High Keratin-7 Expression in Benign Peri-Tumoral Prostatic Glands Is Predictive of Bone Metastasis Onset and Prostate Cancer-Specific Mortality
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Exploratory Cohorts of Patients
2.3. Validation Cohort of Localized PC Patients
2.4. Cell Culture
2.5. Paraffin Processing and Embedding of Cell Line Pellets
2.6. Western Blots Analysis
2.7. Immunohistochemistry (IHC) on FFPE Tissues
2.8. Immunofluorescence (IF) on FFPE Tissues
2.9. IF Staining Digital Image Analyses and Pre-Processing of Scoring Data
2.10. Statistical Analysis of MFI Values from the TMA TF123
3. Results
3.1. Validation of Immunostaining Procedures
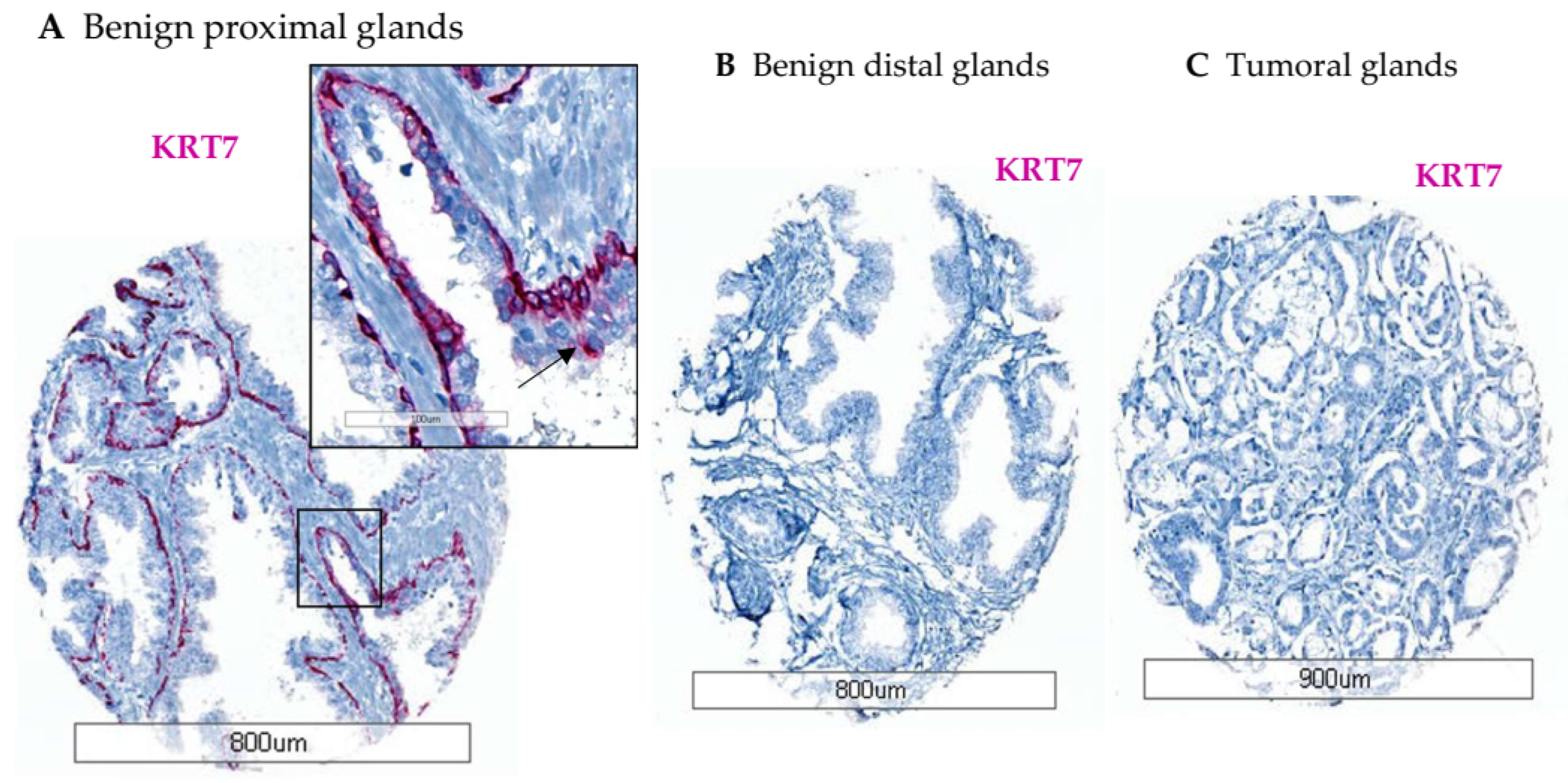
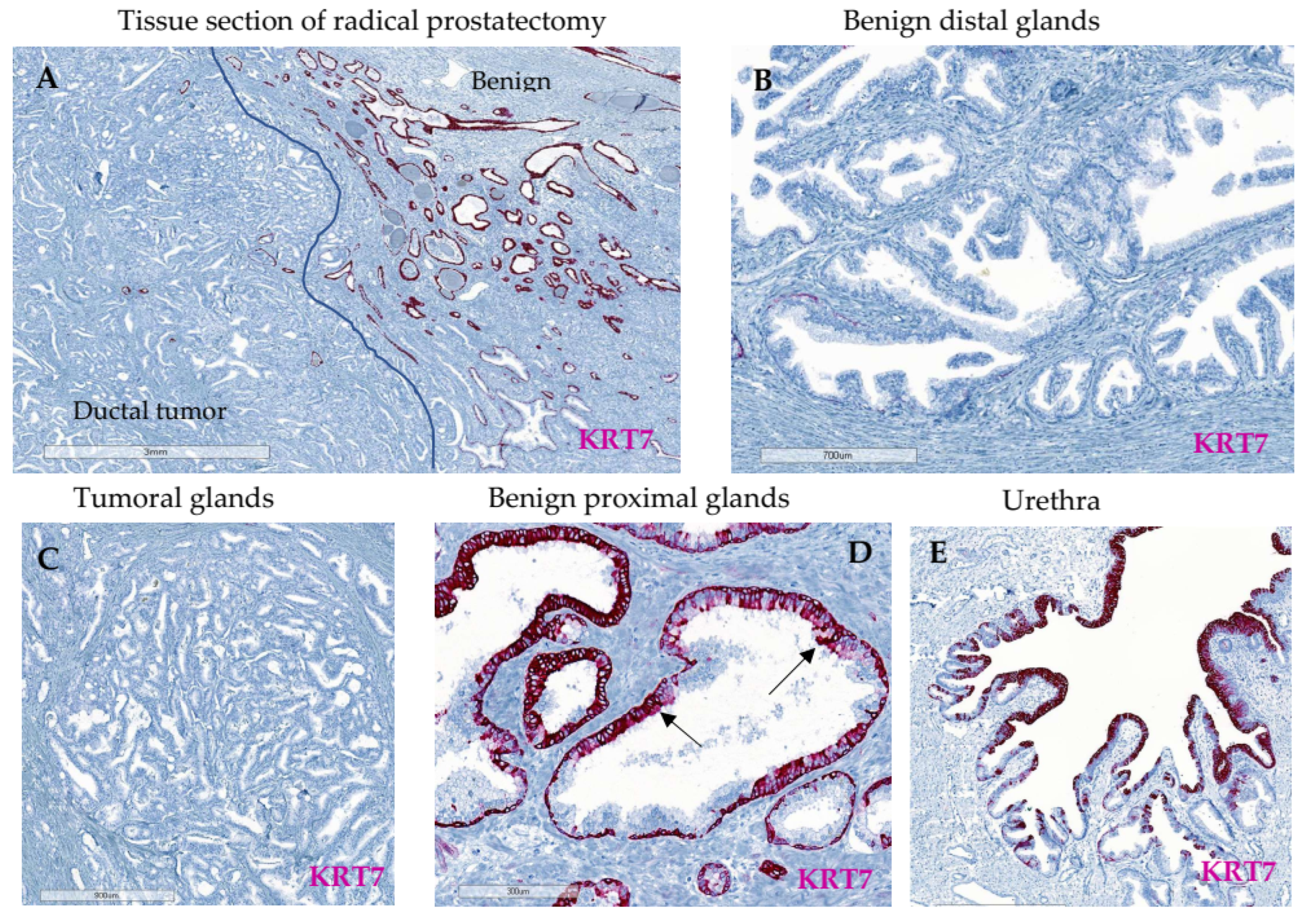
3.2. Characterization of KRT7 Expression in Different Tissue Samples from Exploratory Cohorts
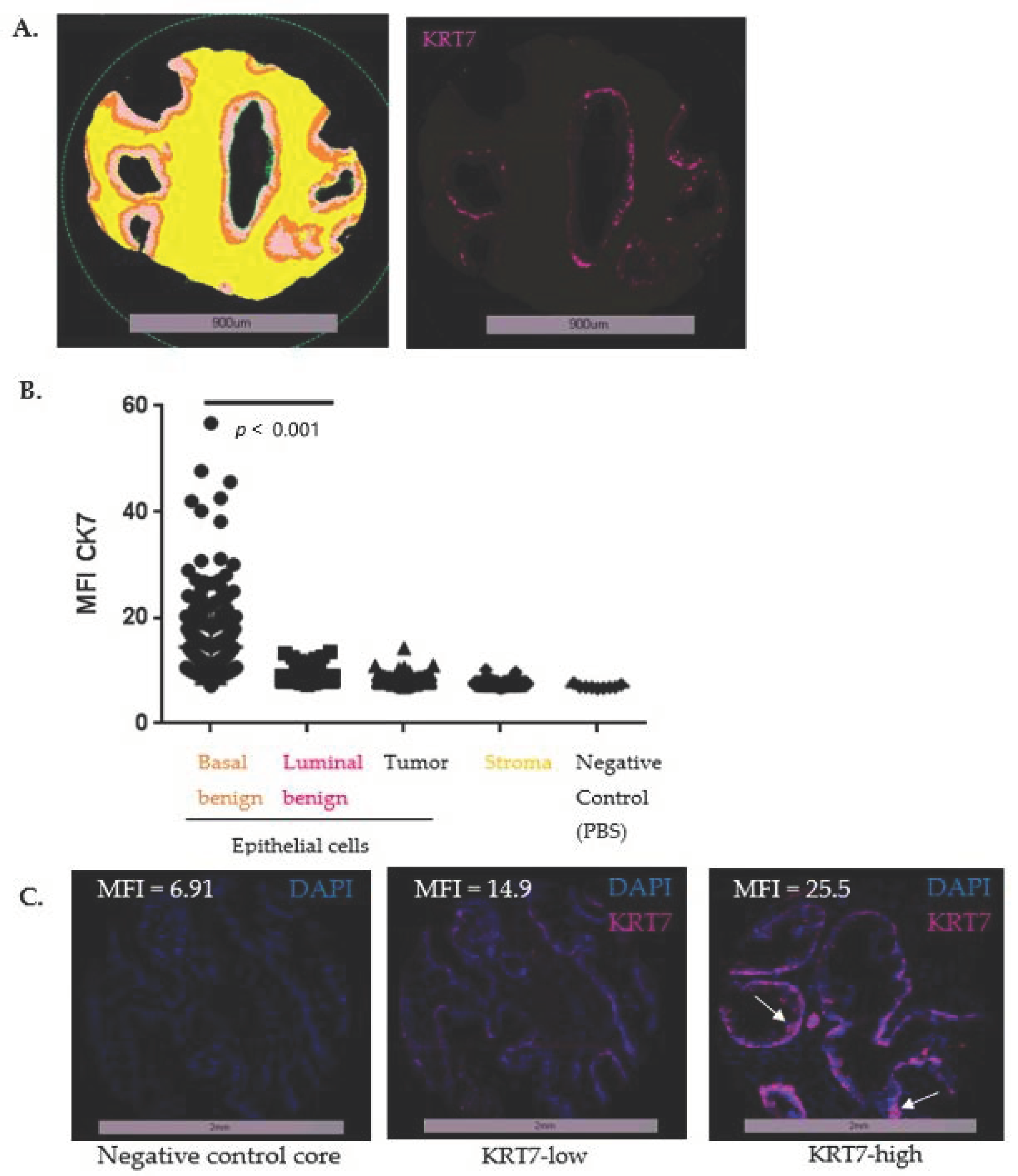
3.2.1. Optimization TMA (n = 18)
3.2.2. Tissue Slides from Healthy Prostate (n = 51)
3.2.3. Whole Tissue Slides from Localized PC (n = 16)
3.2.4. Advanced PC TMA (n = 91)
3.3. Expression of KRT7 in a Cohort of Localized PC (n = 285)
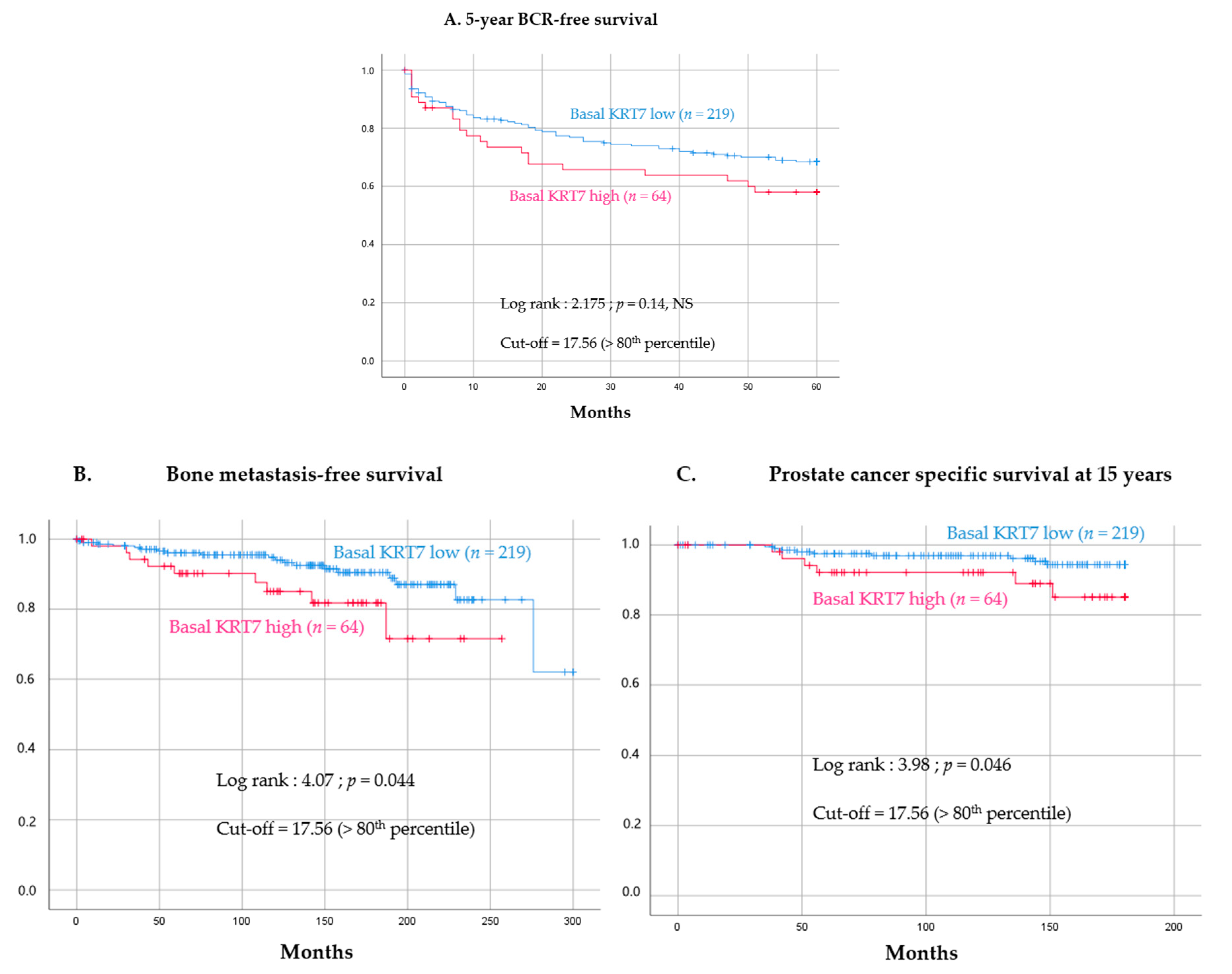
3.4. Prognostic Value of High KRT7 Expression in the Basal Compartment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Karantanos, T.; Corn, P.G.; Thompson, T.C. Prostate Cancer Progression after Androgen Deprivation Therapy: Mechanisms of Castrate Resistance and Novel Therapeutic Approaches. Oncogene 2013, 32, 5501–5511. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.K.; Check, D.P.; Lortet-Tieulent, J.; Laversanne, M.; Jemal, A.; Ferlay, J.; Bray, F.; Cook, M.B.; Devesa, S.S. Prostate Cancer Incidence in 43 Populations Worldwide: An Analysis of Time Trends Overall and by Age Group. Int. J. Cancer 2016, 138, 1388–1400. [Google Scholar] [CrossRef] [PubMed]
- Beesley, L.J.; Morgan, T.M.; Spratt, D.E.; Singhal, U.; Feng, F.Y.; Furgal, A.C.; Jackson, W.C.; Daignault, S.; Taylor, J.M.G. Individual and Population Comparisons of Surgery and Radiotherapy Outcomes in Prostate Cancer Using Bayesian Multistate Models. JAMA Netw. Open 2019, 2, e187765. [Google Scholar] [CrossRef] [Green Version]
- Albertsen, P.C.; Hanley, J.A.; Penson, D.F.; Barrows, G.; Fine, J. 13-Year Outcomes Following Treatment for Clinically Localized Prostate Cancer in a Population Based Cohort. J. Urol. 2007, 177, 932–936. [Google Scholar] [CrossRef]
- Jackson, W.C.; Suresh, K.; Tumati, V.; Allen, S.G.; Dess, R.T.; Salami, S.S.; George, A.; Kaffenberger, S.D.; Miller, D.C.; Hearn, J.W.D.; et al. Intermediate Endpoints After Postprostatectomy Radiotherapy: 5-Year Distant Metastasis to Predict Overall Survival. Eur. Urol. 2018, 74, 413–419. [Google Scholar] [CrossRef]
- D’Amico, A.V.; Whittington, R.; Malkowicz, S.B.; Schultz, D.; Blank, K.; Broderick, G.A.; Tomaszewski, J.E.; Renshaw, A.A.; Kaplan, I.; Beard, C.J.; et al. Biochemical Outcome after Radical Prostatectomy, External Beam Radiation Therapy, or Interstitial Radiation Therapy for Clinically Localized Prostate Cancer. JAMA 1998, 280, 969–974. [Google Scholar] [CrossRef]
- Kattan, M.W.; Eastham, J.A.; Stapleton, A.M.; Wheeler, T.M.; Scardino, P.T. A Preoperative Nomogram for Disease Recurrence Following Radical Prostatectomy for Prostate Cancer. J. Natl. Cancer Inst. 1998, 90, 766–771. [Google Scholar] [CrossRef] [Green Version]
- Cooperberg, M.R.; Pasta, D.J.; Elkin, E.P.; Litwin, M.S.; Latini, D.M.; Du Chane, J.; Carroll, P.R. The University of California, San Francisco Cancer of the Prostate Risk Assessment Score: A Straightforward and Reliable Preoperative Predictor of Disease Recurrence after Radical Prostatectomy. J. Urol. 2005, 173, 1938–1942. [Google Scholar] [CrossRef] [Green Version]
- Dess, R.T.; Suresh, K.; Zelefsky, M.J.; Freedland, S.J.; Mahal, B.A.; Cooperberg, M.R.; Davis, B.J.; Horwitz, E.M.; Terris, M.K.; Amling, C.L.; et al. Development and Validation of a Clinical Prognostic Stage Group System for Nonmetastatic Prostate Cancer Using Disease-Specific Mortality Results From the International Staging Collaboration for Cancer of the Prostate. JAMA Oncol. 2020, 6, 1912–1920. [Google Scholar] [CrossRef]
- Hartman, H.E.; Jackson, W.C. Surrogate Endpoints in Localized Prostate Cancer. Cancer J. 2020, 26, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Gharzai, L.A.; Jiang, R.; Wallington, D.; Jones, G.; Birer, S.; Jairath, N.; Jaworski, E.M.; McFarlane, M.R.; Mahal, B.A.; Nguyen, P.L.; et al. Intermediate Clinical Endpoints for Surrogacy in Localised Prostate Cancer: An Aggregate Meta-Analysis. Lancet Oncol. 2021, 22, 402–410. [Google Scholar] [CrossRef]
- Mazzone, E.; Gandaglia, G.; Ploussard, G.; Marra, G.; Valerio, M.; Campi, R.; Mari, A.; Minervini, A.; Serni, S.; Moschini, M.; et al. Risk Stratification of Patients Candidate to Radical Prostatectomy Based on Clinical and Multiparametric Magnetic Resonance Imaging Parameters: Development and External Validation of Novel Risk Groups. Eur. Urol. 2021, 81, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Trompetter, M.; Smedts, F.; Van der Wijk, J.; Schoots, C.; de Jong, H.-J.; Hopman, A.; De la Rosette, J. Keratin Profiling in the Developing Human Prostate. A Different Approach to Understanding Epithelial Lineage. Anticancer Res. 2008, 28, 237–243. [Google Scholar]
- Oue, N.; Noguchi, T.; Anami, K.; Kitano, S.; Sakamoto, N.; Sentani, K.; Uraoka, N.; Aoyagi, K.; Yoshida, T.; Sasaki, H.; et al. Cytokeratin 7 Is a Predictive Marker for Survival in Patients with Esophageal Squamous Cell Carcinoma. Ann. Surg. Oncol. 2012, 19, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Yang, J. Identification of Novel Biomarkers, MUC5AC, MUC1, KRT7, GAPDH, CD44 for Gastric Cancer. Med. Oncol. 2020, 37, 34. [Google Scholar] [CrossRef] [Green Version]
- Czapiewski, P.; Bobowicz, M.; Pęksa, R.; Skrzypski, M.; Gorczyński, A.; Szczepańska-Michalska, K.; Korwat, A.; Jankowski, M.; Zegarski, W.; Szulgo-Paczkowska, A.; et al. Keratin 7 Expression in Lymph Node Metastases but Not in the Primary Tumour Correlates with Distant Metastases and Poor Prognosis in Colon Carcinoma. Pol. J. Pathol. 2016, 67, 228–234. [Google Scholar] [CrossRef] [Green Version]
- Communal, L.; Roy, N.; Cahuzac, M.; Rahimi, K.; Köbel, M.; Provencher, D.M.; Mes-Masson, A.-M. A Keratin 7 and E-Cadherin Signature Is Highly Predictive of Tubo-Ovarian High-Grade Serous Carcinoma Prognosis. Int. J. Mol. Sci. 2021, 22, 5325. [Google Scholar] [CrossRef]
- Ramaekers, F.; van Niekerk, C.; Poels, L.; Schaafsma, E.; Huijsmans, A.; Robben, H.; Schaart, G.; Vooijs, P. Use of Monoclonal Antibodies to Keratin 7 in the Differential Diagnosis of Adenocarcinomas. Am. J. Pathol. 1990, 136, 641–655. [Google Scholar]
- Goldstein, N.S. Immunophenotypic Characterization of 225 Prostate Adenocarcinomas with Intermediate or High Gleason Scores. Am. J. Clin. Pathol. 2002, 117, 471–477. [Google Scholar] [CrossRef]
- Ceder, J.A.; Aalders, T.W.; Schalken, J.A. Label Retention and Stem Cell Marker Expression in the Developing and Adult Prostate Identifies Basal and Luminal Epithelial Stem Cell Subpopulations. Stem Cell Res. Ther. 2017, 8, 95. [Google Scholar] [CrossRef]
- Sackmann Sala, L.; Boutillon, F.; Menara, G.; De Goyon-Pélard, A.; Leprévost, M.; Codzamanian, J.; Lister, N.; Pencik, J.; Clark, A.; Cagnard, N.; et al. A Rare Castration-Resistant Progenitor Cell Population Is Highly Enriched in Pten-Null Prostate Tumours. J. Pathol. 2017, 243, 51–64. [Google Scholar] [CrossRef] [Green Version]
- Borziak, K.; Finkelstein, J. Comparative Analysis of Public Data Sets to Identify Stemness Markers That Differentiate Liver Cancer Stem Cells. Stud. Health Technol. Inform. 2021, 281, 818–819. [Google Scholar] [CrossRef]
- Clairefond, S.; Péant, B.; Ouellet, V.; Barrès, V.; Tian, Z.; Trudel, D.; Karakiewicz, P.I.; Mes-Masson, A.-M.; Saad, F. PUMA and NOXA Expression in Tumor-Associated Benign Prostatic Epithelial Cells Are Predictive of Prostate Cancer Biochemical Recurrence. Cancers 2020, 12, 3187. [Google Scholar] [CrossRef]
- Clairefond, S.; Ouellet, V.; Péant, B.; Barrès, V.; Karakiewicz, P.I.; Mes-Masson, A.-M.; Saad, F. Expression of ERBB Family Members as Predictive Markers of Prostate Cancer Progression and Mortality. Cancers 2021, 13, 1688. [Google Scholar] [CrossRef]
- Zietarska, M.; Madore, J.; Diallo, J.-S.; Delvoye, N.; Saad, F.; Provencher, D.; Mes-Masson, A.-M. A Novel Method of Cell Embedding for Tissue Microarrays. Histopathology 2010, 57, 323–329. [Google Scholar] [CrossRef]
- Labouba, I.; Le Page, C.; Communal, L.; Kristessen, T.; You, X.; Péant, B.; Barrès, V.; Gannon, P.O.; Mes-Masson, A.-M.; Saad, F. Potential Cross-Talk between Alternative and Classical NF-ΚB Pathways in Prostate Cancer Tissues as Measured by a Multi-Staining Immunofluorescence Co-Localization Assay. PLoS ONE 2015, 10, e0131024. [Google Scholar] [CrossRef]
- Strand, D.W.; Goldstein, A.S. The Many Ways to Make a Luminal Cell and a Prostate Cancer Cell. Endocr. Relat. Cancer 2015, 22, T187–T197. [Google Scholar] [CrossRef] [Green Version]
- Bassily, N.H.; Vallorosi, C.J.; Akdas, G.; Montie, J.E.; Rubin, M.A. Coordinate Expression of Cytokeratins 7 and 20 in Prostate Adenocarcinoma and Bladder Urothelial Carcinoma. Am. J. Clin. Pathol. 2000, 113, 383–388. [Google Scholar] [CrossRef] [Green Version]
- Baures, M.; Dariane, C.; Tika, E.; Puig Lombardi, E.; Barry Delongchamps, N.; Blanpain, C.; Guidotti, J.-E.; Goffin, V. Prostate Luminal Progenitor Cells: From Mouse to Human, from Health to Disease. Nat. Rev. Urol. 2022; in press. [Google Scholar] [CrossRef]
- Adamo, H.H.; Strömvall, K.; Nilsson, M.; Halin Bergström, S.; Bergh, A. Adaptive (TINT) Changes in the Tumor Bearing Organ Are Related to Prostate Tumor Size and Aggressiveness. PLoS ONE 2015, 10, e0141601. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Etheridge, T.; McCormick, J.; Schultz, A.; Khemees, T.A.; Damaschke, N.; Leverson, G.; Woo, K.; Sonn, G.A.; Klein, E.A.; et al. Validation of an Epigenetic Field of Susceptibility to Detect Significant Prostate Cancer from Non-Tumor Biopsies. Clin. Epigenetics 2019, 11, 168. [Google Scholar] [CrossRef] [PubMed]
- Bergström, S.H.; Järemo, H.; Nilsson, M.; Adamo, H.H.; Bergh, A. Prostate Tumors Downregulate Microseminoprotein-Beta (MSMB) in the Surrounding Benign Prostate Epithelium and This Response Is Associated with Tumor Aggressiveness. Prostate 2018, 78, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Gaudreau, P.-O.; Clairefond, S.; Class, C.A.; Boulay, P.-L.; Chrobak, P.; Allard, B.; Azzi, F.; Pommey, S.; Do, K.-A.; Saad, F.; et al. WISP1 Is Associated to Advanced Disease, EMT and an Inflamed Tumor Microenvironment in Multiple Solid Tumors. Oncoimmunology 2019, 8, e1581545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leclerc, B.G.; Charlebois, R.; Chouinard, G.; Allard, B.; Pommey, S.; Saad, F.; Stagg, J. CD73 Expression Is an Independent Prognostic Factor in Prostate Cancer. Clin. Cancer Res. 2016, 22, 158–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosset, A.-A.; Ouellet, V.; Caron, C.; Fragoso, G.; Barrès, V.; Delvoye, N.; Latour, M.; Aprikian, A.; Bergeron, A.; Chevalier, S.; et al. Validation of the Prognostic Value of NF-ΚB P65 in Prostate Cancer: A Retrospective Study Using a Large Multi-Institutional Cohort of the Canadian Prostate Cancer Biomarker Network. PLoS Med. 2019, 16, e1002847. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, L.; Zhou, Z.; Kwon, O.-J.; Zhang, Y.; Nguyen, H.; Dumpit, R.; True, L.; Nelson, P.; Dong, B.; et al. Spatially Restricted Stromal Wnt Signaling Restrains Prostate Epithelial Progenitor Growth through Direct and Indirect Mechanisms. Cell Stem Cell 2019, 24, 753–768.e6. [Google Scholar] [CrossRef]
- Shafer-Weaver, K.A.; Anderson, M.J.; Stagliano, K.; Malyguine, A.; Greenberg, N.M.; Hurwitz, A.A. Cutting Edge: Tumor-Specific CD8+ T Cells Infiltrating Prostatic Tumors Are Induced to Become Suppressor Cells. J. Immunol. 2009, 183, 4848–4852. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Su, Z.; Wei, B.; Liang, Z. KRT7 Overexpression Is Associated with Poor Prognosis and Immune Cell Infiltration in Patients with Pancreatic Adenocarcinoma. Int. J. Gen. Med. 2021, 14, 2677–2694. [Google Scholar] [CrossRef]
- Schaafsma, H.E.; Ramaekers, F.C.; van Muijen, G.N.; Ooms, E.C.; Ruiter, D.J. Distribution of Cytokeratin Polypeptides in Epithelia of the Adult Human Urinary Tract. Histochemistry 1989, 91, 151–159. [Google Scholar] [CrossRef]
- Thorson, P.; Swanson, P.E.; Vollmer, R.T.; Humphrey, P.A. Basal Cell Hyperplasia in the Peripheral Zone of the Prostate. Mod. Pathol. 2003, 16, 598–606. [Google Scholar] [CrossRef] [Green Version]
- Freitas, D.; Andriole, G.L.; Freedland, S.J.; Neto, B.S.; Moreira, D.M. Baseline Basal Cell Hyperplasia Is Not Associated with Baseline Lower Urinary Tract Symptoms, Baseline Clinical Prostatitis or Prostate Cancer in Repeat Biopsies. Urology 2019, 129, 160–164. [Google Scholar] [CrossRef]
- Henry, G.; Malewska, A.; Mauck, R.; Gahan, J.; Hutchinson, R.; Torrealba, J.; Francis, F.; Roehrborn, C.; Strand, D. Molecular Pathogenesis of Human Prostate Basal Cell Hyperplasia. Prostate 2017, 77, 1344–1355. [Google Scholar] [CrossRef]
- Chu, P.; Wu, E.; Weiss, L.M. Cytokeratin 7 and Cytokeratin 20 Expression in Epithelial Neoplasms: A Survey of 435 Cases. Mod. Pathol. 2000, 13, 962–972. [Google Scholar] [CrossRef]
- Genega, E.M.; Hutchinson, B.; Reuter, V.E.; Gaudin, P.B. Immunophenotype of High-Grade Prostatic Adenocarcinoma and Urothelial Carcinoma. Mod. Pathol. 2000, 13, 1186–1191. [Google Scholar] [CrossRef]
- Gheitasi, R.; Sadeghi, E.; Jafari, M. Comparison of Immunohistochemistry Expression of CK7, HMWK and PSA in High-Grade Prostatic Adenocarcinoma and Bladder Transitional Cell Carcinoma. Iran. J. Pathol. 2021, 16, 33–39. [Google Scholar] [CrossRef]
- Cheville, J.C.; Dundore, P.A.; Bostwick, D.G.; Lieber, M.M.; Batts, K.P.; Sebo, T.J.; Farrow, G.M. Transitional Cell Carcinoma of the Prostate: Clinicopathologic Study of 50 Cases. Cancer 1998, 82, 703–707. [Google Scholar] [CrossRef]
- Nagle, R.B.; Brawer, M.K.; Kittelson, J.; Clark, V. Phenotypic Relationships of Prostatic Intraepithelial Neoplasia to Invasive Prostatic Carcinoma. Am. J. Pathol. 1991, 138, 119–128. [Google Scholar]
- Zhang, B.; Ci, X.; Tao, R.; Ni, J.J.; Xuan, X.; King, J.L.; Xia, S.; Li, Y.; Frierson, H.F.; Lee, D.-K.; et al. Klf5 Acetylation Regulates Luminal Differentiation of Basal Progenitors in Prostate Development and Regeneration. Nat. Commun. 2020, 11, 997. [Google Scholar] [CrossRef]
- Che, M.; Chaturvedi, A.; Munro, S.A.; Pitzen, S.P.; Ling, A.; Zhang, W.; Mentzer, J.; Ku, S.-Y.; Puca, L.; Zhu, Y.; et al. Opposing Transcriptional Programs of KLF5 and AR Emerge during Therapy for Advanced Prostate Cancer. Nat. Commun. 2021, 12, 6377. [Google Scholar] [CrossRef]
- Xie, W.; Regan, M.M.; Buyse, M.; Halabi, S.; Kantoff, P.W.; Sartor, O.; Soule, H.; Clarke, N.W.; Collette, L.; Dignam, J.J.; et al. Metastasis-Free Survival Is a Strong Surrogate of Overall Survival in Localized Prostate Cancer. J. Clin. Oncol. 2017, 35, 3097–3104. [Google Scholar] [CrossRef] [PubMed]
- Kotliar, S.N.; Wood, C.G.; Schaeffer, A.J.; Oyasu, R. Transitional Cell Carcinoma Exhibiting Clear Cell Features. A Differential Diagnosis for Clear Cell Adenocarcinoma of the Urinary Tract. Arch. Pathol. Lab. Med. 1995, 119, 79–81. [Google Scholar] [PubMed]
- Polifka, I.; Agaimy, A.; Herrmann, E.; Spath, V.; Trojan, L.; Stöckle, M.; Becker, F.; Ströbel, P.; Wülfing, C.; Schrader, A.J.; et al. High Proliferation Rate and TNM Stage but Not Histomorphological Subtype Are Independent Prognostic Markers for Overall Survival in Papillary Renal Cell Carcinoma. Hum. Pathol. 2019, 83, 212–223. [Google Scholar] [CrossRef]
- Mertz, K.D.; Demichelis, F.; Sboner, A.; Hirsch, M.S.; Dal Cin, P.; Struckmann, K.; Storz, M.; Scherrer, S.; Schmid, D.M.; Strebel, R.T.; et al. Association of Cytokeratin 7 and 19 Expression with Genomic Stability and Favorable Prognosis in Clear Cell Renal Cell Cancer. Int. J. Cancer 2008, 123, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Alsharif, S.; Fallatah, A.; Chung, B.M. Intermediate Filaments as Effectors of Cancer Development and Metastasis: A Focus on Keratins, Vimentin, and Nestin. Cells 2019, 8, 497. [Google Scholar] [CrossRef] [Green Version]
- Werner, S.; Keller, L.; Pantel, K. Epithelial Keratins: Biology and Implications as Diagnostic Markers for Liquid Biopsies. Mol. Aspects Med. 2020, 72, 100817. [Google Scholar] [CrossRef]
- Scott, M.K.D.; Ozawa, M.G.; Chu, P.; Limaye, M.; Nair, V.S.; Schaffert, S.; Koong, A.C.; West, R.; Khatri, P. A Multi-Scale Integrated Analysis Identifies KRT8 as a Pan-Cancer Early Biomarker. Pac. Symp. Biocomput. 2021, 26, 297–308. [Google Scholar]
- Li, M.; Rao, X.; Cui, Y.; Zhang, L.; Li, X.; Wang, B.; Zheng, Y.; Teng, L.; Zhou, T.; Zhuo, W. The Keratin 17/YAP/IL6 Axis Contributes to E-Cadherin Loss and Aggressiveness of Diffuse Gastric Cancer. Oncogene 2021, 41, 770–781. [Google Scholar] [CrossRef]
- Song, H.; Weinstein, H.N.W.; Allegakoen, P.; Wadsworth, M.H.; Xie, J.; Yang, H.; Castro, E.A.; Lu, K.L.; Stohr, B.A.; Feng, F.Y.; et al. Single-Cell Analysis of Human Primary Prostate Cancer Reveals the Heterogeneity of Tumor-Associated Epithelial Cell States. Nat. Commun. 2022, 13, 141. [Google Scholar] [CrossRef]
- Lyon, T.D.; Henry, M.R.; Shah, P.H.; Boorjian, S.A.; Tollefson, M.K.; Frank, I. Development of a Technique for Evaluating the Presence of Malignant Cells in Prostatic Fluid during Robotic Prostatectomy. Urol. Oncol. 2021, 39, 192.e1–192.e6. [Google Scholar] [CrossRef]

| Type of Analyses | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR [95% CI] | p-Value | HR [95% CI] | p-Value | |
| Age at diagnosis | 0.99 [0.926–1.059] | 0.777 | - | - |
| cTNM | 1.304 [0.507–3.355] | 0.581 | - | - |
| Gleason score 0–1 | 21.407 [8.663–52.9] | <0.001 | 27.014 [10.13–72.05] | <0.001 |
| KRT7 MFI expression in basal cells with continuous value | 1.021 [0.973–1.073] | 0.397 | - | - |
| KRT7 MFI expression in basal cells dichotomized with quintiles (high-KRT7 defined as >80th percentile) | 2.238 [1.002–4.999] | 0.049 | 2.907 [1.229–6.875] | 0.014 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dariane, C.; Clairefond, S.; Péant, B.; Communal, L.; Thian, Z.; Ouellet, V.; Trudel, D.; Benzerdjeb, N.; Azzi, F.; Méjean, A.; et al. High Keratin-7 Expression in Benign Peri-Tumoral Prostatic Glands Is Predictive of Bone Metastasis Onset and Prostate Cancer-Specific Mortality. Cancers 2022, 14, 1623. https://doi.org/10.3390/cancers14071623
Dariane C, Clairefond S, Péant B, Communal L, Thian Z, Ouellet V, Trudel D, Benzerdjeb N, Azzi F, Méjean A, et al. High Keratin-7 Expression in Benign Peri-Tumoral Prostatic Glands Is Predictive of Bone Metastasis Onset and Prostate Cancer-Specific Mortality. Cancers. 2022; 14(7):1623. https://doi.org/10.3390/cancers14071623
Chicago/Turabian StyleDariane, Charles, Sylvie Clairefond, Benjamin Péant, Laudine Communal, Zhe Thian, Véronique Ouellet, Dominique Trudel, Nazim Benzerdjeb, Feryel Azzi, Arnaud Méjean, and et al. 2022. "High Keratin-7 Expression in Benign Peri-Tumoral Prostatic Glands Is Predictive of Bone Metastasis Onset and Prostate Cancer-Specific Mortality" Cancers 14, no. 7: 1623. https://doi.org/10.3390/cancers14071623
APA StyleDariane, C., Clairefond, S., Péant, B., Communal, L., Thian, Z., Ouellet, V., Trudel, D., Benzerdjeb, N., Azzi, F., Méjean, A., Timsit, M.-O., Baurès, M., Guidotti, J.-E., Goffin, V., Karakiewicz, P. I., Mes-Masson, A.-M., & Saad, F. (2022). High Keratin-7 Expression in Benign Peri-Tumoral Prostatic Glands Is Predictive of Bone Metastasis Onset and Prostate Cancer-Specific Mortality. Cancers, 14(7), 1623. https://doi.org/10.3390/cancers14071623








