Oestrogen Receptor Isoforms May Represent a Therapeutic Target in Oesophageal Adenocarcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Treatment Experiments
2.3. MTS Viability Assay
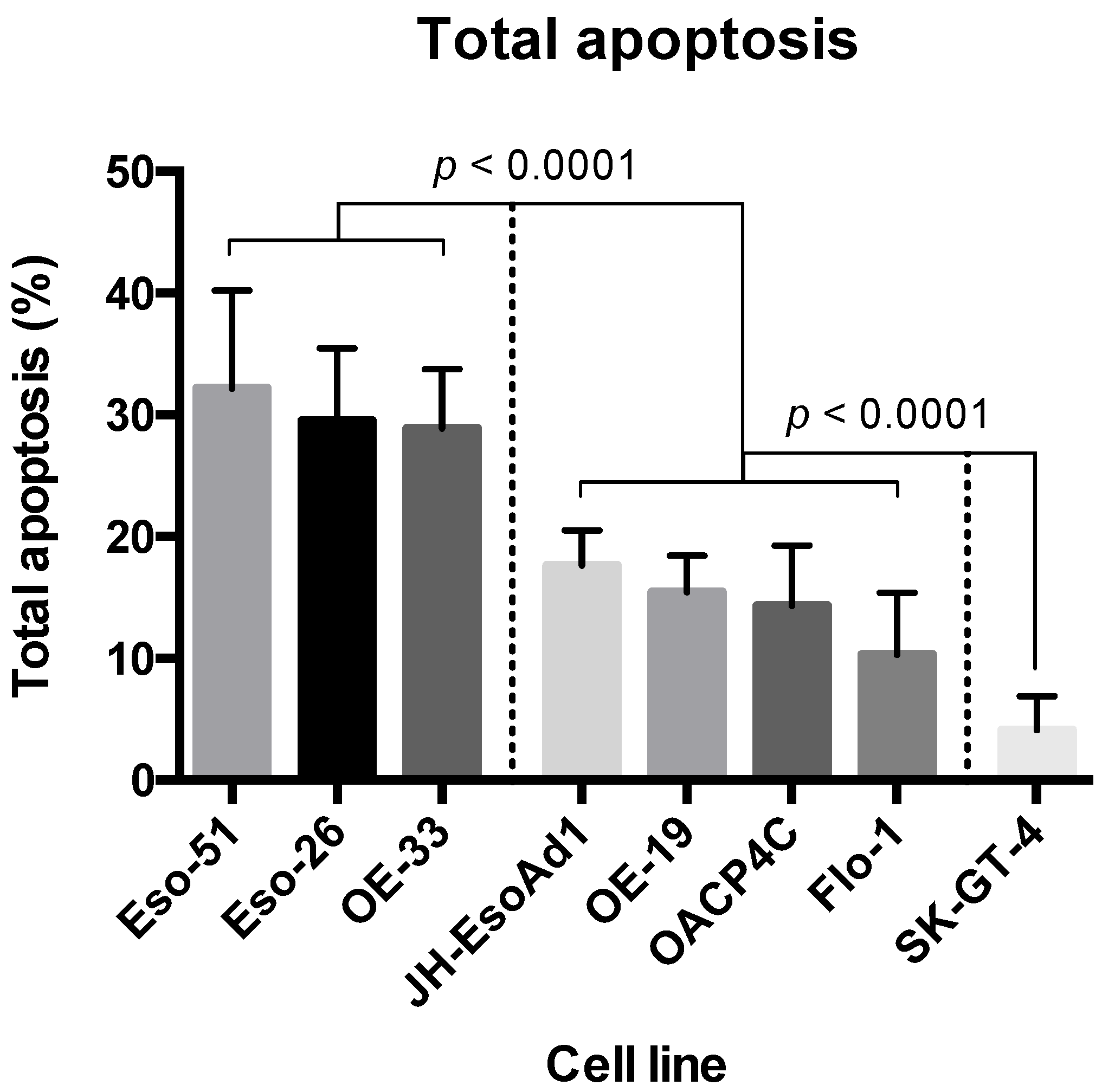
2.4. Apoptosis Assay
3. Protein Analysis
4. Results
4.1. Oestrogen Receptor Expression Patterns in Oesophageal Adenocarcinoma Cell Lines
4.2. Cytotoxicity of 4-Hydroxytamoxifen in Oesophageal Adenocarcinoma Cell Lines
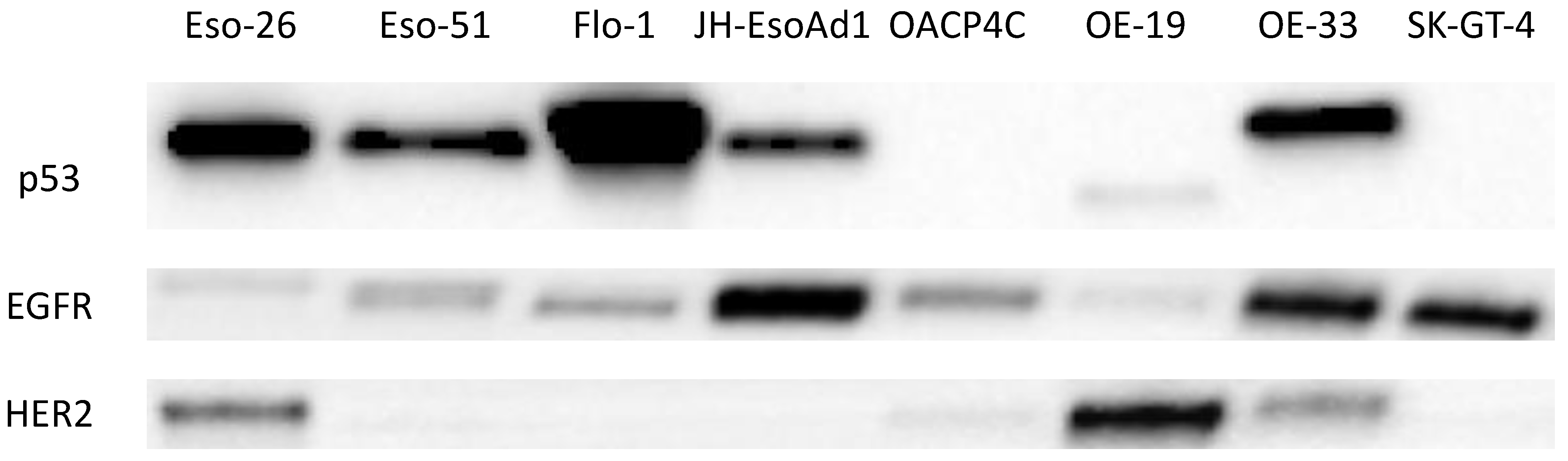
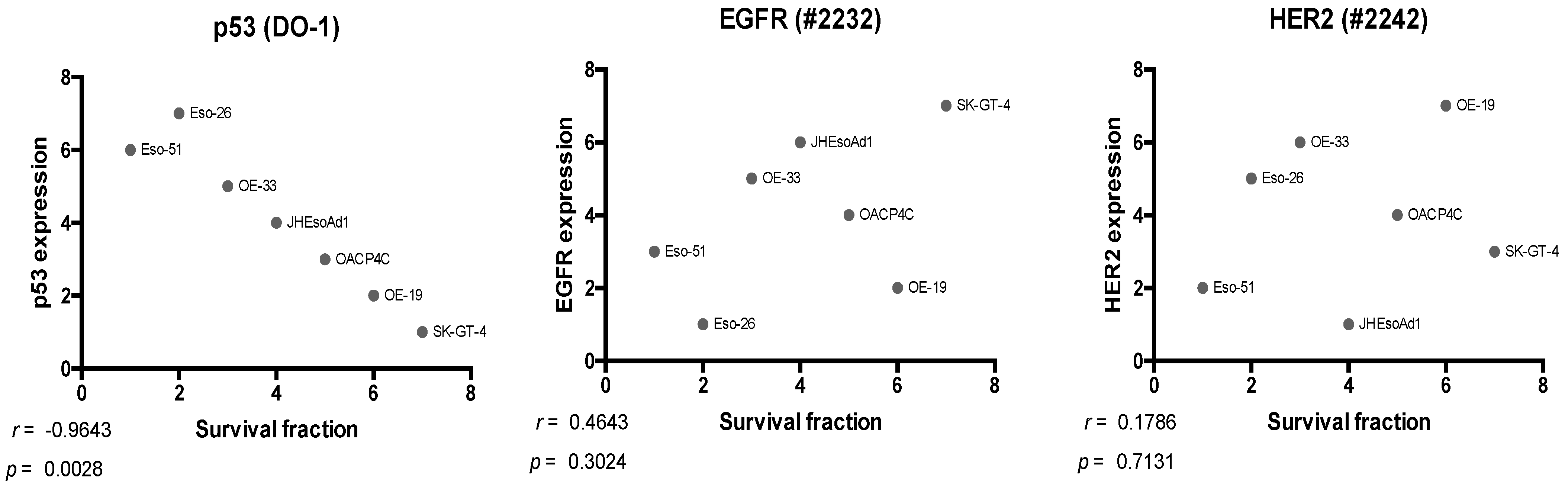
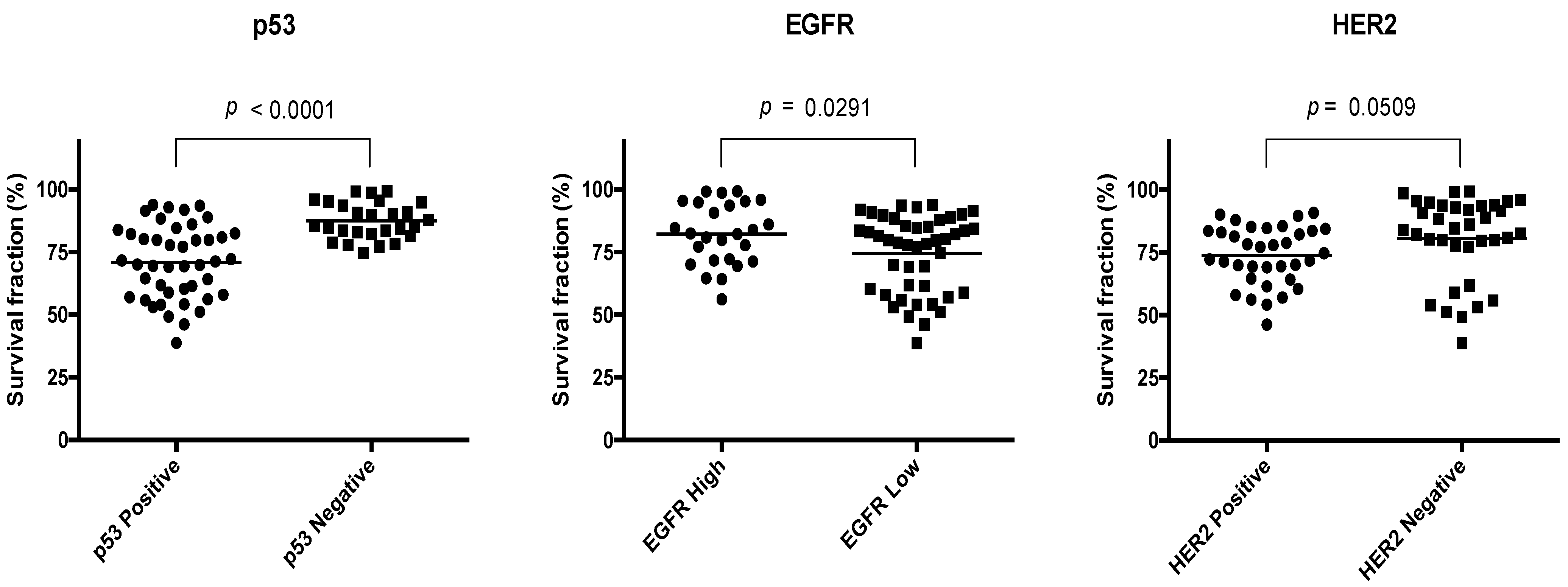
4.3. Expression of EGFR, HER2, and p53
4.4. Relationship between ER Expression and Response to Tamoxifen
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kauppila, J.H.; Mattsson, F.; Brusselaers, N.; Lagergren, J. Prognosis of oesophageal adenocarcinoma and squamous cell carcinoma following surgery and no surgery in a nationwide Swedish cohort study. BMJ Open 2018, 8, e021495. [Google Scholar] [CrossRef] [PubMed]
- Australian Institute of Health and Welfare. Cancer Survival and Prevalence in Australia: Period Estimates from 1982 to 2010; AIHW: Canberra, Australia, 2012. [Google Scholar]
- Pohl, H.; Welch, H.G. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J. Natl. Cancer Inst. 2005, 97, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Coleman, H.G.; Xie, S.-H.; Lagergren, J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology 2018, 154, 390–405. [Google Scholar] [CrossRef]
- Nagaraja, V.; Eslick, G.D. Forthcoming prognostic markers for esophageal cancer: A systematic review and meta-analysis. J. Gastrointest. Oncol. 2014, 5, 67–76. [Google Scholar] [CrossRef]
- Then, E.O.; Lopez, M.; Saleem, S.; Gayam, V.; Sunkara, T.; Culliford, A.; Gaduputi, V. Esophageal Cancer: An Updated Surveillance Epidemiology and End Results Database Analysis. World J. Oncol. 2020, 11, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.A.; Kennedy, E.B.; Catenacci, D.V.; Deighton, D.C.; Goodman, A.K.; Malhotra, N.K.; Willett, C.; Stiles, B.; Sharma, P.; Tanget, L.; et al. Treatment of Locally Advanced Esophageal Carcinoma: ASCO Guideline. J. Clin. Oncol. 2020, 38, 2677–2694, Erratum in: J. Clin. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Due, S.L.; Watson, D.I.; Hussey, D.J. Oestrogen receptors: A potential therapeutic target in oesophageal adenocarcinoma? ANZ J. Surg. 2021, 91, 1390–1396. [Google Scholar] [CrossRef]
- Chandanos, E.; Lindblad, M.; Jia, C.; Rubio, C.A.; Ye, W.; Lagergren, J. Tamoxifen exposure and risk of oesophageal and gastric adenocarcinoma: A population-based cohort study of breast cancer patients in Sweden. Br. J. Cancer 2006, 95, 118–122. [Google Scholar] [CrossRef]
- Akgun, H.; Lechago, J.; Younes, M. Estrogen receptor-beta is expressed in Barrett’s metaplasia and associated adenocarcinoma of the esophagus. Anticancer. Res. 2002, 22, 1459–1461. [Google Scholar]
- Tiffin, N.; Suvarna, S.K.; Trudgill, N.J.; Riley, S.A. Sex hormone receptor immunohistochemistry staining in Barrett’s oesophagus and adenocarcinoma. Histopathology 2003, 42, 95–96. [Google Scholar] [CrossRef]
- Liu, L.; Chirala, M.; Younes, M. Expression of estrogen receptor-beta isoforms in Barrett’s metaplasia, dysplasia and esophageal adenocarcinoma. Anticancer Res. 2004, 24, 2919–2924. [Google Scholar] [PubMed]
- Kalayarasan, R.; Ananthakrishnan, N.; Kate, V.; Basu, D. Estrogen and progesterone receptors in esophageal carcinoma. Dis. Esophagus 2008, 21, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Due, S.L.; Watson, D.I.; Bastian, I.; Ding, G.Q.; Sukocheva, O.A.; Astill, D.S.; Vat, L.; Hussey, D.J. Tamoxifen enhances the cytotoxicity of conventional chemotherapy in esophageal adenocarcinoma cells. Surg. Oncol. 2016, 25, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Sukocheva, O.A.; Wee, C.; Ansar, A.; Hussey, D.J.; Watson, D.I. Effect of estrogen on growth and apoptosis in esophageal adenocarcinoma cells. Dis. Esophagus 2013, 26, 628–635. [Google Scholar] [CrossRef]
- Thomas, C.; Gustafsson, J. The different roles of ER subtypes in cancer biology and therapy. Nat. Cancer 2011, 11, 597–608. [Google Scholar] [CrossRef]
- Konduri, S.D.; Medisetty, R.; Liu, W.; Kaipparettu, B.A.; Srivastava, P.; Brauch, H.; Fritz, P.; Swetzig, W.M.; Gardner, A.E.; Khan, S.A.; et al. Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc. Natl. Acad. Sci. USA 2010, 107, 15081–15086. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Morrison, G.; Gillihan, R.; Guo, J.; Ward, R.M.; Fu, X.; Botero, M.F.; Healy, N.A.; Hilsenbeck, S.G.; Phillips, G.L.; et al. Different mechanisms for resistance to trastuzumab versus lapatinib in HER2-positive breast cancers—Role of estrogen receptor and HER2 reactivation. Breast Cancer Res. 2011, 13, R121. [Google Scholar] [CrossRef]
- Berthois, Y.; Katzenellenbogen, J.A. Phenol red in tissue culture media is a weak estrogen: Implications concerning the study of estrogen-responsive cells in culture. Proc. Natl. Acad. Sci. USA 1986, 83, 2496–2500. [Google Scholar] [CrossRef]
- Cao, Z.; West, C.; Norton-Wenzel, C.S.; Rej, R.; Davis, F.B.; Davis, P.J. Effects of Resin or Charcoal Treatment on Fetal Bovine Serum and Bovine Calf Serum. Endocr. Res. 2009, 34, 101–108. [Google Scholar] [CrossRef]
- Seeger, H.; Huober, J.; Wallwiener, D.; Mueck, A.O. Inhibition of human breast cancer cell proliferation with estradiol metabolites is as effective as with tamoxifen. Horm. Metab. Res. 2004, 36, 277–280. [Google Scholar] [CrossRef]
- Kisanga, E.R.; Gjerde, J.; Gonzaga, A.G.; Pigatto, F.; Pesci-Feltri, A.; Robertson, C.; Serrano, D.; Pelosi, G.; DeCensi, A.; Lien, E.A. Tamoxifen and Metabolite Concentrations in Serum and Breast Cancer Tissue during Three Dose Regimens in a Randomized Preoperative Trial. Clin. Cancer Res. 2004, 10, 2336–2343. [Google Scholar] [CrossRef] [PubMed]
- Bekele, R.T.; Venkatraman, G.; Liu, R.-Z.; Tang, X.; Mi, S.; Benesch, M.; Mackey, J.R.; Godbout, R.; Curtis, J.M.; McMullen, T.P.W.; et al. Oxidative stress contributes to the tamoxifen-induced killing of breast cancer cells: Implications for tamoxifen therapy and resistance. Sci. Rep. 2016, 6, 21164. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Malerva, L.; Park, J.; Zou, L.; Hu, Y.; Moradpour, Z.; Pearlberg, J.; Sawyer, J.; Stevens, H.; Harlow, E.; LaBaer, J. High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc. Natl. Acad. Sci. USA 2011, 108, 2058–2063. [Google Scholar] [CrossRef] [PubMed]
- Wlodkowic, D.; Telford, W.; Skommer, J.; Darzynkiewicz, Z. Apoptosis and Beyond: Cytometry in Studies of Programmed Cell Death. Methods Cell Biol. 2011, 103, 55–98. [Google Scholar] [CrossRef]
- Mattes, M.J. Apoptosis assays with lymphoma cell lines: Problems and pitfalls. Br. J. Cancer 2007, 96, 928–936. [Google Scholar] [CrossRef][Green Version]
- Colella, A.D.; Chegenii, N.; Tea, M.N.; Gibbins, I.L.; Williams, K.A.; Chataway, T.K. Comparison of Stain-Free gels with traditional immunoblot loading control methodology. Anal. Biochem. 2012, 430, 108–110. [Google Scholar] [CrossRef]
- Dulak, A.M.; Stojanov, P.; Peng, S.; Lawrence, M.S.; Fox, C.; Stewart, C.; Bandla, S.; Imamura, Y.; Schumacher, S.E.; Shefler, E.; et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 2013, 45, 478–486. [Google Scholar] [CrossRef]
- Secrier, M.; Li, X.; De, S.N.; Eldridge, M.; Contino, G.; Bornschein, J.; MacRae, S.; Grehan, N.; O’Donovan, M.; Miremadi, A.; et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat. Genet. 2016, 48, 1131–1141. [Google Scholar] [CrossRef]
- Smith, S.D.; Enge, M.; Bao, W.; Thullberg, M.; Costa, T.D.; Olofsson, H.; Gashi, B.; Selivanova, G.; Strömblad, S. Protein kinase Cα (PKCα) regulates p53 localization and melanoma cell survival downstream of integrin αv in three-dimensional collagen and in vivo. J. Biol. Chem. 2012, 287, 29336–29347. [Google Scholar] [CrossRef]
- Slováčková, J.; Grochova, D.; Navrátilová, J.; Šmarda, J.; Smardova, J. Transactivation by temperature-dependent p53 mutants in yeast and human cells. Cell Cycle 2010, 9, 2141–2148. [Google Scholar] [CrossRef][Green Version]
- Bao, W.; Strömblad, S. Integrin alphav-mediated inactivation of p53 controls a MEK1-dependent melanoma cell survival pathway in three-dimensional collagen. J. Cell Biol. 2004, 167, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Mosoyan, G.; Nagi, C.; Marukian, S.; Teixeira, A.; Simonian, A.; Resnick-Silverman, L.; DiFeo, A.; Johnston, D.; Reynolds, S.R.; Roses, D.F.; et al. Multiple Breast Cancer Cell-Lines Derived from a Single Tumor Differ in Their Molecular Characteristics and Tumorigenic Potential. PLoS ONE 2013, 8, e55145. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, J.; Khan, S.; Zhang, Y.; Zhu, K.; Zhou, E.; Gong, M.; Liu, B.; Kan, Q.; Zhang, Q. Selective synergistic anticancer effects of cisplatin and oridonin against human p53-mutant esophageal squamous carcinoma cells. Anti-Cancer Drugs 2021, 33, e444–e452. [Google Scholar] [CrossRef] [PubMed]
- Eichelmann, A.-K.; Mayne, G.; Chiam, K.; Due, S.; Bastian, I.; Butz, F.; Wang, T.; Sykes, P.; Clemons, N.; Liu, D.; et al. Mutant p53 Mediates Sensitivity to Cancer Treatment Agents in Oesophageal Adenocarcinoma Associated with MicroRNA and SLC7A11 Expression. Int. J. Mol. Sci. 2021, 22, 5547. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Konduri, S.D.; Bansal, S.; Nayak, B.K.; Rajasekaran, S.A.; Karuppayil, S.M.; Rajasekaran, A.K.; Das, G.M. Estrogen Receptor-α Binds p53 Tumor Suppressor Protein Directly and Represses Its Function. J. Biol. Chem. 2006, 281, 9837–9840. [Google Scholar] [CrossRef]
- Fichter, C.D.; Herz, C.; Münch, C.; Opitz, O.G.; Werner, M.; Lassmann, S. Occurrence of multipolar mitoses and association with Aurora-A/-B kinases and p53 mutations in aneuploid esophageal carcinoma cells. BMC Cell Biol. 2011, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Mahidhara, R.S.; De Oliveira, P.E.Q.; Kohout, J.; Beer, D.G.; Lin, J.; Watkins, S.; Robbins, P.D.; Hughes, S.J. Altered trafficking of Fas and subsequent resistance to Fas-mediated apoptosis occurs by a wild-type p53 independent mechanism in esophageal adenocarcinoma1. J. Surg. Res. 2005, 123, 302–311. [Google Scholar] [CrossRef]
- Liu, D.S.H.; Read, M.; Cullinane, C.; Azar, W.J.; Fennell, C.M.; Montgomery, K.G.; Haupt, S.; Haupt, Y.; Wiman, K.G.; Duong, C.P.; et al. APR-246 potently inhibits tumour growth and overcomes chemoresistance in preclinical models of oesophageal adenocarcinoma. Gut 2015, 64, 1506–1516. [Google Scholar] [CrossRef]
- Inoue, K.; Fry, E.A. Aberrant Splicing of Estrogen Receptor, HER2, and CD44 Genes in Breast Cancer. Genet. Epigenetics 2015, 7, GEG.S35500-32. [Google Scholar] [CrossRef]
- Ascenzi, P.; Bocedi, A.; Marino, M. Structure-function relationship of estrogen receptor alpha and beta: Impact on human health. Mol. Aspects Med. 2006, 27, 299–402. [Google Scholar] [CrossRef]
- Zhao, C.; Matthews, J.; Tujague, M.; Wan, J.; Ström, A.; Toresson, G.; Lam, E.W.; Cheng, G.; Gustafsson, J.A.; Dahlman-Wright, K. Estrogen receptor beta2 negatively regulates the transactivation of estrogen receptor alpha in human breast cancer cells. Cancer Res. 2007, 67, 3955–3962. [Google Scholar] [CrossRef] [PubMed]
- Le Romancer, M.; Poulard, C.; Cohen, P.; Sentis, S.; Renoir, J.M.; Corbo, L. Cracking the estrogen receptor’s posttranslational code in breast tumors. Endocr. Rev. 2011, 32, 597–622. [Google Scholar] [CrossRef] [PubMed]
- Pink, J.J.; Wu, S.-Q.; Wolf, D.M.; Bilimoria, M.M.; Jordan, V.C. A Novel 80 kDa Human Estrogen Receptor Containing a Duplication of Exons 6 and 7. Nucleic Acids Res. 1996, 24, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Schafer, J.M.; Lee, E.S.; O’Regan, R.M.; Yao, K.; Jordan, V.C. Rapid development of tamoxifen-stimulated mutant p53 breast tumors (T47D) in athymic mice. Clin. Cancer Res. 2000, 6, 4373–4380. [Google Scholar] [PubMed]
- Powell, C.E.; Soto, A.M.; Sonnenschein, C. Identification and characterization of membrane estrogen receptor from MCF7 estrogen-target cells. J. Steroid Biochem. Mol. Biol. 2001, 77, 97–108. [Google Scholar] [CrossRef]
- Salomonsson, M.; Häggblad, J.; O’Malley, B.W.; Sitbon, G.M. The human estrogen receptor hormone binding domain dimerizes independently of ligand activation. J. Steroid Biochem. Mol. Biol. 1994, 48, 447–452. [Google Scholar] [CrossRef]
- Tamrazi, A.; Carlson, K.E.; Daniels, J.R.; Hurth, K.M.; Katzenellenbogen, J.A. Estrogen receptor dimerization: Ligand binding regulates dimer affinity and dimer dissociation rate. Mol. Endocrinol. 2002, 16, 2706–2719. [Google Scholar] [CrossRef]
- Moore, J.T.; McKee, D.D.; Slentz-Kesler, K.; Moore, L.B.; Jones, S.A.; Horne, E.L.; Su, J.L.; Kliewer, S.A.; Lehmann, J.M.; Willson, T.M. Cloning and characterization of human estrogen receptor beta isoforms. Biochem. Biophys. Res. Commun. 1998, 247, 75–78. [Google Scholar] [CrossRef]
- Leung, Y.K.; Mak, P.; Hassan, S.; Ho, S.M. Estrogen receptor (ER)-beta isoforms: A key to understanding ER-beta signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 13162–13167. [Google Scholar] [CrossRef]
- Aichler, M.; Motschmann, M.; Jütting, U.; Luber, B.; Becker, K.; Ott, K.; Lordick, F.; Langer, R.; Feith, M.; Siewert, J.R.; et al. Epidermal growth factor receptor (EGFR) is an independent adverse prognostic factor in esophageal adenocarcinoma patients treated with cisplatin-based neoadjuvant chemotherapy. Oncotarget 2014, 5, 6620–6632. [Google Scholar] [CrossRef][Green Version]
- Fichter, C.D.; Timme, S.; Braun, J.A.; Gudernatsch, V.; Schöpflin, A.; Bogatyreva, L.; Geddert, H.; Faller, G.; Klimstra, D.; Tang, L.; et al. EGFR, HER2 and HER3 dimerization patterns guide targeted inhibition in two histotypes of esophageal cancer. Int. J. Cancer 2014, 135, 1517–1530. [Google Scholar] [CrossRef] [PubMed]
- Sukocheva, O.A.; Li, B.; Due, S.L.; Hussey, D.J.; Watson, D.I. Androgens and esophageal cancer: What do we know? World J. Gastroenterol. 2015, 21, 6146–6156. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, J.J.; van Marion, R.; Beer, D.G.; Lin, L.; Chaves, P.; Ribeiro, C.; Pereira, A.D.; Roque, L.; Darnton, S.J.; Altorki, N.K.; et al. Verification and unmasking of widely used human esophageal adenocarcinoma cell lines. J. Natl. Cancer Inst. 2010, 102, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.J.; Nambu, Y.; Soldes, O.S.; Hamstra, D.; Rehemtulla, A.; Iannettoni, M.D.; Orringer, M.B.; Beer, D.G. Fas/APO-1 (CD95) is not translocated to the cell membrane in esophageal adenocarcinoma. Cancer Res. 1997, 57, 5571–5578. [Google Scholar] [PubMed]
- Alvarez, H.; Koorstra, J.B.; Hong, S.M.; Boonstra, J.J.; Dinjens, W.N.; Foratiere, A.A.; Wu, T.T.; Montgomery, E.; Eshleman, J.R.; Maitra, A. Establishment and characterization of a bona fide Barrett esophagus-associated adenocarcinoma cell line. Cancer Biol. Ther. 2008, 7, 1753–1755. [Google Scholar]
- De Both, N.J.; Wijnhoven, B.P.L.; Sleddens, H.F.B.M.; Tilanus, H.W.; Dinjens, W.N.M. Establishment of cell lines from adenocarcinomas of the esophagus and gastric cardia growing in vivo and in vitro. Virchows Arch. 2001, 438, 451–456. [Google Scholar] [CrossRef]
- Rockett, J.C.; Larkin, K.; Darnton, S.J.; Morris, A.G.; Matthews, H.R. Five newly established oesophageal carcinoma cell lines: Phenotypic and immunological characterization. Br. J. Cancer 1997, 75, 258–263. [Google Scholar] [CrossRef]
- Altorki, N.; Schwartz, G.K.; Blundell, M.; Davis, B.M.; Kelsen, D.P.; Albino, A.P. Characterization of cell lines established from human gastric-esophageal adenocarcinomas. Biologic phenotype and invasion potential. Cancer 1993, 72, 649–657. [Google Scholar] [CrossRef]
- Soule, H.D.; Vazguez, J.; Long, A.; Albert, S.; Brennan, M. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 1973, 51, 1409–1416. [Google Scholar] [CrossRef]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, 452–457. [Google Scholar] [CrossRef]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Bouaoun, L.; Sonkin, D.; Ardin, M.; Hollstein, M.; Byrnes, G.; Zavadil, J.; Olivier, M. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum. Mutat. 2016, 37, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Butz, F.; Eichelmann, A.-K.; Mayne, G.C.; Wang, T.; Bastian, I.; Chiam, K.; Marri, S.; Sykes, P.J.; Wijnhoven, B.P.; Toxopeus, E.; et al. MicroRNA Profiling in Oesophageal Adenocarcinoma Cell Lines and Patient Serum Samples Reveals a Role for miR-451a in Radiation Resistance. Int. J. Mol. Sci. 2020, 21, 8898. [Google Scholar] [CrossRef] [PubMed]
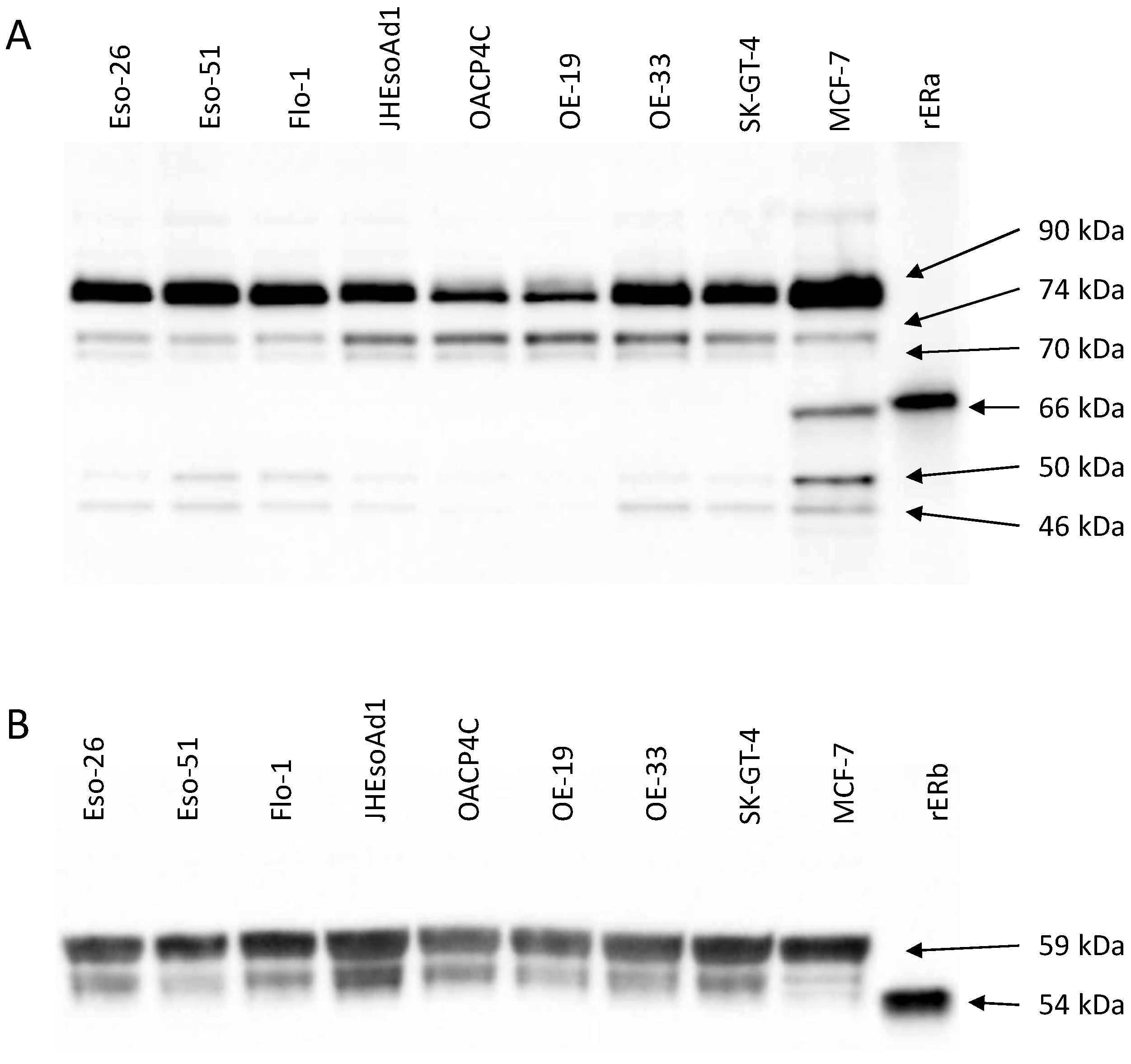
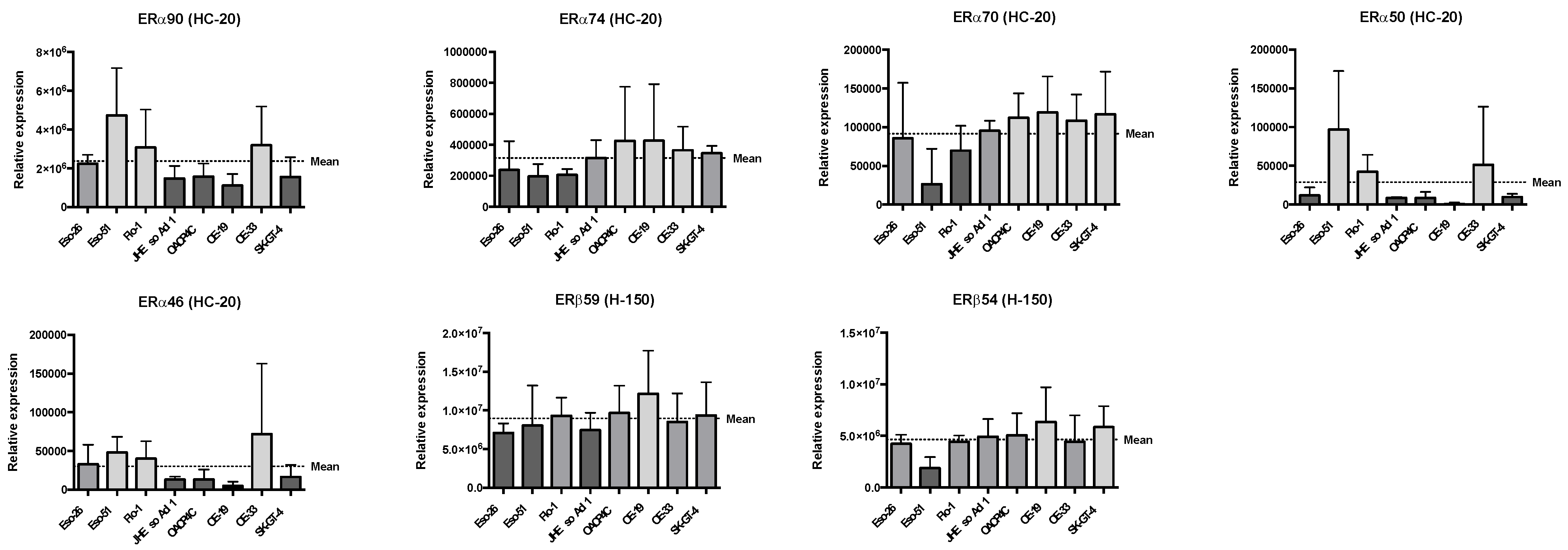
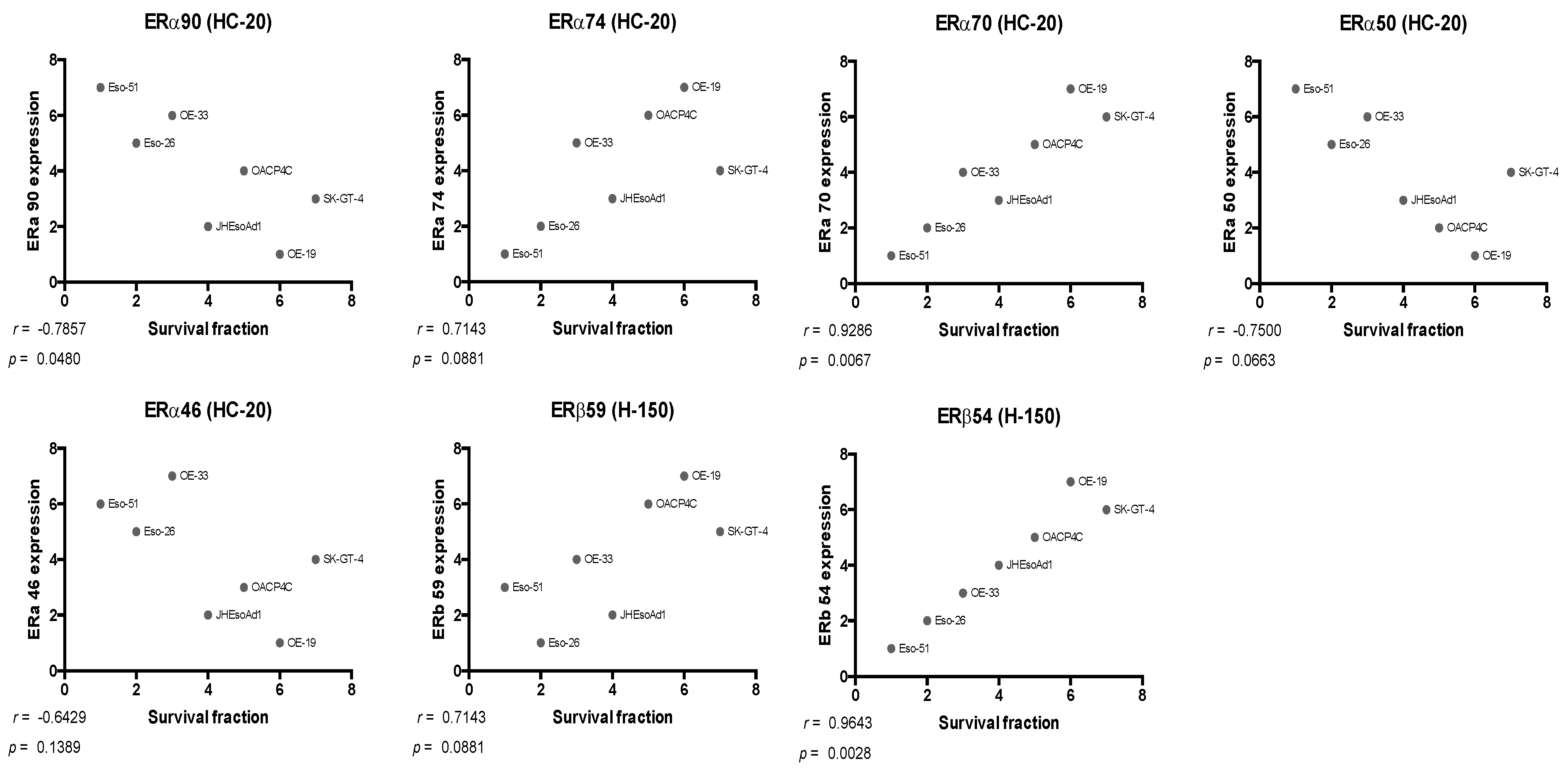
| ERα74 | ρ = −0.619 | |||||
| p = 0.115 | ||||||
| ERα70 | ρ = −0.738 | ρ = 0.929 | ||||
| p = 0.046 | p = 0.002 | |||||
| ERα50 | ρ = 0.929 | ρ = −0.738 | ρ = −0.762 | |||
| p = 0.002 | p = 0.046 | p = 0.037 | ||||
| ERα46 | ρ = 0.952 | ρ = −0.548 | ρ = −0.619 | ρ = 0.952 | ||
| p = 0.001 | p = 0.171 | p = 0.115 | p = 0.001 | |||
| ERβ59 | ρ = −0.405 | ρ = 0.690 | ρ = 0.714 | ρ = −0.548 | ρ = −0.405 | |
| p = 0.327 | p = 0.069 | p = 0.058 | p = 0.171 | p = 0.327 | ||
| ERβ54 | ρ = −0.857 | ρ = 0.762 | ρ = 0.881 | ρ = −0.857 | ρ = −0.786 | ρ = 0.786 |
| p = 0.011 | p = 0.037 | p = 0.007 | p = 0.011 | p = 0.028 | p = 0.028 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Due, S.L.; Watson, D.I.; Bastian, I.; Eichelmann, A.-K.; Hussey, D.J. Oestrogen Receptor Isoforms May Represent a Therapeutic Target in Oesophageal Adenocarcinoma. Cancers 2022, 14, 1891. https://doi.org/10.3390/cancers14081891
Due SL, Watson DI, Bastian I, Eichelmann A-K, Hussey DJ. Oestrogen Receptor Isoforms May Represent a Therapeutic Target in Oesophageal Adenocarcinoma. Cancers. 2022; 14(8):1891. https://doi.org/10.3390/cancers14081891
Chicago/Turabian StyleDue, Steven L., David I. Watson, Isabell Bastian, Ann-Kathrin Eichelmann, and Damian J. Hussey. 2022. "Oestrogen Receptor Isoforms May Represent a Therapeutic Target in Oesophageal Adenocarcinoma" Cancers 14, no. 8: 1891. https://doi.org/10.3390/cancers14081891
APA StyleDue, S. L., Watson, D. I., Bastian, I., Eichelmann, A.-K., & Hussey, D. J. (2022). Oestrogen Receptor Isoforms May Represent a Therapeutic Target in Oesophageal Adenocarcinoma. Cancers, 14(8), 1891. https://doi.org/10.3390/cancers14081891






