D.; Hasan, M.; Samaras, D.; Kurc, T.; Saltz, J.H. Characterizing Immune Responses in Whole Slide Images of Cancer With Digital Pathology and Pathomics. Curr. Pathobiol. Rep. 2020, 8, 133–148. [Google Scholar] [CrossRef]
- Amgad, M.; Elfandy, H.; Hussein, H.; Atteya, L.A.; Elsebaie, M.A.; Abo Elnasr, L.S.; Sakr, R.A.; Salem, H.S.; Ismail, A.F.; Saad, A.M.J.B. Structured crowdsourcing enables convolutional segmentation of histology images. Bioinformatics 2019, 35, 3461–3467. [Google Scholar] [CrossRef]
- Amgad, M.; Sarkar, A.; Srinivas, C.; Redman, R.; Ratra, S.; Bechert, C.J.; Calhoun, B.C.; Mrazeck, K.; Kurkure, U.; Cooper, L.A. Joint region and nucleus segmentation for characterization of tumor infiltrating lymphocytes in breast cancer. In Medical Imaging 2019: Digital Pathology; International Society for Optics and Photonics: Bellingham, WA, USA, 2019. [Google Scholar]
- Barnes, M.; Sarkar, A.; Redman, R.; Bechert, C.; Srinivas, C. Development of a histology-based digital pathology image analysis algorithm for assessment of tumor infiltrating lymphocytes in HER2+ breast cancer. Cancer Res. 2018, 78. [Google Scholar] [CrossRef]
- Amgad, M.; Salgado, R.; Cooper, L.A.D. MuTILs: Explainable, multiresolution computational scoring of Tumor-Infiltrating Lymphocytes in breast carcinomas using clinical guidelines. medRxiv 2022. [Google Scholar] [CrossRef]
- El Bairi, K.; Haynes, H.R.; Blackley, E.; Fineberg, S.; Shear, J.; Turner, S.; de Freitas, J.R.; Sur, D.; Amendola, L.C.; Gharib, M.; et al. The tale of TILs in breast cancer: A report from The International Immuno-Oncology Biomarker Working Group. NPJ Breast Cancer 2021, 7, 150. [Google Scholar] [CrossRef]
- Kilmartin, D.; O’Loughlin, M.; Andreu, X.; Bagó-Horváth, Z.; Bianchi, S.; Chmielik, E.; Cserni, G.; Figueiredo, P.; Floris, G.; Foschini, M.P.; et al. Intra-Tumour Heterogeneity Is One of the Main Sources of Inter-Observer Variation in Scoring Stromal Tumour Infiltrating Lymphocytes in Triple Negative Breast Cancer. Cancers 2021, 13, 4410. [Google Scholar] [CrossRef]
- Thagaard, J.; Stovgaard, E.S.; Vognsen, L.G.; Hauberg, S.; Dahl, A.; Ebstrup, T.; Doré, J.; Vincentz, R.E.; Jepsen, R.K.; Roslind, A.; et al. Automated Quantification of sTIL Density with H&E-Based Digital Image Analysis Has Prognostic Potential in Triple-Negative Breast Cancers. Cancers 2021, 13, 3050. [Google Scholar] [CrossRef]
- Graham, S.; Vu, Q.D.; Raza, S.E.A.; Azam, A.; Tsang, Y.W.; Kwak, J.T.; Rajpoot, N. Hover-Net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 2019, 58, 101563. [Google Scholar] [CrossRef]
- Dudgeon, S.; Wen, S.; Hanna, M.; Gupta, R.; Amgad, M.; Sheth, M.; Marble, H.; Huang, R.; Herrmann, M.; Szu, C.; et al. A pathologist-annotated dataset for validating artificial intelligence: A project description and pilot study. J. Pathol. Inform. 2021, 12, 45. [Google Scholar] [CrossRef]
- Nederlof, I.; De Bortoli, D.; Bareche, Y.; Nguyen, B.; de Maaker, M.; Hooijer, G.K.J.; Buisseret, L.; Kok, M.; Smid, M.; Van den Eynden, G.; et al. Comprehensive evaluation of methods to assess overall and cell-specific immune infiltrates in breast cancer. Breast Cancer Res. 2019, 21, 151. [Google Scholar] [CrossRef]
- AbdulJabbar, K.; Raza, S.E.A.; Rosenthal, R.; Jamal-Hanjani, M.; Veeriah, S.; Akarca, A.; Lund, T.; Moore, D.A.; Salgado, R.; Al Bakir, M.; et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nature Med. 2020, 26, 1054–1062. [Google Scholar] [CrossRef]
- Kolberg-Liedtke, C.; Gluz, O.; Heinisch, F.; Feuerhake, F.; Kreipe, H.; Clemens, M.; Nuding, B.; Malter, W.; Reimer, T.; Wuerstlein, R.; et al. Association of TILs with clinical parameters, Recurrence Score® results, and prognosis in patients with early HER2-negative breast cancer (BC)-a translational analysis of the prospective WSG PlanB trial. Breast Cancer Res. 2020, 22, 47. [Google Scholar] [CrossRef]
- Hornychova, H.; Melichar, B.; Tomšová, M.; Mergancová, J.; Urminská, H.; Ryška, A. Tumor-infiltrating lymphocytes predict response to neoadjuvant chemotherapy in patients with breast carcinoma. Cancer Investig. 2008, 26, 1024–1031. [Google Scholar] [CrossRef]
- Loi, S.; Michiels, S.; Adams, S.; Loibl, S.; Budczies, J.; Denkert, C.; Salgado, R. The journey of tumor-infiltrating lymphocytes as a biomarker in breast cancer: Clinical utility in an era of checkpoint inhibition. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2021, 32, 1236–1244. [Google Scholar] [CrossRef]
- Mahmoud, S.M.; Paish, E.C.; Powe, D.G.; Macmillan, R.D.; Grainge, M.J.; Lee, A.H.; Ellis, I.O.; Green, A.R. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011, 29, 1949–1955. [Google Scholar] [CrossRef]
- Mohammed, Z.; Going, J.; Edwards, J.; Elsberger, B.; Doughty, J.; McMillan, D. The relationship between components of tumour inflammatory cell infiltrate and clinicopathological factors and survival in patients with primary operable invasive ductal breast cancer. Br. J. Cancer 2012, 107, 864–873. [Google Scholar] [CrossRef]
- Seo, A.; Lee, H.; Kim, E.; Kim, H.; Jang, M.; Lee, H.; Kim, Y.; Kim, J.; Park, S. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br. J. Cancer 2013, 109, 2705–2713. [Google Scholar] [CrossRef]
- Basu, A.; Ramamoorthi, G.; Jia, Y.; Faughn, J.; Wiener, D.; Awshah, S.; Kodumudi, K.; Czerniecki, B.J. Chapter Six—Immunotherapy in breast cancer: Current status and future directions. Adv. Cancer Res. 2019, 143, 295–349. [Google Scholar]
- Dieci, M.V.; Criscitiello, C.; Goubar, A.; Viale, G.; Conte, P.; Guarneri, V.; Ficarra, G.; Mathieu, M.C.; Delaloge, S.; Curigliano, G.; et al. Prognostic value of tumor-infiltrating lymphocytes on residual disease after primary chemotherapy for triple-negative breast cancer: A retrospective multicenter study. Ann. Oncol. 2014, 25, 611–618. [Google Scholar] [CrossRef]
- Garaud, S.; Buisseret, L.; Solinas, C.; Gu-Trantien, C.; de Wind, A.; Van den Eynden, G.; Naveaux, C.; Lodewyckx, J.N.; Boisson, A.; Duvillier, H.; et al. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight 2019, 5, e129641. [Google Scholar] [CrossRef]
- Gu-Trantien, C.; Loi, S.; Garaud, S.; Equeter, C.; Libin, M.; De Wind, A.; Ravoet, M.; Le Buanec, H.; Sibille, C.; Manfouo-Foutsop, G. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Investig. 2013, 123, 2873–2892. [Google Scholar] [CrossRef] [PubMed]
- Vanhersecke, L.; Brunet, M.; Guégan, J.-P.; Rey, C.; Bougouin, A.; Cousin, S.; Le Moulec, S.; Besse, B.; Loriot, Y.; Larroquette, M.; et al. Mature tertiary lymphoid structures predict immune checkpoint inhibitor efficacy in solid tumors independently of PD-L1 expression. Nat. Cancer 2021, 2, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Colbeck, E.J.; Ager, A.; Gallimore, A.; Jones, G.W. Tertiary lymphoid structures in cancer: Drivers of antitumor immunity, immunosuppression, or bystander sentinels in disease? Front. Immunol. 2017, 8, 1830. [Google Scholar] [CrossRef] [PubMed]
- Cortés, J.; Lipatov, O.; Im, S.-A.; Gonçalves, A.; Lee, K.; Schmid, P.; Tamura, K.; Testa, L.; Witzel, I.; Ohtani, S. KEYNOTE-119: Phase III study of pembrolizumab (pembro) versus single-agent chemotherapy (chemo) for metastatic triple negative breast cancer (mTNBC). Ann. Oncol. 2019, 30, v859–v860. [Google Scholar] [CrossRef]
- Hammerl, D.; Smid, M.; Timmermans, A.M.; Sleijfer, S.; Martens, J.W.M.; Debets, R. Breast cancer genomics and immuno-oncological markers to guide immune therapies. Semin. Cancer Biol. 2018, 52, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Nanda, R.; Liu, M.C.; Yau, C.; Asare, S.; Hylton, N.; Veer, L.V.t.; Perlmutter, J.; Wallace, A.M.; Chien, A.J.; Forero-Torres, A. Pembrolizumab plus standard neoadjuvant therapy for high-risk breast cancer (BC): Results from I-SPY 2. J. Clin. Oncol. 2017, 35, 506. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.-H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Thomas, A.; Routh, E.D.; Pullikuth, A.; Jin, G.; Su, J.; Chou, J.W.; Hoadley, K.A.; Print, C.; Knowlton, N.; Black, M.A.; et al. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology 2018, 7, e1490854. [Google Scholar] [CrossRef]
- Xu, Z.; Verma, A.; Naveed, U.; Bakhoum, S.F.; Khosravi, P.; Elemento, O. Deep learning predicts chromosomal instability from histopathology images. iScience 2021, 24, 102394. [Google Scholar] [CrossRef]
- Karn, T.; Jiang, T.; Hatzis, C.; Sänger, N.; El-Balat, A.; Rody, A.; Holtrich, U.; Becker, S.; Bianchini, G.; Pusztai, L. Association between genomic metrics and immune infiltration in triple-negative breast cancer. JAMA Oncol. 2017, 3, 1707–1711. [Google Scholar] [CrossRef]
- McGranahan, N.; Furness, A.J.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
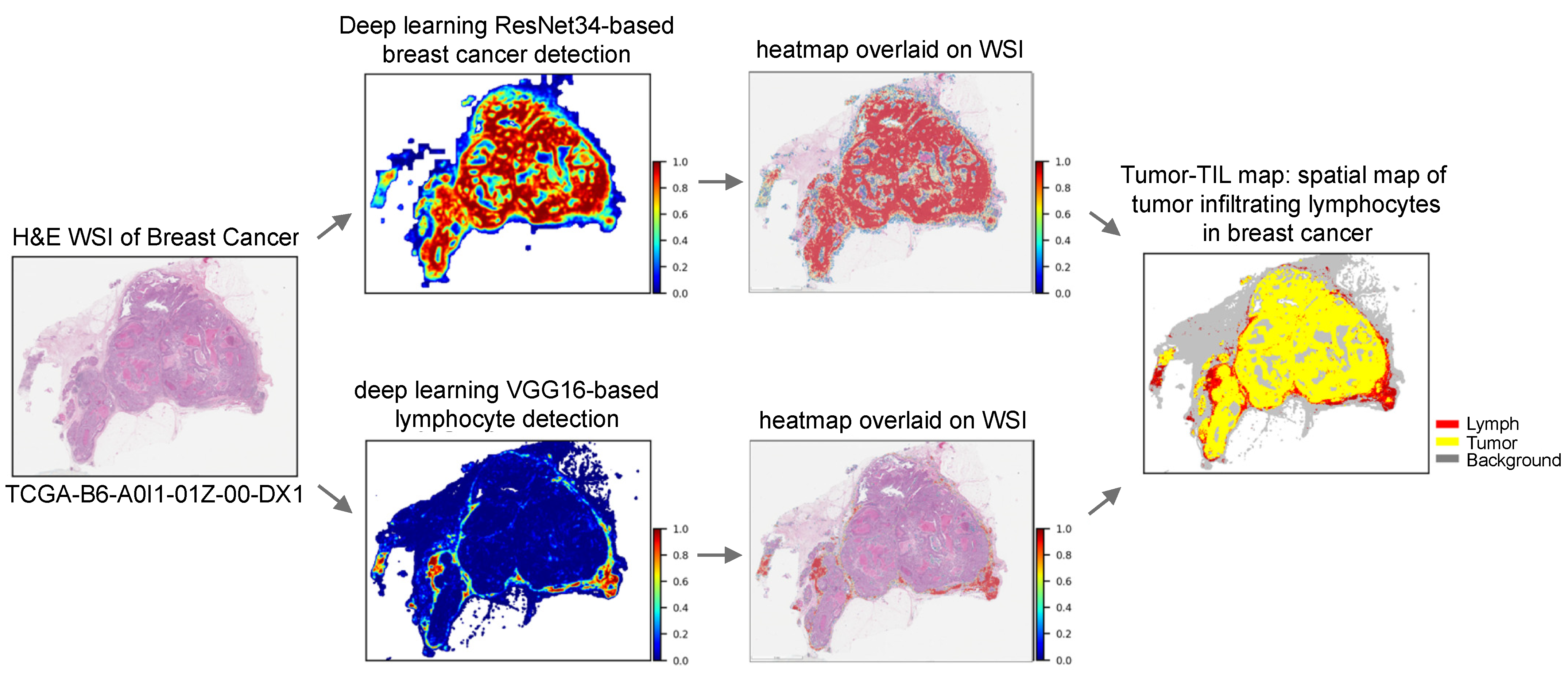
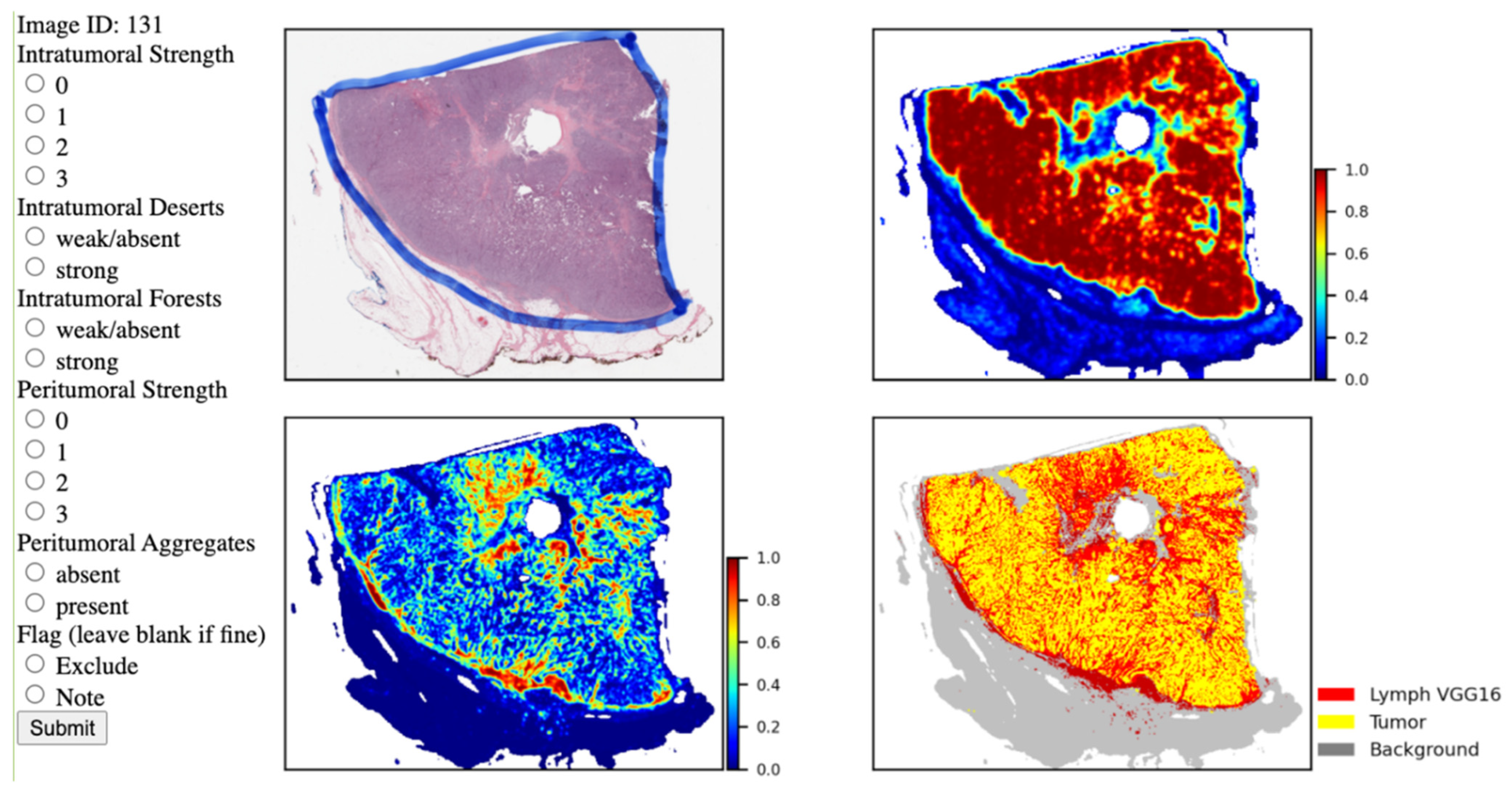

| Variation | TCGA BRCA (n = 935) | UNC CBCS (n = 1081) | p-Value |
|---|---|---|---|
| Race | |||
| African American | 140 (15.0%) | 577 (53.4%) | <0.001 |
| Non-African American | 711 (76.0%) | 504 (46.6%) | |
| Missing | 84 (9.0%) | 0 (0%) | |
| Stage | |||
| Stage I | 162 (17.3%) | 417 (38.6%) | <0.001 |
| Stage II | 545 (58.3%) | 499 (46.2%) | |
| Stage III | 213 (22.8%) | 138 (12.8%) | |
| Stage IV | 15 (1.6%) | 27 (2.5%) | |
| Grade | |||
| 1 | N/A | 199 (18.4%) | N/A |
| 2 | N/A | 377 (34.9%) | |
| 3 | N/A | 478 (44.2%) | |
| Missing | 935 (100%) | 27 (2.5%) | |
| ER by IHC | |||
| Negative | 197 (21.1%) | 249 (23.0%) | 0.676 |
| Positive | 692 (74.0%) | 831 (76.9%) | |
| Missing | 46 (4.9%) | 1 (0.1%) | |
| PAM50 Subtype | |||
| Basal | 167 (17.9%) | 245 (22.7%) | 0.0383 |
| Her2 | 76 (8.1%) | 82 (7.6%) | |
| LumA | 489 (52.3%) | 553 (51.2%) | |
| LumB | 203 (21.7%) | 201 (18.6%) | |
| Percent Infiltration (%) | |||
| Mean (SD) | 7.7 (10.4) | 5.6 (10.2) | <0.001 |
| Median [Min, Max] | 3.8 [0, 64.2] | 1.9 [0, 88.9] | |
| TIL Class | |||
| Low Infiltration | 648 (69.3%) | 806 (74.6%) | 0.01 |
| High Infiltration | 287 (30.7%) | 275 (25.4%) | |
| “TIL Sensitive” | |||
| No | 477 (51.0%) | 599 (55.4%) | 0.217 |
| Yes | 430 (46.0%) | 481 (44.5%) | |
| Missing | 28 (3.0%) | 1 (0.1%) | |
| Progression Events | |||
| Number with Event | 119 (12.7%) | 164 (15.1%) | 0.131 |
| Median days to event [Min,Max] | 791 (21,5117) | 1048.5 (161,3274) | 0.0288 |
| Scoring Category | Score Range | Score | Criteria |
|---|---|---|---|
| Intratumoral Strength | 0–3 | 0 | No patches positive for TILs present within the tumor region |
| 1 | Low intratumoral TIL infiltration (weak) with scant/limited TIL-positive patches within tumor region | ||
| 2 | Intermediate level of TIL infiltration (moderate) with variable spatial distribution (e.g., high density of TILs in some regions and no TILs in others, but not limited to a single focus) | ||
| 3 | High intratumoral TIL infiltration (strong) with numerous and diffuse TIL-positive patches | ||
| Intratumoral Deserts | 0–1 | 0 | small regions of tumor without TIL patches (absent) OR focal areas tumor with little to no TIL patches (weak) (≤10% of tumor area) |
| 1 | significant intratumoral regions completely clear of TIL infiltrate (≥25% of tumor area) | ||
| Intratumoral Forests | 0–1 | 0 | TIL infiltrate either not present (absent) or TIL-positive patches are evenly distributed alongside TIL-negative tumor patches (weak) with 1–2 small confluent groups of TILs |
| 1 | Confluent groups of TILs spanning ≥10% of tumor area present (strong) | ||
| Peritumoral Strength | 0–3 | 0 | No patches positive for TILs present in the region of the invasive boundary (absent) |
| 1 | Low peritumoral TIL infiltration with scant/scattered TIL-positive patches at the boundary of tumor and adjacent normal tissues (weak) | ||
| 2 | Intermediate level of TIL infiltration (moderate) | ||
| 3 | High peritumoral TIL infiltration (marked) with nearly confluent accumulation of TIL-positive patches surrounding the tumor at the invasive boundary | ||
| Tertiary Lymphoid Aggregates | 0–1 | 0 | No significant presence of TIL clusters or minimal focal aggregates beyond the peritumoral region |
| 1 | Multifocal lymphoid aggregates exist beyond the peritumoral region |
| Feature | TCGA BRCA (n = 430) | UNC CBCS (n = 481) | p-Value |
|---|---|---|---|
| Percent Infiltration (%) | |||
| Mean (SD) | 10 (11.9) | 7.89 (12) | 0.00725 |
| Median [Min, Max] | 5.4 [0, 64.2] | 3.3 [0, 88.9] | |
| TIL Class | |||
| Low | 247 (57.4%) | 298 (62.0%) | 0.187 |
| High | 183 (42.6%) | 183 (38.0%) | |
| Intratumoral Strength | |||
| 0 | 4 (0.9%) | 11 (2.3%) | <0.001 |
| 1 | 247 (57.4%) | 282 (58.6%) | |
| 2 | 113 (26.3%) | 64 (13.3%) | |
| 3 | 42 (9.8%) | 34 (7.1%) | |
| Unscored | 24 (5.6%) | 90 (18.7%) | |
| TIL Forests | |||
| Absent | 188 (43.7%) | 260 (54.1%) | <0.001 |
| Present | 218 (50.7%) | 131 (27.2%) | |
| Unscored | 24 (5.6%) | 90 (18.7%) | |
| TIL Deserts | |||
| Absent | 260 (60.5%) | 191 (39.7%) | <0.001 |
| Present | 146 (34.0%) | 200 (41.6%) | |
| Unscored | 24 (5.6%) | 90 (18.7%) | |
| Peritumoral Strength | |||
| 0 | 9 (2.1%) | 3 (0.6%) | 0.365 |
| 1 | 149 (34.7%) | 138 (28.7%) | |
| 2 | 146 (34.0%) | 149 (31.0%) | |
| 3 | 102 (23.7%) | 101 (21.0%) | |
| Unscored | 24 (5.6%) | 90 (18.7%) | |
| Peritumoral Aggregates | |||
| Absent | 87 (20.2%) | 10 (2.1%) | <0.001 |
| Present | 319 (74.2%) | 381 (79.2%) | |
| Unscored | 24 (5.6%) | 90 (18.7%) | |
| Metric | Level | TCGA BRCA Hazard Ratio | UNC CBCS Hazard Ratio | High-Risk Feature (Score) |
|---|---|---|---|---|
| Intratumoral strength | 1 | 0.62 | 0.69 | Low intratumoral strength (0–1) |
| 2 | 0.27 | 0.34 | ||
| 3 | 0.19 | 0.28 | ||
| TIL deserts | Present | 2.0 ** | 1.2 | TIL deserts present (1) |
| TIL forests | Present | 0.56 * | 0.65 | TIL forests absent (0) |
| Peritumoral strength | 1 | 2.07 | 0.49 | Low peritumoral strength (0–1) |
| 2 | 1.14 | 0.41 | ||
| 3 | 0.61 | 0.17 * | ||
| Lymphoid aggregates | Present | 0.78 | 0.42 | Lymphoid aggregates absent (0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fassler, D.J.; Torre-Healy, L.A.; Gupta, R.; Hamilton, A.M.; Kobayashi, S.; Van Alsten, S.C.; Zhang, Y.; Kurc, T.; Moffitt, R.A.; Troester, M.A.; et al. Spatial Characterization of Tumor-Infiltrating Lymphocytes and Breast Cancer Progression. Cancers 2022, 14, 2148. https://doi.org/10.3390/cancers14092148
Fassler DJ, Torre-Healy LA, Gupta R, Hamilton AM, Kobayashi S, Van Alsten SC, Zhang Y, Kurc T, Moffitt RA, Troester MA, et al. Spatial Characterization of Tumor-Infiltrating Lymphocytes and Breast Cancer Progression. Cancers. 2022; 14(9):2148. https://doi.org/10.3390/cancers14092148
Chicago/Turabian StyleFassler, Danielle J., Luke A. Torre-Healy, Rajarsi Gupta, Alina M. Hamilton, Soma Kobayashi, Sarah C. Van Alsten, Yuwei Zhang, Tahsin Kurc, Richard A. Moffitt, Melissa A. Troester, and et al. 2022. "Spatial Characterization of Tumor-Infiltrating Lymphocytes and Breast Cancer Progression" Cancers 14, no. 9: 2148. https://doi.org/10.3390/cancers14092148
APA StyleFassler, D. J., Torre-Healy, L. A., Gupta, R., Hamilton, A. M., Kobayashi, S., Van Alsten, S. C., Zhang, Y., Kurc, T., Moffitt, R. A., Troester, M. A., Hoadley, K. A., & Saltz, J. (2022). Spatial Characterization of Tumor-Infiltrating Lymphocytes and Breast Cancer Progression. Cancers, 14(9), 2148. https://doi.org/10.3390/cancers14092148






