Context-Aware Saliency Guided Radiomics: Application to Prediction of Outcome and HPV-Status from Multi-Center PET/CT Images of Head and Neck Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Multi-Center Data Set
2.2. PET/CT Imaging
2.3. Clinical Parameters and Outcome Information
2.4. Tumor Segmentation
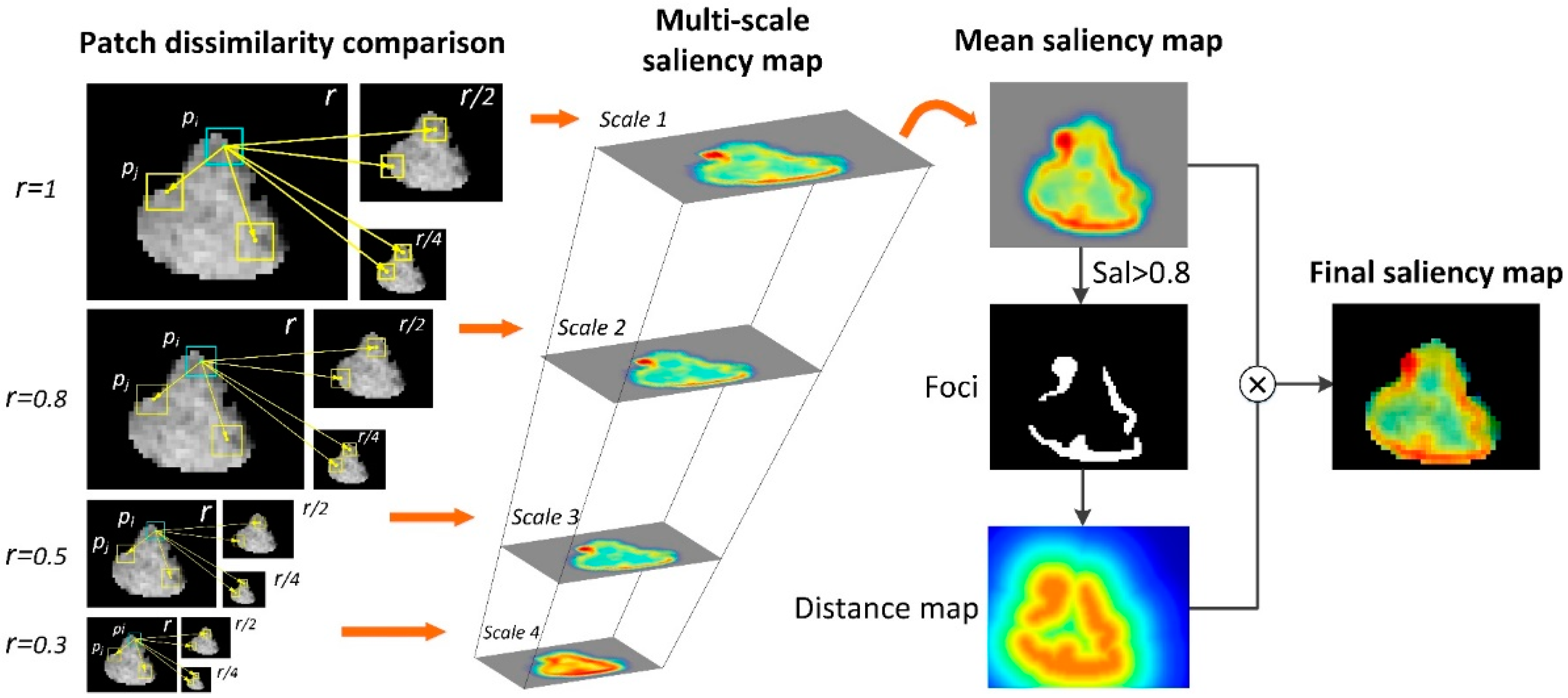
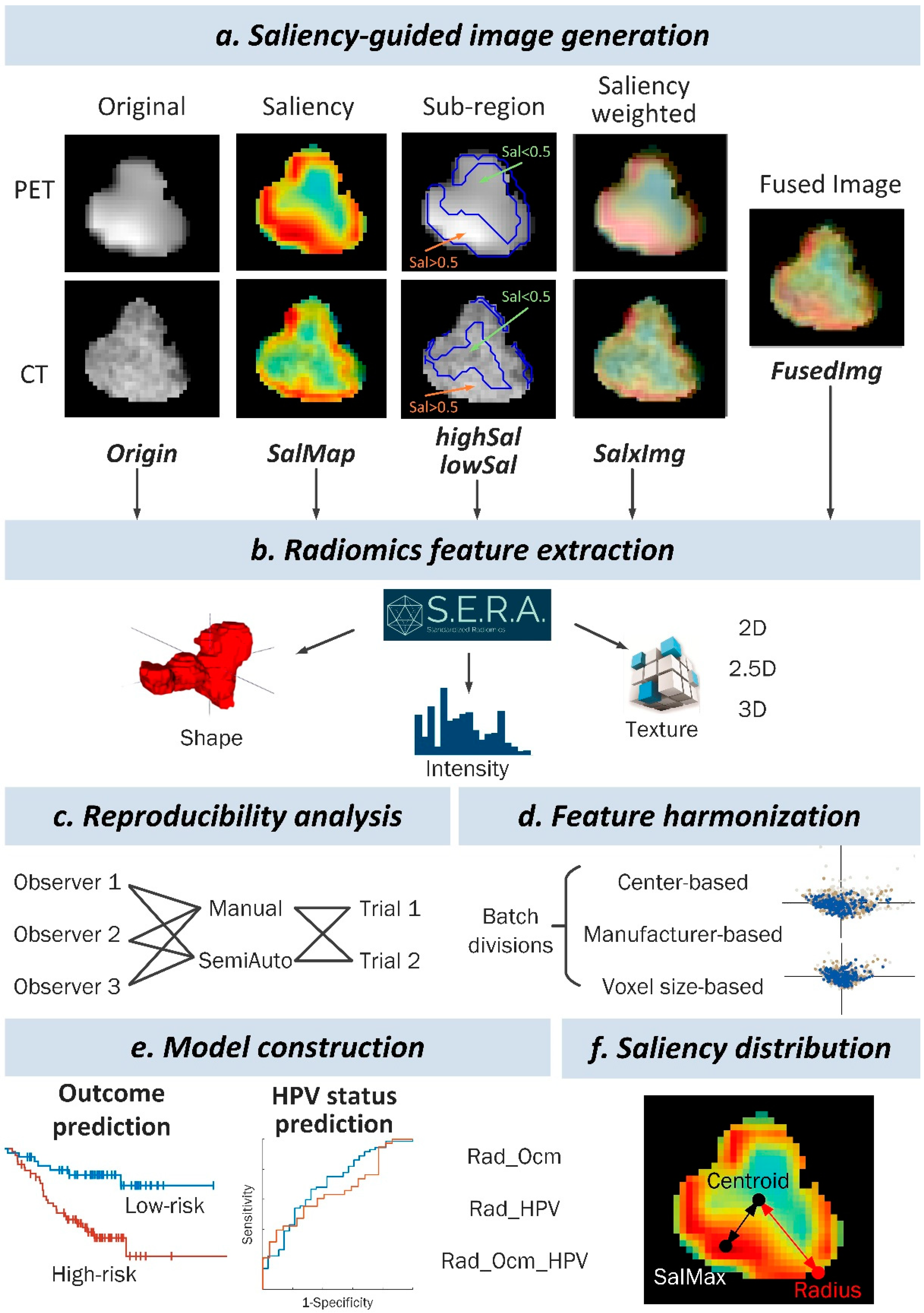
2.5. Context-Aware Saliency Map Generation
2.6. Feature Extraction
2.7. Reproducibility Analysis
2.8. Feature Harmonization
2.9. Model Construction
3. Results
3.1. Patient Characteristics
3.2. Feature Screening
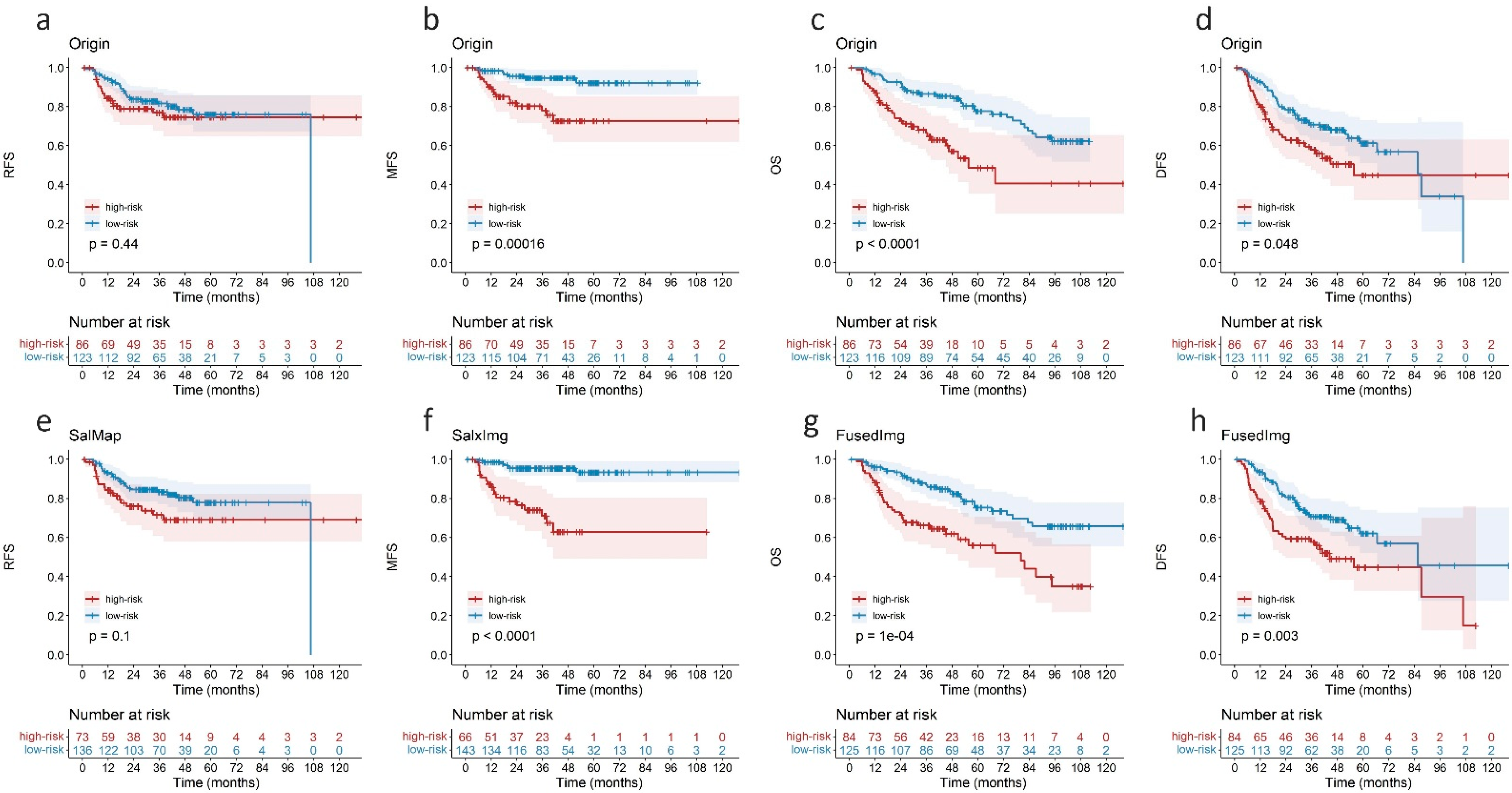
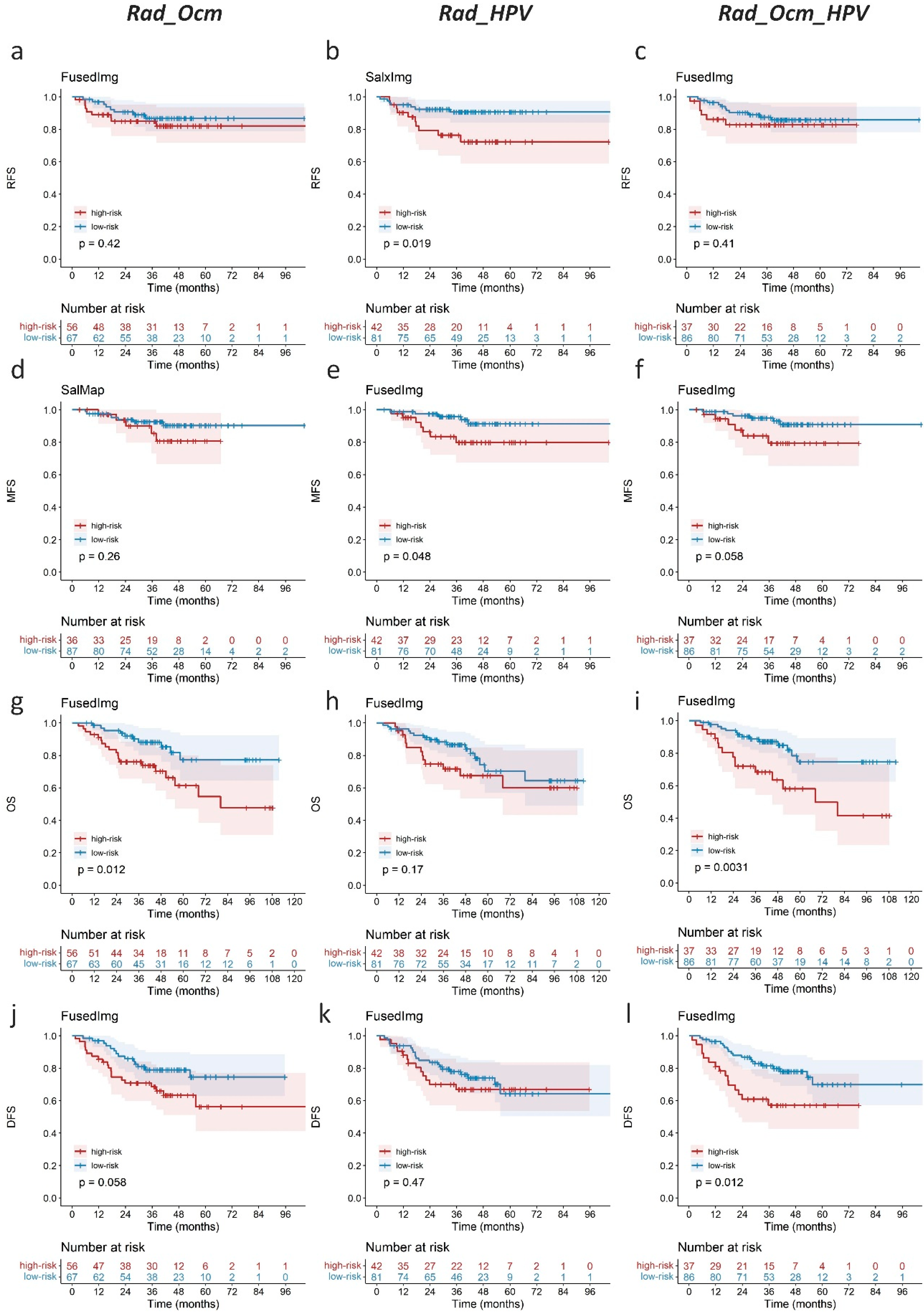
3.3. Performance of Saliency Guided Prognostic Model
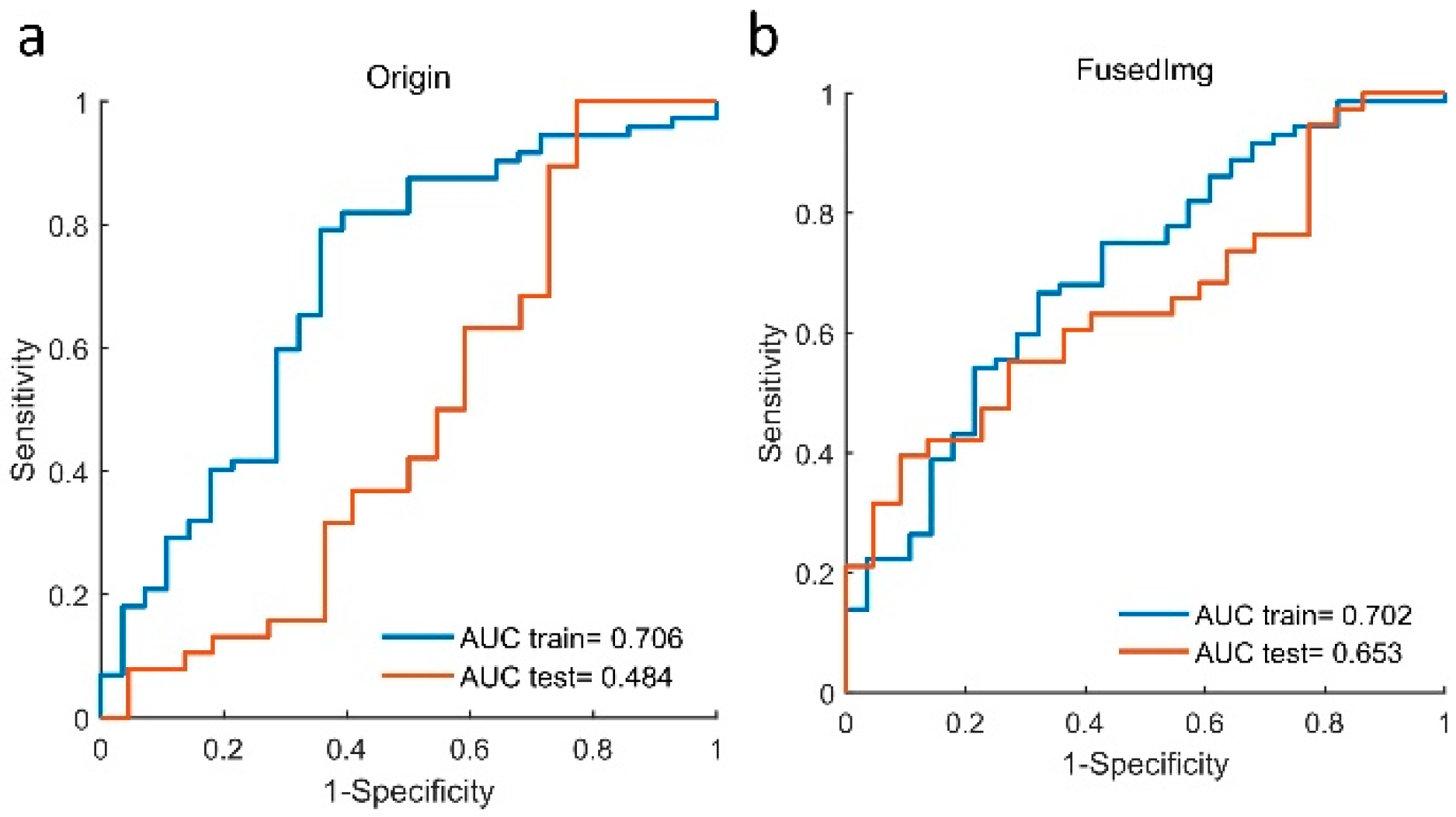
3.4. Complementary Prognostic Value of Radiomics Score of HPV Status
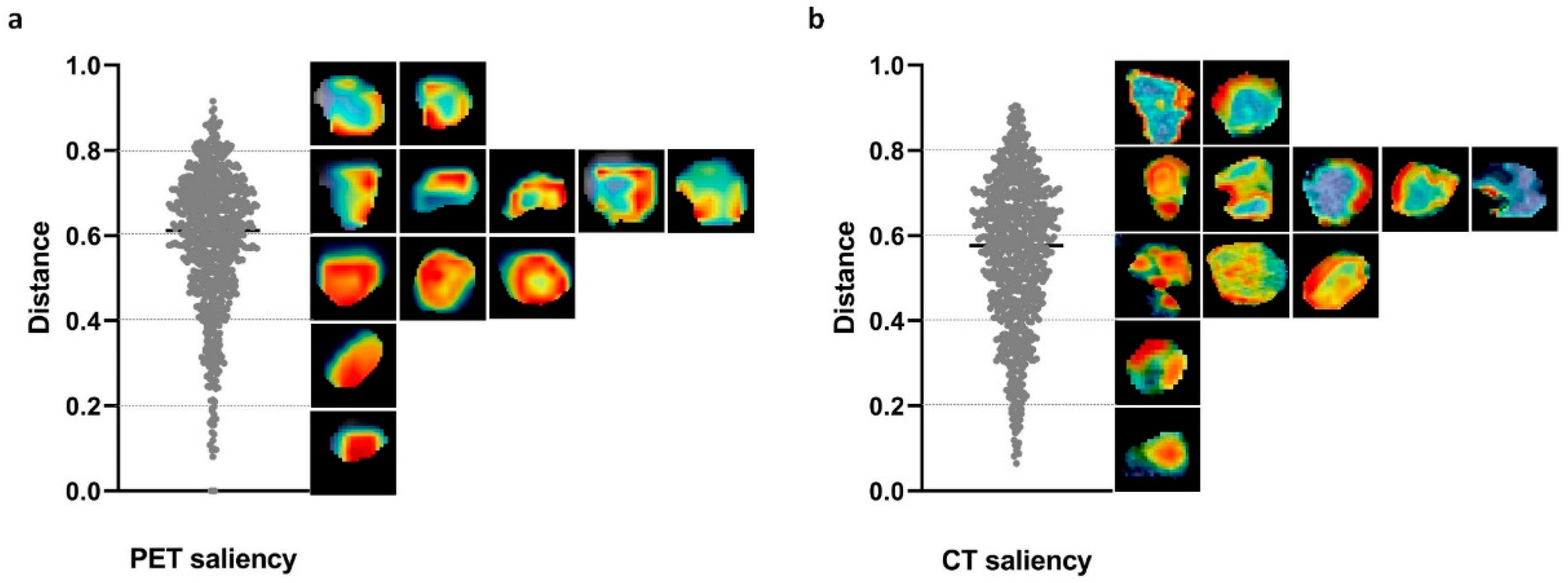
3.5. Saliency Distribution Localization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar]
- Mroz, E.A.; Rocco, J.W. Intra-tumor heterogeneity in head and neck cancer and its clinical implications. World J. Otorhinolaryngol. Head Neck Surg. 2016, 2, 60–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canning, M.; Guo, G.; Yu, M.; Myint, C.; Groves, M.W.; Byrd, J.K.; Cui, Y. Heterogeneity of the Head and Neck Squamous Cell Carcinoma Immune Landscape and Its Impact on Immunotherapy. Front. Cell Dev. Biol. 2019, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Reuze, S.; Schernberg, A.; Orlhac, F.; Sun, R.; Chargari, C.; Dercle, L.; Deutsch, E.; Buvat, I.; Robert, C. Radiomics in Nuclear Medicine Applied to Radiation Therapy: Methods, Pitfalls, and Challenges. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1117–1142. [Google Scholar] [CrossRef] [PubMed]
- Mody, M.D.; Rocco, J.W.; Yom, S.S.; Haddad, R.I.; Saba, N.F. Head and neck cancer. Lancet 2021, 398, 2289–2299. [Google Scholar] [CrossRef]
- Seiwert, T.Y.; Foster, C.C.; Blair, E.A.; Karrison, T.G.; Agrawal, N.; Melotek, J.M.; Portugal, L.; Brisson, R.J.; Dekker, A.; Kochanny, S.; et al. OPTIMA: A phase II dose and volume de-escalation trial for human papillomavirus-positive oropharyngeal cancer. Ann. Oncol. 2019, 30, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Di Stasio, D.; Montella, M.; Romano, A.; Colella, G.; Serpico, R.; Lucchese, A. High-Definition Ultrasound Characterization of Squamous Carcinoma of the Tongue: A Descriptive Observational Study. Cancers 2022, 14, 564. [Google Scholar] [CrossRef]
- Cicciù, M.; Cervino, G.; Fiorillo, L.; D’Amico, C.; Oteri, G.; Troiano, G.; Zhurakivska, K.; Lo Muzio, L.; Herford, A.S.; Crimi, S.; et al. Early Diagnosis on Oral and Potentially Oral Malignant Lesions: A Systematic Review on the VELscope(®) Fluorescence Method. Dent. J. 2019, 7, 93. [Google Scholar] [CrossRef] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Haider, S.P.; Burtness, B.; Yarbrough, W.G.; Payabvash, S. Applications of radiomics in precision diagnosis, prognostication and treatment planning of head and neck squamous cell carcinomas. Cancers Head Neck 2020, 5, 6. [Google Scholar] [CrossRef]
- Bruixola, G.; Remacha, E.; Jimenez-Pastor, A.; Dualde, D.; Viala, A.; Monton, J.V.; Ibarrola-Villava, M.; Alberich-Bayarri, A.; Cervantes, A. Radiomics and radiogenomics in head and neck squamous cell carcinoma: Potential contribution to patient management and challenges. Cancer Treat. Rev. 2021, 99, 102263. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.B.; Yuan, Q.Y.; Wang, Q.S.; Ma, J.H.; Feng, Q.J.; Chen, W.F.; Rahmim, A.; Lu, L.J. Radiomics Analysis of PET and CT Components of PET/CT Imaging Integrated with Clinical Parameters: Application to Prognosis for Nasopharyngeal Carcinoma. Mol. Imaging Biol. 2019, 21, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Gerashchenko, T.S.; Denisov, E.V.; Litviakov, N.V.; Zavyalova, M.V.; Vtorushin, S.V.; Tsyganov, M.M.; Perelmuter, V.M.; Cherdyntseva, N.V. Intratumor heterogeneity: Nature and biological significance. Biochemestry 2013, 78, 1201–1215. [Google Scholar] [CrossRef] [PubMed]
- Differding, S.; Hanin, F.X.; Grégoire, V. PET imaging biomarkers in head and neck cancer. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 613–622. [Google Scholar] [CrossRef]
- De Vries, E.G.E.; Kist de Ruijter, L.; Lub-de Hooge, M.N.; Dierckx, R.A.; Elias, S.G.; Oosting, S.F. Integrating molecular nuclear imaging in clinical research to improve anticancer therapy. Nat. Rev. Clin. Oncol. 2019, 16, 241–255. [Google Scholar] [CrossRef]
- Mu, W.; Liang, Y.; Hall, L.O.; Tan, Y.; Balagurunathan, Y.; Wenham, R.; Wu, N.; Tian, J.; Gillies, R.J. 18F-FDG PET/CT Habitat Radiomics Predicts Outcome of Patients with Cervical Cancer Treated with Chemoradiotherapy. Radiol. Artif. Intell. 2020, 2, e190218. [Google Scholar] [CrossRef]
- Wu, J.; Gensheimer, M.F.; Dong, X.; Rubin, D.L.; Napel, S.; Diehn, M.; Loo, B.W., Jr.; Li, R. Robust Intratumor Partitioning to Identify High-Risk Subregions in Lung Cancer: A Pilot Study. Int. J. Radiat. Oncol. Biol. Phys. 2016, 95, 1504–1512. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Lv, W.B.; Feng, H.; Du, D.Y.; Yuan, Q.Y.; Wang, Q.S.; Dai, Z.H.; Yang, W.; Feng, Q.J.; Ma, J.H.; et al. Subregional Radiomics Analysis of PET/CT Imaging with Intratumor Partitioning: Application to Prognosis for Nasopharyngeal Carcinoma. Mol. Imaging Biol. 2020, 22, 1414–1426. [Google Scholar] [CrossRef]
- Cong, R.; Lei, J.; Fu, H.; Cheng, M.M.; Lin, W.; Huang, Q. Review of Visual Saliency Detection with Comprehensive Information. IEEE Trans. Circ. Syst. Vid. 2019, 29, 2941–2959. [Google Scholar] [CrossRef] [Green Version]
- Itti, L.; Koch, C. Computational modelling of visual attention. Nat. Rev. Neurosci. 2001, 2, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Walther, D.; Koch, C. Modeling attention to salient proto-objects. Neural. Netw. 2006, 19, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zhang, L. Saliency Detection: A Spectral Residual Approach. In Proceedings of the IEEE Conference Comput. Vis. Pattern Recognit., Minneapolis, MN, USA, 17–22 June 2007. [Google Scholar]
- Goferman, S.; Zelnik-Manor, L.; Tal, A. Context-Aware Saliency Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 1915–1926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwanenburg, A. Radiomics in nuclear medicine: Robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2638–2655. [Google Scholar] [CrossRef]
- Orlhac, F.; Eertink, J.J.; Cottereau, A.S.; Zijlstra, J.M.; Thieblemont, C.; Meignan, M.A.; Boellaard, R.; Buvat, I. A guide to ComBat harmonization of imaging biomarkers in multicenter studies. J. Nucl. Med. 2021, 63, 172–179. [Google Scholar] [CrossRef]
- Lv, W.B.; Ashrafinia, S.; Ma, J.H.; Lu, L.J.; Rahmim, A. Multi-Level Multi-Modality Fusion Radiomics: Application to PET and CT Imaging for Prognostication of Head and Neck Cancer. IEEE J. Biomed. Health Inform. 2020, 24, 2268–2277. [Google Scholar] [CrossRef]
- Lv, W.B.; Feng, H.; Du, D.Y.; Ma, J.H.; Lu, L.J. Complementary Value of Intra- and Peri-Tumoral PET/CT Radiomics for Outcome Prediction in Head and Neck Cancer. IEEE Access 2021, 9, 81818–81827. [Google Scholar] [CrossRef]
- McNitt-Gray, M.; Napel, S.; Jaggi, A.; Mattonen, S.A.; Hadjiiski, L.; Muzi, M.; Goldgof, D.; Balagurunathan, Y.; Pierce, L.A.; Kinahan, P.E.; et al. Standardization in Quantitative Imaging: A Multicenter Comparison of Radiomic Features from Different Software Packages on Digital Reference Objects and Patient Data Sets. Tomography 2020, 6, 118–128. [Google Scholar] [CrossRef]
- Lei, M.; Varghese, B.; Hwang, D.; Cen, S.; Lei, X.; Desai, B.; Azadikhah, A.; Oberai, A.; Duddalwar, V. Benchmarking Various Radiomic Toolkit Features While Applying the Image Biomarker Standardization Initiative toward Clinical Translation of Radiomic Analysis. J. Digit. Imaging 2021, 34, 1156–1170. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Bradshaw, T.J.; Boellaard, R.; Dutta, J.; Jha, A.K.; Jacobs, P.; Li, Q.; Liu, C.; Sitek, A.; Saboury, B.; Scott, P.J.H.; et al. Nuclear Medicine and Artificial Intelligence: Best Practices for Algorithm Development. J. Nucl. Med. 2021, in press. [CrossRef] [PubMed]
- Amini, M.; Nazari, M.; Shiri, I. Multi-level multi-modality (PET and CT) fusion radiomics: Prognostic modeling for non-small cell lung carcinoma. Phys. Med. Biol. 2021, 66, 205017. [Google Scholar] [CrossRef] [PubMed]
- Vuong, D.; Tanadini-Lang, S.; Wu, Z.; Marks, R.; Unkelbach, J.; Hillinger, S.; Eboulet, E.I.; Thierstein, S.; Peters, S.; Pless, M.; et al. Radiomics Feature Activation Maps as a New Tool for Signature Interpretability. Front. Oncol. 2020, 10, 578895. [Google Scholar] [CrossRef] [PubMed]
- Saito, N.; Nadgir, R.N.; Nakahira, M.; Takahashi, M.; Uchino, A.; Kimura, F.; Truong, M.T.; Sakai, O. Posttreatment CT and MR Imaging in Head and Neck Cancer: What the Radiologist Needs to Know. RadioGraphics 2012, 32, 1261–1282. [Google Scholar] [CrossRef] [Green Version]
- De Felice, F.; de Vincentiis, M.; Valentini, V.; Musio, D.; Mezi, S.; Lo Mele, L.; Terenzi, V.; D’Aguanno, V.; Cassoni, A.; Di Brino, M.; et al. Follow-up program in head and neck cancer. Crit. Rev. Oncol. Hematol. 2017, 113, 151–155. [Google Scholar] [CrossRef]
- Cheng, N.M.; Chang, J.T.; Huang, C.G.; Tsan, D.L.; Ng, S.H.; Wang, H.M.; Liao, C.T.; Lin, C.Y.; Hsu, C.L.; Yen, T.C. Prognostic value of pretreatment 18F-FDG PET/CT and human papillomavirus type 16 testing in locally advanced oropharyngeal squamous cell carcinoma. Eur. J. Nucl. Med. Mol. Imaging 2012, 39, 1673–1684. [Google Scholar] [CrossRef]
- Haider, S.P.; Mahajan, A.; Zeevi, T.; Baumeister, P.; Reichel, C.; Sharaf, K.; Forghani, R.; Kucukkaya, A.S.; Kann, B.H.; Judson, B.L.; et al. PET/CT radiomics signature of human papilloma virus association in oropharyngeal squamous cell carcinoma. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2978–2991. [Google Scholar] [CrossRef]
- Huang, C.; Cintra, M.; Brennan, K.; Zhou, M.; Colevas, A.D.; Fischbein, N.; Zhu, S.; Gevaert, O. Development and validation of radiomic signatures of head and neck squamous cell carcinoma molecular features and subtypes. EBioMedicine 2019, 45, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Orlhac, F.; Frouin, F.; Nioche, C.; Ayache, N.; Buvat, I. Validation of A Method to Compensate Multicenter Effects Affecting CT Radiomics. Radiology 2019, 291, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Orlhac, F.; Boughdad, S.; Philippe, C.; Stalla-Bourdillon, H.; Nioche, C.; Champion, L.; Soussan, M.; Frouin, F.; Frouin, V.; Buvat, I. A Postreconstruction Harmonization Method for Multicenter Radiomic Studies in PET. J. Nucl. Med. 2018, 59, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Da-Ano, R.; Visvikis, D.; Hatt, M. Harmonization strategies for multicenter radiomics investigations. Phys. Med. Biol. 2020, 65, 24TR02. [Google Scholar] [CrossRef] [PubMed]
- Daano, R.; Masson, I.; Lucia, F.; Doré, M.; Robin, P.; Alfieri, J.; Rousseau, C.; Mervoyer, A.; Md, C.; Castelli, J.; et al. Performance comparison of modified ComBat for harmonization of radiomic features for multicenter studies. Sci. Rep. 2020, 10, 10248. [Google Scholar] [CrossRef] [PubMed]
- Tixier, F.; Jaouen, V.; Hognon, C.; Gallinato, O.; Colin, T.; Visvikis, D. Evaluation of conventional and deep learning based image harmonization methods in radiomics studies. Phys. Med. Biol. 2021, 66, 245009. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | All | Training | Internal Validation | External Testing | OPC HPV Training | OPC HPV Testing | OPC Testing |
|---|---|---|---|---|---|---|---|
| No. | 806 | 500 | 97 | 209 | 100 | 60 | 123 |
| Age, median (range) | 60 (18–91) | 58 (18–90) | 57 (20–82) | 63 (26–90) | 62 (34–81) | 60 (44–76) | 63 (38–90) |
| Sex, no. (%) | |||||||
| Male | 654 (81%) | 404 (81%) | 78 (80%) | 163 (78%) | 82 (82%) | 47 (78%) | 93 (76%) |
| Female | 161 (19%) | 96 (19%) | 19 (20%) | 46 (22%) | 18 (18%) | 13 (22%) | 30 (24%) |
| Smoking, no. (%) | |||||||
| Non-smoker | 122 (15%) | 87 (17%) | 25 (26%) | 10 (5%) | 16 (16%) | 2 (3%) | 3 (2%) |
| Former-smoker | 107 (13%) | 70 (14%) | 27 (28%) | 10 (5%) | 3 (%) | 2 (3%) | 5 (4%) |
| Current-smoker | 207 (26%) | 153 (30%) | 45 (46%) | 9 (4%) | 10 (%) | - | 3 (2%) |
| TNM Stage, no. (%) | |||||||
| I | 28 (3%) | 7 (2%) | 3 (3%) | 18 (9%) | 2 (%) | 3 (5%) | 4 (3%) |
| II | 60 (8%) | 36 (7%) | 8 (8%) | 16 (8%) | 8 (%) | 3 (5%) | 7 (6%) |
| III | 148 (18%) | 106 (21%) | 13 (14%) | 29 (13%) | 14 (%) | 7 (12%) | 14 (11%) |
| IV | 570 (71%) | 351 (70%) | 73 (75%) | 146 (70%) | 76 (%) | 47 (78%) | 98 (80%) |
| Site, no. (%) | |||||||
| Nasopharynx | 37 (5%) | 29 (6%) | - | 8 (4%) | - | - | - |
| Oropharynx | 507 (63%) | 317 (63%) | 67 (69%) | 123 (60%) | 100 (100%) | 60 (100%) | 123 (100%) |
| Hypopharynx | 29 (3%) | 18 (4%) | 2 (2%) | 9 (4%) | - | - | - |
| Larynx | 144 (18%) | 85 (17%) | 6 (7%) | 53 (25%) | - | - | - |
| Oral cavity | 57 (7%) | 29 (6%) | 17 (18%) | 11 (5%) | - | - | - |
| Others | 32 (4%) | 22 (4%) | 5 (5%) | 5 (2%) | - | - | - |
| Therapy, no. (%) | |||||||
| Surgery | 35 (4%) | 1 (1%) | 9 (9%) | 25 (12%) | - | 4 (7%) | 10 (8%) |
| RT | 141 (18%) | 69 (13%) | 3 (3%) | 69 (33%) | 16 (%) | 20 (33%) | 29 (24%) |
| Surgery + RT | 42 (5%) | 30 (6%) | 7 (7%) | 5 (2%) | - | - | - |
| CRT | 508 (63%) | 334 (67%) | 67 (69%) | 107 (51%) | 77 (%) | 35 (58%) | 82 (67%) |
| Surgery + CRT | 80 (10%) | 66 (13%) | 11 (12%) | 3 (2%) | 7 (%) | 1 (2%) | 2 (1%) |
| HPV status, no. (%) | |||||||
| Positive | 115 (14%) | 76 (15%) | - | 39 (18%) | 72 (%) | 38 (63%) | 38 (31%) |
| Negative | 86 (11%) | 55 (11%) | - | 31 (14%) | 28 (%) | 22 (37%) | 22 (18%) |
| Grade, no. (%) | |||||||
| 1 | 17 (2%) | 15 (3%) | 2 (2%) | - | 2 (%) | - | - |
| 2 | 212 (26%) | 148 (30%) | 47 (48%) | 17 (8%) | 8 (%) | 2 (3%) | 8 (7%) |
| 3 | 139 (17%) | 104 (21%) | 24 (25%) | 11 (5%) | 17 (%) | 2 (3%) | 3 (2%) |
| Outcome, no. (%) | |||||||
| Recurrence | 215 (27%) | 134 (27%) | 36 (37%) | 45 (22%) | 12 (%) | 7 (12%) | 18 (15%) |
| Metastasis | 168 (21%) | 115 (23%) | 27 (28%) | 26 (12%) | 13 (%) | 4 (7%) | 12 (10%) |
| Death | 244 (30%) | 151 (30%) | 27 (28%) | 66 (32%) | 10 (%) | 16 (27%) | 29 (24%) |
| Model Configuration | External Testing C-Index | ||||||
|---|---|---|---|---|---|---|---|
| Image Type | Modality | Harmonization Method | Num of Features | RFS | MFS | OS | DFS |
| Origin | CT | None | 3 | 0.559 | 0.739 | 0.659 | 0.582 |
| (0.459–0.654) | (0.637–0.843) | (0.587–0.739) | (0.508–0.661) | ||||
| SalMap | Clin + CT | None | 8 | 0.621 | 0.759 | 0.677 | 0.636 |
| (0.518–0.721) | (0.668–0.848) | (0.604–0.756) | (0.562–0.713) | ||||
| highSal | CT | Scanner-based | 20 | 0.575 | 0.676 | 0.600 | 0.563 |
| (0.469–0.679) | (0.572–0.787) | (0.524–0.681) | (0.483–0.645) | ||||
| lowSal | CT | Scanner-based | 4 | 0.604 | 0.778 | 0.663 | 0.612 |
| (0.499–0.705) | (0.689–0.866) | (0.599–0.732) | (0.539–0.687) | ||||
| SalxImg | PET + CT | Voxel size-based | 2 | 0.616 | 0.785 | 0.675 | 0.633 |
| (0.537–0.688) | (0.712–0.864) | (0.614–0.740) | (0.579–0.688) | ||||
| FusedImg | PET + CT | Voxel size-based | 3 | 0.595 | 0.746 | 0.685 | 0.641 |
| (0.506–0.690) | (0.669–0.837) | (0.626–0.761) | (0.575–0.718) | ||||
| Rad_Ocm | Rad_HPV | Rad_Ocm_HPV | ||||
|---|---|---|---|---|---|---|
| C-Index | Model | C-Index | Model | C-Index | Model | |
| RFS | 0.603 | FusedImg | 0.641 | SalxImg | 0.616 | FusedImg |
| (0.460–0.737) | (0.544–0.749) | (0.491–0.731) | ||||
| MFS | 0.668 | SalMap | 0.687 | FusedImg | 0.683 | FusedImg |
| (0.505–0.822) | (0.546–0.853) | (0.544–0.843) | ||||
| OS | 0.671 | FusedImg | 0.627 | FusedImg | 0.702 | FusedImg |
| (0.553–0.781) | (0.529–0.738) | (0.599–0.807) | ||||
| DFS | 0.651 | FusedImg | 0.615 | FusedImg | 0.684 | FusedImg |
| (0.546–0.752) | (0.533–0.717) | (0.589–0.780) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, W.; Xu, H.; Han, X.; Zhang, H.; Ma, J.; Rahmim, A.; Lu, L. Context-Aware Saliency Guided Radiomics: Application to Prediction of Outcome and HPV-Status from Multi-Center PET/CT Images of Head and Neck Cancer. Cancers 2022, 14, 1674. https://doi.org/10.3390/cancers14071674
Lv W, Xu H, Han X, Zhang H, Ma J, Rahmim A, Lu L. Context-Aware Saliency Guided Radiomics: Application to Prediction of Outcome and HPV-Status from Multi-Center PET/CT Images of Head and Neck Cancer. Cancers. 2022; 14(7):1674. https://doi.org/10.3390/cancers14071674
Chicago/Turabian StyleLv, Wenbing, Hui Xu, Xu Han, Hao Zhang, Jianhua Ma, Arman Rahmim, and Lijun Lu. 2022. "Context-Aware Saliency Guided Radiomics: Application to Prediction of Outcome and HPV-Status from Multi-Center PET/CT Images of Head and Neck Cancer" Cancers 14, no. 7: 1674. https://doi.org/10.3390/cancers14071674
APA StyleLv, W., Xu, H., Han, X., Zhang, H., Ma, J., Rahmim, A., & Lu, L. (2022). Context-Aware Saliency Guided Radiomics: Application to Prediction of Outcome and HPV-Status from Multi-Center PET/CT Images of Head and Neck Cancer. Cancers, 14(7), 1674. https://doi.org/10.3390/cancers14071674






