Radiomics/Radiogenomics in Lung Cancer: Basic Principles and Initial Clinical Results
Abstract
Simple Summary
Abstract
1. Introduction
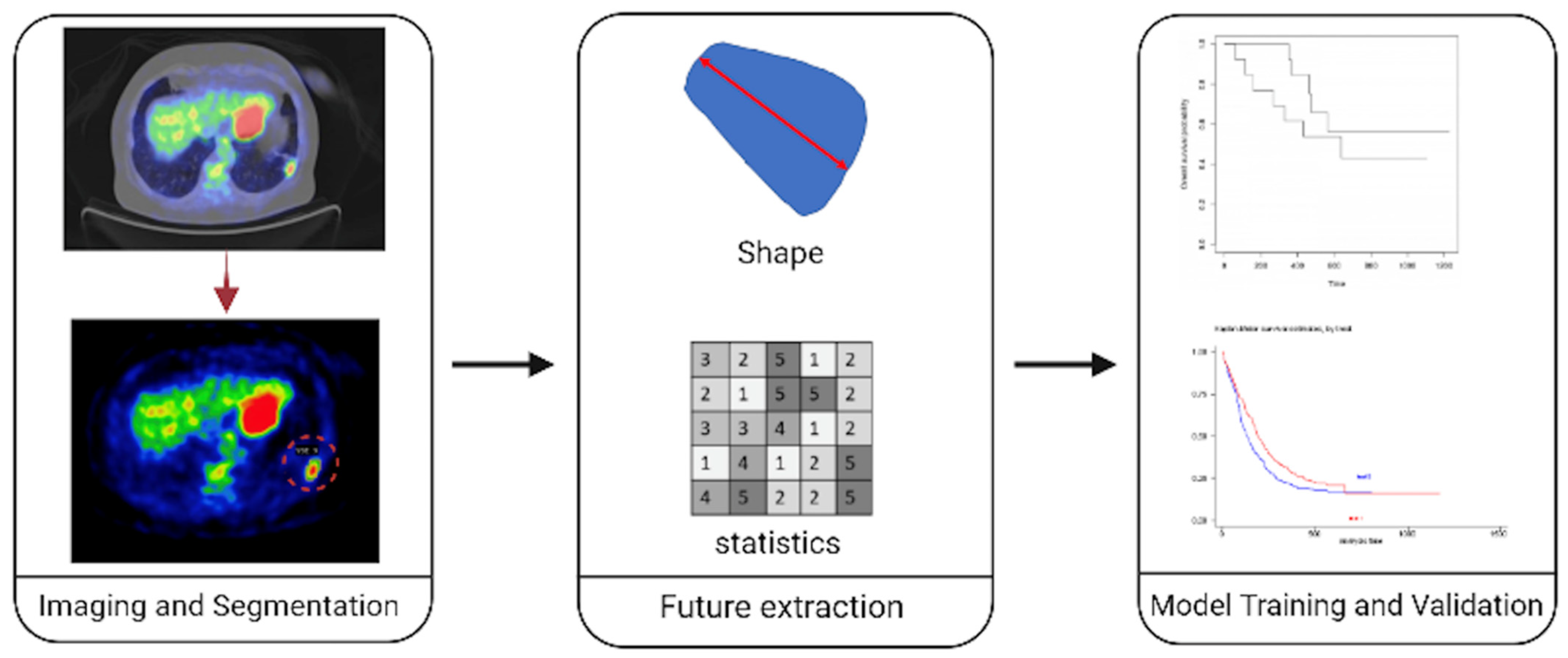
2. Radiomics Pipeline
2.1. Defining the Problem at the Clinical Level
2.2. Identification of Proper Data Sources
2.3. Data Acquisition and Standardisation
Tumour Segmentation
2.4. Feature Computation
2.5. Feature Selection
2.6. Model Selection/Assessment
3. Clinical Applications of Radiomics/Radiogenomics in Lung Cancer
4. Definition of the Clinical Problem
5. Stage II NSCLC
6. Locally Advanced-Stage III and Metastatic-Stage IV NSCLC
7. Discussion
7.1. Challenges
7.2. How to Move to Radiomics 2.0
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M.; Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Fujita, M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 2018, 109, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Story, M.D.; Durante, M. Radiogenomics. Med. Phys. 2018, 45, e1111–e1122. [Google Scholar] [CrossRef]
- Papanikolaou, N.; Matos, C.; Koh, D.M. How to develop a meaningful radiomic signature for clinical use in oncologic patients. Cancer Imaging 2020, 20, 33. [Google Scholar] [CrossRef]
- de Farias, E.C.; di Noia, C.; Han, C.; Sala, E.; Castelli, M.; Rundo, L. Impact of GAN-based lesion-focused medical image super-resolution on the robustness of radiomic features. Sci. Rep. 2021, 11, 21361. [Google Scholar] [CrossRef] [PubMed]
- Santinha, J.; Matos, C.; Figueiredo, M.; Papanikolaou, N. Improving performance and generalizability in radiogenomics: A pilot study for prediction of IDH1/2 mutation status in gliomas with multicentric data. J. Med. Imaging (Bellingham) 2021, 8, 031905. [Google Scholar] [CrossRef]
- Galavis, P.E.; Hollensen, C.; Jallow, N.; Paliwal, B.; Jeraj, R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. 2010, 49, 1012–1016. [Google Scholar] [CrossRef]
- Cook, G.J.R.; Azad, G.; Owczarczyk, K.; Siddique, M.; Goh, V. Challenges and Promises of PET Radiomics. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1083–1089. [Google Scholar] [CrossRef]
- Incoronato, M.; Aiello, M.; Infante, T.; Cavaliere, C.; Grimaldi, A.M.; Mirabelli, P.; Monti, S.; Salvatore, M. Radiogenomic Analysis of Oncological Data: A Technical Survey. Int. J. Mol. Sci. 2017, 18, 805. [Google Scholar] [CrossRef]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Lee, H.Y.; Park, H.; Schiebler, M.L.; van Beek, E.J.R.; Ohno, Y.; Seo, J.B.; Leung, A. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. Eur. J. Radiol. 2017, 86, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Labby, Z.E.; Armato, S.G.; Dignam, J.J.; Straus, C.; Kindler, H.L.; Nowak, A.K. Lung volume measurements as a surrogate marker for patient response in malignant pleural mesothelioma. J. Thorac. Oncol. 2013, 8, 478–486. [Google Scholar] [CrossRef]
- Rizzo, S.; Petrella, F.; Passaro, A.; de Marinis, F.; Bellomi, M. Proposals for revisions of the classification of lung cancers with multiple pulmonary sites: The radiologist’s, thoracic surgeon’s and oncologist’s point of view. J. Thorac. Dis. 2016, 8, E805–E808. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Hyun, S.H.; Lee, K.S.; Kim, B.T.; Kim, J.; Shim, Y.M.; Ahn, M.J.; Kim, T.S.; Yi, C.A.; Chung, M.J. Volume-based parameter of 18)F-FDG PET/CT in malignant pleural mesothelioma: Prediction of therapeutic response and prognostic implications. Ann. Surg. Oncol. 2010, 17, 2787–2794. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, H.Y.; Ahn, M.J.; Park, K.; Ahn, J.S.; Sun, J.M.; Lee, K.S. Volume-based growth tumor kinetics as a prognostic biomarker for patients with EGFR mutant lung adenocarcinoma undergoing EGFR tyrosine kinase inhibitor therapy: A case control study. Cancer Imaging 2016, 16, 5. [Google Scholar] [CrossRef]
- Bhargava, R.; Madabhushi, A. Emerging Themes in Image Informatics and Molecular Analysis for Digital Pathology. Annu. Rev. Biomed. Eng. 2016, 18, 387–412. [Google Scholar] [CrossRef]
- Zhou, M.; Leung, A.; Echegaray, S.; Gentles, A.; Shrager, J.B.; Jensen, K.C.; Berry, G.J.; Plevritis, S.K.; Rubin, D.L.; Napel, S.; et al. Non-Small Cell Lung Cancer Radiogenomics Map Identifies Relationships between Molecular and Imaging Phenotypes with Prognostic Implications. Radiology 2018, 286, 307–315. [Google Scholar] [CrossRef]
- Rizzo, S.; Petrella, F.; Buscarino, V.; De Maria, F.; Raimondi, S.; Barberis, M.; Fumagalli, C.; Spitaleri, G.; Rampinelli, C.; De Marinis, F.; et al. CT Radiogenomic Characterization of EGFR, K-RAS, and ALK Mutations in Non-Small Cell Lung Cancer. Eur. Radiol. 2016, 26, 32–42. [Google Scholar] [CrossRef]
- Lo Gullo, R.; Daimiel, I.; Morris, E.A.; Pinker, K. Combining molecular and imaging metrics in cancer: Radiogenomics. Insights Imaging 2020, 11, 1. [Google Scholar] [CrossRef]
- Gevaert, O.; Xu, J.; Hoang, C.D.; Leung, A.N.; Xu, Y.; Quon, A.; Rubin, D.L.; Napel, S.; Plevritis, S.K. Non-small cell lung cancer: Identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology 2012, 264, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.J.; Ganeshan, B.; Miles, K.A.; Campbell, D.H.; Cheung, P.Y.; Frank, S.; Korn, R.L. Noninvasive image texture analysis differentiates K-ras mutation from pan-wildtype NSCLC and is prognostic. PLoS ONE 2014, 9, e100244. [Google Scholar] [CrossRef]
- Lv, J.; Zhang, H.; Ma, J.; Ma, Y.; Gao, G.; Song, Z.; Yang, Y. Comparison of CT radiogenomic and clinical characteristics between EGFR and KRAS mutations in lung adenocarcinomas. Clin. Radiol. 2018, 73, 590.e591–590.e598. [Google Scholar] [CrossRef] [PubMed]
- Rios Velazquez, E.; Parmar, C.; Liu, Y.; Coroller, T.P.; Cruz, G.; Stringfield, O.; Ye, Z.; Makrigiorgos, M.; Fennessy, F.; Mak, R.H.; et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res. 2017, 77, 3922–3930. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.F.; Parmar, C.; Kim, J.; Huynh, E.; Mak, R.H.; Aerts, H.J.W.L. Impact of experimental design on PET radiomics in predicting somatic mutation status. Eur. J. Radiol. 2017, 97, 8–15. [Google Scholar] [CrossRef]
- Yip, S.S.; Kim, J.; Coroller, T.P.; Parmar, C.; Velazquez, E.R.; Huynh, E.; Mak, R.H.; Aerts, H.J. Associations Between Somatic Mutations and Metabolic Imaging Phenotypes in Non-Small Cell Lung Cancer. J. Nucl. Med. 2017, 58, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.J.; Lee, C.T.; Jheon, S.H.; Park, J.S.; Chung, J.H. Radiologic Characteristics of Surgically Resected Non-Small Cell Lung Cancer With ALK Rearrangement or EGFR Mutations. Ann. Thorac. Surg. 2016, 101, 473–480. [Google Scholar] [CrossRef]
- Ozkan, E.; West, A.; Dedelow, J.A.; Chu, B.F.; Zhao, W.; Yildiz, V.O.; Otterson, G.A.; Shilo, K.; Ghosh, S.; King, M.; et al. CT Gray-Level Texture Analysis as a Quantitative Imaging Biomarker of Epidermal Growth Factor Receptor Mutation Status in Adenocarcinoma of the Lung. AJR Am. J. Roentgenol. 2015, 205, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.; Grossmann, P.; Tan, Y.; Oxnard, G.R.; Rizvi, N.; Schwartz, L.H.; Zhao, B. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Sci. Rep. 2016, 6, 33860. [Google Scholar] [CrossRef]
- de Jong, E.E.C.; van Elmpt, W.; Rizzo, S.; Colarieti, A.; Spitaleri, G.; Leijenaar, R.T.H.; Jochems, A.; Hendriks, L.E.L.; Troost, E.G.C.; Reymen, B.; et al. Applicability of a prognostic CT-based radiomic signature model trained on stage I-III non-small cell lung cancer in stage IV non-small cell lung cancer. Lung Cancer 2018, 124, 6–11. [Google Scholar] [CrossRef]
- Gevaert, O.; Echegaray, S.; Khuong, A.; Hoang, C.D.; Shrager, J.B.; Jensen, K.C.; Berry, G.J.; Guo, H.H.; Lau, C.; Plevritis, S.K.; et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci. Rep. 2017, 7, 41674. [Google Scholar] [CrossRef] [PubMed]
- Sacconi, B.; Anzidei, M.; Leonardi, A.; Boni, F.; Saba, L.; Scipione, R.; Anile, M.; Rengo, M.; Longo, F.; Bezzi, M.; et al. Analysis of CT features and quantitative texture analysis in patients with lung adenocarcinoma: A correlation with EGFR mutations and survival rates. Clin. Radiol. 2017, 72, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, E.C.; Frankland, M.P.; Johnson, T.F.; Antic, S.L.; Chen, H.; Chen, S.C.; Karwoski, R.A.; Walker, R.; Landman, B.A.; Clay, R.D.; et al. Assessing the inter-observer variability of Computer-Aided Nodule Assessment and Risk Yield (CANARY) to characterise lung adenocarcinomas. PLoS ONE 2018, 13, e0198118. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, B.; Bianconi, F.; Palumbo, I.; Fravolini, M.L.; Minestrini, M.; Nuvoli, S.; Stazza, M.L.; Rondini, M.; Spanu, A. Value of shape and texture features from 18F-FDG PET/CT to discriminate between benign and malignant solitary pulmonary nodules: An experimental evaluation. Diagnostics 2020, 10, 696. [Google Scholar] [CrossRef]
- Caruso, D.; Zerunian, M.; Daffina, J.; Polici, M.; Polidori, M.; Tipaldi, M.A.; Ronconi, E.; Pucciarelli, F.; Lucertini, E.; Rossi, M.; et al. Radiomics and functional imaging in lung cancer: The importance of radiological heterogeneity beyond FDG PET/CT and lung biopsy. Eur. J. Radiol. 2021, 142, 109874. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, X.L.; Zhang, T.; Wang, J.; Zhang, T.; Tian, R. Use of radiomics based on (18)F-FDG PET/CT and machine learning methods to aid clinical decision-making in the classification of solitary pulmonary lesions: An innovative approach. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 2904–2913. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Li, H.; Zhao, Y.; Liu, Z.; Sun, K.; Zhu, X.; Qi, Q.; An, B.; Shen, D.; et al. Genomic characterisation of pulmonary subsolid nodules: Mutational landscape and radiological features. Eur. Respir. J. 2020, 55, 1901409. [Google Scholar] [CrossRef]
- Li, M.; Zhang, L.; Tang, W.; Ma, P.Q.; Zhou, L.N.; Jin, Y.J.; Qi, L.L.; Wu, N. Quantitative features of dual-energy spectral computed tomography for solid lung adenocarcinoma with. Transl. Lung Cancer Res. 2019, 8, 401–412. [Google Scholar] [CrossRef]
- Tsutani, Y.; Shimada, Y.; Ito, H.; Miyata, Y.; Ikeda, N.; Nakayama, H.; Okada, M. Identification of High-Risk of Recurrence in Clinical Stage I Non-Small Cell Lung Cancer. Front. Oncol. 2021, 11, 622742. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J.; Wu, Y.L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Grove, O.; Berglund, A.E.; Schabath, M.B.; Aerts, H.J.W.L.; Dekker, A.; Wang, H.; Velazquez, E.R.; Lambin, P.; Gu, Y.; Balagurunathan, Y.; et al. Correction: Quantitative Computed Tomographic Descriptors Associate Tumor Shape Complexity and Intratumor Heterogeneity with Prognosis in Lung Adenocarcinoma. PLoS ONE 2021, 16, e0248541. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, Z.; He, L.; Chen, X.; Pan, D.; Ma, Z.; Liang, C.; Tian, J. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016, 281, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, S.; Maldonado, F.; Rajagopalan, S.; Karwoski, R.A.; DePew, Z.S.; Bartholmai, B.J.; Peikert, T.; Robb, R.A. Noninvasive risk stratification of lung adenocarcinoma using quantitative computed tomography. J. Thorac. Oncol. 2014, 9, 1698–1703. [Google Scholar] [CrossRef]
- Depeursinge, A.; Yanagawa, M.; Leung, A.N.; Rubin, D.L. Predicting adenocarcinoma recurrence using computational texture models of nodule components in lung CT. Med. Phys. 2015, 42, 2054–2063. [Google Scholar] [CrossRef]
- Parmar, C.; Leijenaar, R.T.; Grossmann, P.; Rios Velazquez, E.; Bussink, J.; Rieveld, D.; Rietbergen, M.M.; Haibe-Kains, B.; Lambin, P.; Aerts, H.J. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Sci. Rep. 2015, 5, 11044. [Google Scholar] [CrossRef]
- Coroller, T.P.; Grossmann, P.; Hou, Y.; Rios Velazquez, E.; Leijenaar, R.T.; Hermann, G.; Lambin, P.; Haibe-Kains, B.; Mak, R.H.; Aerts, H.J. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 2015, 114, 345–350. [Google Scholar] [CrossRef]
- Coroller, T.P.; Agrawal, V.; Narayan, V.; Hou, Y.; Grossmann, P.; Lee, S.W.; Mak, R.H.; Aerts, H.J. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother. Oncol. 2016, 119, 480–486. [Google Scholar] [CrossRef]
- Coroller, T.P.; Agrawal, V.; Huynh, E.; Narayan, V.; Lee, S.W.; Mak, R.H.; Aerts, H.J.W.L. Radiomic-Based Pathological Response Prediction from Primary Tumors and Lymph Nodes in NSCLC. J. Thorac. Oncol. 2017, 12, 467–476. [Google Scholar] [CrossRef]
- Borcoman, E.; Kanjanapan, Y.; Champiat, S.; Kato, S.; Servois, V.; Kurzrock, R.; Goel, S.; Bedard, P.; Le Tourneau, C. Novel patterns of response under immunotherapy. Ann. Oncol. 2019, 30, 385–396. [Google Scholar] [CrossRef]
- Haanen, J.B.A.G.; Carbonnel, F.; Robert, C.; Kerr, K.M.; Peters, S.; Larkin, J.; Jordan, K.; Committee, E.G. Management of toxicities from immunotherapy: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28, iv119–iv142. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Choi, J.H.; Park, J.H.; Byun, B.H.; Lim, I.; Kim, B.I.; Choi, C.W.; Lim, S.M. Differences of 18F-FDG PET tumor texture parameters according to the presence of epidermal growth factor receptor mutation in non-small cell lung cancer. J. Nucl. Med. Conf. Soc. Nucl. Med. Mol. Imaging Annu. Meet. 2015, 56 (Suppl. 3), 1393. [Google Scholar]
- Sorensen, J.; Erasmus, J., Jr.; Shroff, G.; Rao, A.; Stingo, F.; Court, L.; Gold, K.A.; Lee, J.; Heymach, J.V.; Swisher, S.; et al. Combining CT texture analysis with semantic imaging descriptions for the radiogenomic detection of EGFR and KRAS mutations in NSCLC. J. Thorac. Oncol. 2015, 2, S700. [Google Scholar]
- Jia, T.; Xiong, J.; Li, X.; Ma, J.; Ren, Y.; Xu, Z.; Cai, X.; Zhang, J.; Zhao, J.; Fu, X. Detecting epidermal growth factor receptor mutation status in patients with lung adenocarcinoma using radiomics and random forest. J. Thorac. Oncol. 2017, 12, S1860. [Google Scholar] [CrossRef][Green Version]
- Clay, R.; Johnson, T.; Bartholmai, B.J.; Karwoski, R.; Maldonado, F.; Rajagopalan, S.; Peikert, T. Canary (computer aided nodule analysis and risk yield) as a radiologic biomarker to predict tumor response to EGFR-targeted therapy in adenocarcinoma of the lung. Am. J. Respir. Crit. Care Med. 2018, 197, A4684. [Google Scholar]
- Zhang, L.; Chen, B.; Liu, X.; Song, J.; Fang, M.; Hu, C.; Dong, D.; Li, W.; Tian, J. Quantitative Biomarkers for Prediction of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer. Transl. Oncol. 2018, 11, 94–101. [Google Scholar] [CrossRef]
- Sawan, P.; Plodkowski, A.J.; Li, A.E.; Li, B.T.; Drilon, A.; Capanu, M.; Ginsberg, M.S. CT features of HER2-mutant lung adenocarcinomas. Clin. Imaging 2018, 51, 279–283. [Google Scholar] [CrossRef]
- Halpenny, D.F.; Riely, G.J.; Hayes, S.; Yu, H.; Zheng, J.; Moskowitz, C.S.; Ginsberg, M.S. Are there imaging characteristics associated with lung adenocarcinomas harboring ALK rearrangements? Lung Cancer 2014, 86, 190–194. [Google Scholar] [CrossRef]
- Tang, C.; Hobbs, B.; Amer, A.; Li, X.; Behrens, C.; Canales, J.R.; Cuentas, E.P.; Villalobos, P.; Fried, D.; Chang, J.Y.; et al. Development of an Immune-Pathology Informed Radiomics Model for Non-Small Cell Lung Cancer. Sci. Rep. 2018, 8, 1922. [Google Scholar] [CrossRef]
- Jia, T.Y.; Xiong, J.F.; Li, X.Y.; Yu, W.; Xu, Z.Y.; Cai, X.W.; Ma, J.C.; Ren, Y.C.; Larsson, R.; Zhang, J.; et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur. Radiol. 2019, 29, 4742–4750. [Google Scholar] [CrossRef]
- Li, S.; Ding, C.; Zhang, H.; Song, J.; Wu, L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med. Phys. 2019, 46, 4545–4552. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.; Sun, G.; Fan, L.; Wang, Y.; Xia, Y.; Guan, Y.; Li, Q.; Zhang, D.; Liu, S.; Li, Z. Radiomics signature: A potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer 2019, 132, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Dong, X.; Wang, J.; Li, W.; Gu, Z.; Gao, D.; Zhong, N.; Guan, Y. Computed Tomography-Based Radiomics Signature: A Potential Indicator of Epidermal Growth Factor Receptor Mutation in Pulmonary Adenocarcinoma Appearing as a Subsolid Nodule. Oncologist 2019, 24, e1156–e1164. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kong, C.; Xu, W.; Yang, S.; Shi, D.; Zhang, J.; Du, M.; Wang, S.; Bai, Y.; Zhang, T.; et al. Decoding tumor mutation burden and driver mutations in early stage lung adenocarcinoma using CT-based radiomics signature. Thorac. Cancer 2019, 10, 1904–1912. [Google Scholar] [CrossRef]
- Bak, S.H.; Park, H.; Lee, H.Y.; Kim, Y.; Kim, H.L.; Jung, S.H.; Kim, H.; Kim, J.; Park, K. Imaging genotyping of functional signaling pathways in lung squamous cell carcinoma using a radiomics approach. Sci. Rep. 2018, 8, 3284. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.; Mahadevaiah, G.; Dekker, A. An Approach Toward Automatic Classification of Tumor Histopathology of Non-Small Cell Lung Cancer Based on Radiomic Features. Tomography 2016, 2, 374–377. [Google Scholar] [CrossRef]
- Wu, W.; Parmar, C.; Grossmann, P.; Quackenbush, J.; Lambin, P.; Bussink, J.; Mak, R.; Aerts, H.J. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front. Oncol. 2016, 6, 71. [Google Scholar] [CrossRef]
- Zhu, X.; Dong, D.; Chen, Z.; Fang, M.; Zhang, L.; Song, J.; Yu, D.; Zang, Y.; Liu, Z.; Shi, J.; et al. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur. Radiol. 2018, 28, 2772–2778. [Google Scholar] [CrossRef]
- Linning, E.; Lu, L.; Li, L.; Yang, H.; Schwartz, L.H.; Zhao, B. Radiomics for Classifying Histological Subtypes of Lung Cancer Based on Multiphasic Contrast-Enhanced Computed Tomography. J. Comput. Assist. Tomogr. 2019, 43, 300–306. [Google Scholar] [CrossRef]
- Liu, J.; Cui, J.; Liu, F.; Yuan, Y.; Guo, F.; Zhang, G. Multi-subtype classification model for non-small cell lung cancer based on radiomics: SLS model. Med. Phys. 2019, 46, 3091–3100. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, J.; Tian, Y.; Yuan, S.; Li, X. Correlation between radiomic features based on contrast-enhanced computed tomography images and Ki-67 proliferation index in lung cancer: A preliminary study. Thorac. Cancer 2018, 9, 1235–1240. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Feng, Z.; Liang, Q.; Li, M.; Deng, J.; Ma, M.; Wang, W.; Liu, J.; Liu, P.; Rong, P. Machine learning-based radiomics strategy for prediction of cell proliferation in non-small cell lung cancer. Eur. J. Radiol. 2019, 118, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Song, S.H.; Park, H.; Lee, G.; Lee, H.Y.; Sohn, I.; Kim, H.S.; Lee, S.H.; Jeong, J.Y.; Kim, J.; Lee, K.S.; et al. Imaging Phenotyping Using Radiomics to Predict Micropapillary Pattern within Lung Adenocarcinoma. J. Thorac. Oncol. 2017, 12, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Fang, M.; Dong, D.; Wei, X.; Liu, L.; Xu, X.; Jiang, X.; Tian, J.; Liu, Z. A Radiomics Signature in Preoperative Predicting Degree of Tumor Differentiation in Patients with Non-small Cell Lung Cancer. Acad. Radiol. 2018, 25, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- She, Y.; Zhang, L.; Zhu, H.; Dai, C.; Xie, D.; Xie, H.; Zhang, W.; Zhao, L.; Zou, L.; Fei, K.; et al. The predictive value of CT-based radiomics in differentiating indolent from invasive lung adenocarcinoma in patients with pulmonary nodules. Eur. Radiol. 2018, 28, 5121–5128. [Google Scholar] [CrossRef]
- Yang, B.; Guo, L.; Lu, G.; Shan, W.; Duan, L.; Duan, S. Radiomic signature: A non-invasive biomarker for discriminating invasive and non-invasive cases of lung adenocarcinoma. Cancer Manag. Res. 2019, 11, 7825–7834. [Google Scholar] [CrossRef]
- Fornacon-Wood, I.; Faivre-Finn, C.; O’Connor, J.P.B.; Price, G.J. Radiomics as a personalised medicine tool in lung cancer: Separating the hope from the hype. Lung Cancer 2020, 146, 197–208. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anagnostopoulos, A.K.; Gaitanis, A.; Gkiozos, I.; Athanasiadis, E.I.; Chatziioannou, S.N.; Syrigos, K.N.; Thanos, D.; Chatziioannou, A.N.; Papanikolaou, N. Radiomics/Radiogenomics in Lung Cancer: Basic Principles and Initial Clinical Results. Cancers 2022, 14, 1657. https://doi.org/10.3390/cancers14071657
Anagnostopoulos AK, Gaitanis A, Gkiozos I, Athanasiadis EI, Chatziioannou SN, Syrigos KN, Thanos D, Chatziioannou AN, Papanikolaou N. Radiomics/Radiogenomics in Lung Cancer: Basic Principles and Initial Clinical Results. Cancers. 2022; 14(7):1657. https://doi.org/10.3390/cancers14071657
Chicago/Turabian StyleAnagnostopoulos, Athanasios K., Anastasios Gaitanis, Ioannis Gkiozos, Emmanouil I. Athanasiadis, Sofia N. Chatziioannou, Konstantinos N. Syrigos, Dimitris Thanos, Achilles N. Chatziioannou, and Nikolaos Papanikolaou. 2022. "Radiomics/Radiogenomics in Lung Cancer: Basic Principles and Initial Clinical Results" Cancers 14, no. 7: 1657. https://doi.org/10.3390/cancers14071657
APA StyleAnagnostopoulos, A. K., Gaitanis, A., Gkiozos, I., Athanasiadis, E. I., Chatziioannou, S. N., Syrigos, K. N., Thanos, D., Chatziioannou, A. N., & Papanikolaou, N. (2022). Radiomics/Radiogenomics in Lung Cancer: Basic Principles and Initial Clinical Results. Cancers, 14(7), 1657. https://doi.org/10.3390/cancers14071657







