Clonality, Mutation and Kaposi Sarcoma: A Systematic Review
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
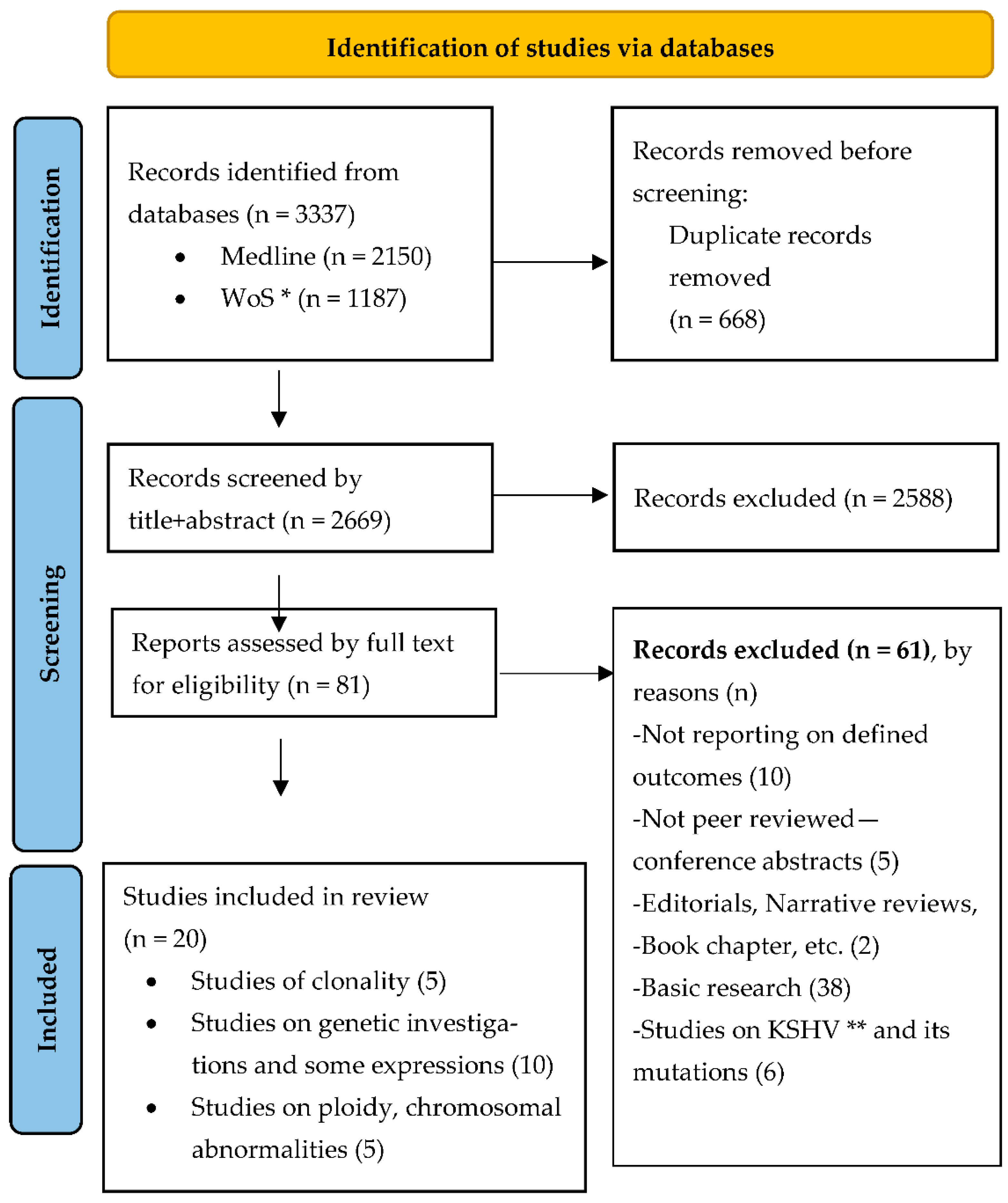
2.1. Literature Search and Study Selection
2.2. Data Extraction
2.3. Assessment of Risk of Bias of Included Studies
2.4. Synthesis of Results
3. Results
| Included Studies (All Case Series) Reporting Outcomes on Clonality | |||||
|---|---|---|---|---|---|
| Study 1 | Study 2 | Study 3 | Study 4 | Study 5 | |
| Authors | Delabesse et al. [23] | Ding et al. [24] | Gil et al. [25] | Rabkin et al. [26] | Rabkin et al. [27] |
| Year (Country) | 1997 (France) | 2015 (China) | 1998 (USA) | 1995 (USA) | 1997 (Zambia and USA) |
| Sample Description (Ca+Ct) | 7 + 6 ♀ skin biopsies Age not reported. | 14 + 1 ♀ Mean age 48.4 years (range 27–71) | 12 ♀, 4 had multiple biopsies. Mean age 49 years (range 27–89). | 3 ♀ +number Ct not described Age not reported | 10 ♀, 5 lesion samples + 1 Ct for each Median age 26 years (range 20–35). |
| Case Classification | Clinically: 4 Classic KS, 3 AIDS SG: 1 macular, 3 plaque, 3 nodular | Clinically: 6 Classic KS, 8 AIDS SG: 2 macular, 11 plaque, 1 nodular Other: all cases HHV-8 + | Clinically: 2 Classic KS and 10 AIDS (8 advanced disease, 3 history of glucocorticoid use) SG: not reported. | Clinically: all HIV type 1-sero+ SG: all nodular KS lesions | Clinically: all HIV positive (8 serological, 2 clinical diagnosis) SG: multiple nodular KS |
| Study Results | |||||
| Description of Investigation Used | Punch biopsies HP: HES MA: DNA extraction Clonality AS: PCR amplification, HUMARA | HP: Surgical tissues, standard histology MA: IHCS of the primary mcAB. Clonality AS: PCR amplification, HUMARA detection of single-nucleotide polymorphism sites in PGK | HP: Cutaneous tumour biopsies. HES. Review performed by, to DNA results, blinded pathologist. MA: DNA extraction. Clonality AS: Secondary PCR products analysed by electrophoresis + autoradiography | HP: Cutaneous biopsies, HES MA: DNA extraction. Clonality AS: X chromosome inactivation assay (HUMARA) | HP: Cutaneous biopsies, HES MA: DNA extraction. Clonality AS: X chromosome inactivation assay (HUMARA) |
| Main Results | Descriptive: All 7 patients were heterozygous for HUMARA Clonality: All polyclonal pattern of inactivation | Descriptive: 2 Ca failed to amplify HUMARA, 11 analysed HUMARA, 5 KS PGK Clonality: 1Ca+1Ct polyclonal pattern, rest monoclonal Ca with no significant differences between groups (p > 0.05) | Descriptive: 41 different regions from 24 biopsies were studied. Clonality: 5 Ca clonal, 2 Ca inactivate, 7 Ca polyclonal pattern of inactivation, 2 Ca both clonal/polyclonal inactivation | Descriptive: All 3 patients heterozygous androgen receptor Clonality: 2 of 3 Ca monoclonal pattern | Descriptive: 2 Ca excluded (homozygous HUMARA), 8 Ca (40 tumours, 32 studied) Clonality: 23 (85%) tumours had unbalanced methylation (predominance one allele), 6 Ca (23 tumours) concordant methylation (≤0.00001). |
| Conclusions | It is a polyclonal cell proliferation. | Suggest a clonal neoplasm. | Suggest a clonal neoplasm, but polyclonal inactivation pattern observed may be premalignant stage or false negative results. | Suggest a clonal neoplasm (at least in AIDS Ca). | Data indicate monoclonal cancer. |
| Included Studies Reporting Outcomes on Mutations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Study 1 | Study 2 | Study 3 | Study 4 | Study 5 | Study 6 | Study 7 | Study 8 | Study 9 | Study 10 | |
| Authors | Nicolaides et al. [28] | Kiuru-Kuhlefelt et al. [29] | Cerimele et al. [22] | Tornesello et al. [21] | Huang et al. [30] | Guttman-Yassky et al. [31] | Feller et al. [32] | Cordiali-Fei et al. [33] | Li, Jian J. [34] | Scinicariello et al. [35] |
| Year (Country) | 1994 (USA) | 2000 (Finland + USA) | 1984 (Italy) | 2009 (Italy + Uganda + Greece) | 1993 (USA) | 2012 (USA + Israel) | 2014 (USA) | 2014 (Italy) | 1997 (USA) | 1994 (USA) |
| Study Design | Case series | Case series | Case series | Case control | Case series | Case series | Case series | Case series | Case series | Case series |
| Sample Description (Ca + Ct) | 31 Ca No demographics reported. | 12 Ca 10 ♂, 2 ♀ Age: NR | (65) 22 + 220 49 ♂, 16 ♀ Age: range 57–82 years | 67 + 150 No data disaggregated for ♂♀ Median age: 75 (29.5–47) years | 38 + 10 No demographics reported. | 9 + 4 6 ♂, 7 ♀ Mean age: Ca 69.5 (20–86) years | 24 + 17 21 ♂, 3 ♀ Mean age: Ca 59 (26–86) years | 3 Ca same family 2 ♂, 1 ♀ Age: 63, 64, 40 years | 15 + 5 No demographics reported. | 17 Ca No demographics reported. |
| Case Classification | Clinically: 24 HIV+, 7 HIV− SG: 16 nodular, 6 patch, 9 plaque | Clinically: 6 ♂ HIV+ or AIDS | Clinically: all endemic KS (Sardinia). | Clinically: 33 classic, 2 iatrogenic, 19 epidemic and 13 epidemic Other: all HHV8+ | Clinically: 31 AIDS, 7 classic | Clinically: All classic SG: 4 early, 2 mixed and 3 nodular | Clinically: 6 HIV+, rest unknown SG: 9 patch, 7 plaque, 8 nodular | Clinically: All HIV− Other: all HHV-8+ | Clinically: All AIDS | Clinically: 10 Classic, 7 HIV+ |
| Study Results | ||||||||||
| Description of Investigation Used | GenA: PCR-SSCP (Orita procedure) to detect mutations | GenA: PCR HHV-8 sequence specific primers, aCGH, digital image analysis, FISH, IHCS | GenA: THLA-ABC typing with 162 antisera. HLA-DR and MT typing using 47 antisera. StatA: significance calculated using X’ test+Wolf’s relative risk test. | HP: Cutaneous biopsies GenA: TP53 genotype at codon 72 StatA: Fisher’s exact or X’ test for comparison of Ca/Ct. Student’s t test for age differences | HP: Biopsy/autopsy samples. GenA: R T-PCR, DNA sequencing, ICHS, Southern blot hybridization | HP: Punch biopsies GenA: Gene chip analysis, DNA microarray analysis, IHCS, Immunoflouresence+, obtaining expression profile LEC and BEC gene signature | HP: All sections reviewed by dermatopathologists GenA: FISH, IHCS | Not reported | GenA: RT-PCR + PCR-SSCP in immunoperoxidase stains | GenA: DNA+ PCR amplification, HPV DNA detection, p53 direct sequencing with (ɣ32P) ATP end-labelled primers |
| Main Results | Genetics: 10 Kras overexpression (3 Kras amplification, 7 various mutations Kras exon) | Genetics: 4 recurrent again at 11q13; 4 containingFGF4 and INT2 (expression of FGF4 and INT2 was found in 9 and 3 Ca respectively). | Genetics: No differences in A, B, C antigen frequency (DR5 72.7% Ca, 23.1% Ct, p < 0.0001; DR3 9.1% Ca 53.6% Ct, p < 0.01; DR5 36.4% Ca 18.1% Ct, p not significant) | Genetics: African Ca: PHoZ 50%, PHtZ 31.8%, AHoZ 18.2%, (p = 0.1872) Caucasian Ca: PHoZ 6.7, PHtZ 55.6, AHoZ 37.8%, (p = 0.0567). Stratified by HIV: No significant differences in alleles | Genetics: Int-2 expressed in 21 Ca (55.2%), NASalt in 8. Most variations in int-2 cDNA located in exon 1 (four in exons 2 and 3) | Genetics: Gene expression level markers gradually increased from normal through all KS stages, particularly LEC genes. | Genetics: c-myc amplification in all Ca, IHCS positive for c-Myc in 13 Ca (54%) | Genetics: IL-6 promoter polymorphism G-174C (2 ♂ HtZ, ♀ HoZ) | Genetics: 4 Ca p53 in nuclei+cytoplasm, 5 MDM2 in the nuclei (2 IHCS+ p53, 3 IHCS− p53 protein) | Genetics: 4 Ca (23%) HPV DNA detected (1 AIDS), 5 Ca (24%) p53 HtZ (none in HPV +) |
| Conclusions | Suggests K-ras mutation plays a significant role in KS oncogenesis. | No evidence of HHV-8 integration to genome. | Preliminary evidence of structural chromosome rearrangement. | p53 polymorphism at codon 72 does not represent a RF for KS. | Int-2 expression may play a role in KS oncogenesis. | Suggests local expression of chemokines/growth factors (no clonal exp.) as oncogenesis. | No amplification of the c-myc gene detected. | Suggests that EBV can cause HHV-8 reactivation in predisposed Ca causing KS. | Suggest p53 may be involved in AIDS KS pathogenesis. | Indicate role of HPV to KS pathogenesis and p53 alteration to malignancy progression. |
| Included Studies (All Case Series) Reporting Chromosomal Aberrations | |||||
|---|---|---|---|---|---|
| Study 1 | Study 2 | Study 3 | Study 4 | Study 5 | |
| Authors | Kaaya et al. [36] | Kaaya et al. [37] | Bisceglia et al. [38] | Pyakurel et al. [39] | Reizis et al. [40] |
| Year (Country) | 2000 (Sweden + Tanzania) | 1992 (Sweden + Tanzania) | 1991 (UK + Italy) | 2006 (Sweden + Germany + Tanzania) | 1995 (Israel) |
| Sample Description (Ca + Ct) | 32 Ca (12 Ca ploidy analysis) 22 ♂, 10 ♀ Age: range 8–68 years | 20 Ca 17 ♂, 3 ♀ Mean age: 40 (up to 83) years | 96 Ca (66 analysed, 143 biopsies) 69 ♂, 27 ♀ Mean age: 69 (10–89) years | 27 + 1 Mean age: males 37.5 (23–65) years, females not reported | 39 Ca Mean age: Iatrogenic Ca 68 (54–80) years |
| Case Classification | Clinically: 8 endemic, 24 AIDS SG: 24 nodular, 4 patch, 4 plaque Other: 12 Ploidy analysed Ca (6 endemic, 6 AIDS) | Clinically: 10 endemic, 10 AIDS SG: 17 nodular skin, and 3 generalised lymph node lesions | Clinically: 93% sporadic, 6% AIDS, 1% Hepatitis B | Clinically: 9 Endemic, 18 AIDS SG: 18 nodular, 9 patch | Clinically: 31 classic, 8 iatrogenic (steroid induced) |
| Study Results | |||||
| Description of Investigation Used | HP: Surgical tissues Mol.An: HHV-8 DNA PCR, IHCS, Ploidy by DNA flow cytometry, apoptotic cells (TUNEL-assay) | HP: Surgical tissues. Mol.An: IHCS, DNA measurements | HP: slides classified by histological criteria Mol.An: Mitoses count, Flow Cytometry (DNA aneuploidy ≥1 GO/Gl peak modal channel number) | HP: Surgical tissues Mol.An: Ligation-mediated PCR+DNA labelling, aCGH, FISH | HP: tissue blocks Mol.An: Flowcytometry |
| Main Results | Ploidy: All 12 diploid cellular DNA content, and low numbers of cells (1.6–8.9%) in S and G2 phase. Ploidy values similar normal cells in non-involved tissue of the same section. In contrast the malignant cell line (KS Y-1) showed a near triploid, aneuploid DNA content and a moderate proliferation rate (13% cells in S + G2 phase). | Ploidy: 70% cells contained DNA values ≥2.5, but not greater than 5C level. Both clinical types with euploid DNA pattern. | Ploidy: 6 lesions (5.8%) DNA aneuploid with a clustering around a DNA index of 1.5 (range 1.4–1.6). Increasing mitotic counts and S-phase plus G2-phase cells were seen with progression of the phase and pattern of disease. Nodular and spindle cell forms had the highest mitotic counts and S-phase plus G2-phase cells. | Chromosomal results: 20 (87%) Ca only recurrent aberration loss of Y chromosome One patch Ca showed in addition loss of Xq. Nodular showed recurrent copy number changes in chromosomes 16, 17, 21, X, Y, and other random changes. | Ploidy: 28 classic Ca showed a diploid pattern. Of 8 iatrogenic, 7 were aneuploid and 1 diploid. |
| Conclusions | Represents a diploid, probably reactive, cell proliferation, which progressively increases the expression of antiapoptotic factors (cellular and viral). | Corroborates previous suggestions that KS could represent a reactive process, rather than a clonal proliferation. | Suggest a low level of DNA aneuploidy, but flow cytometry does not solve the dilemma of whether KS is a hyperplastic or neoplastic process. | Support the view that KS (in males) develops into a clonal tumour yet initially is a hyperplastic reactive cell proliferation. | Iatrogenic KS mostly aneuploid pattern, classic KS diploid pattern on flow cytometry. |
- Clinical and morphological description of clonality studies: 34 (74%) of the 46 cases were HIV associated, 12 (16%) classic KS forms of all 17 (37%) studied nodular lesions, 3 (7%) were plaque lesion samples, and the 26 (57%) remaining cases were from 2 studies [25,27], which did not specify morphological characteristics of the lesions.Clinical and morphological description of mutation studies: in total, 84 (29%) HIV associated cases were included, 146 (52%) classic KS and 2 (1%) cases of iatrogenic KS. For 49 (17%) cases, no clinical information was available since Nicolaides et al. [28], comprising 31 cases, did not describe their clinical characteristics and Feller et al. [32] described only 6 of 18 included cases. All together reported that 20 (7%) were nodular lesions, 27 (10%) plaque lesions and 17 (6%) patch lesions, but these data were missing for 217 (77%) cases since 7 [21,22,29,30,33,34,35] of the 10 included studies did not specify the morphological characteristics of the lesions. Two studies reported separately for HHV8 positivity, Tornesello et al. [21] with all 67 cases reported as positive and Cordiali-Fei et al. [33] detecting in all the three cases of the studied family high titres of anti-HHV-8 (type A virus) antibodies.
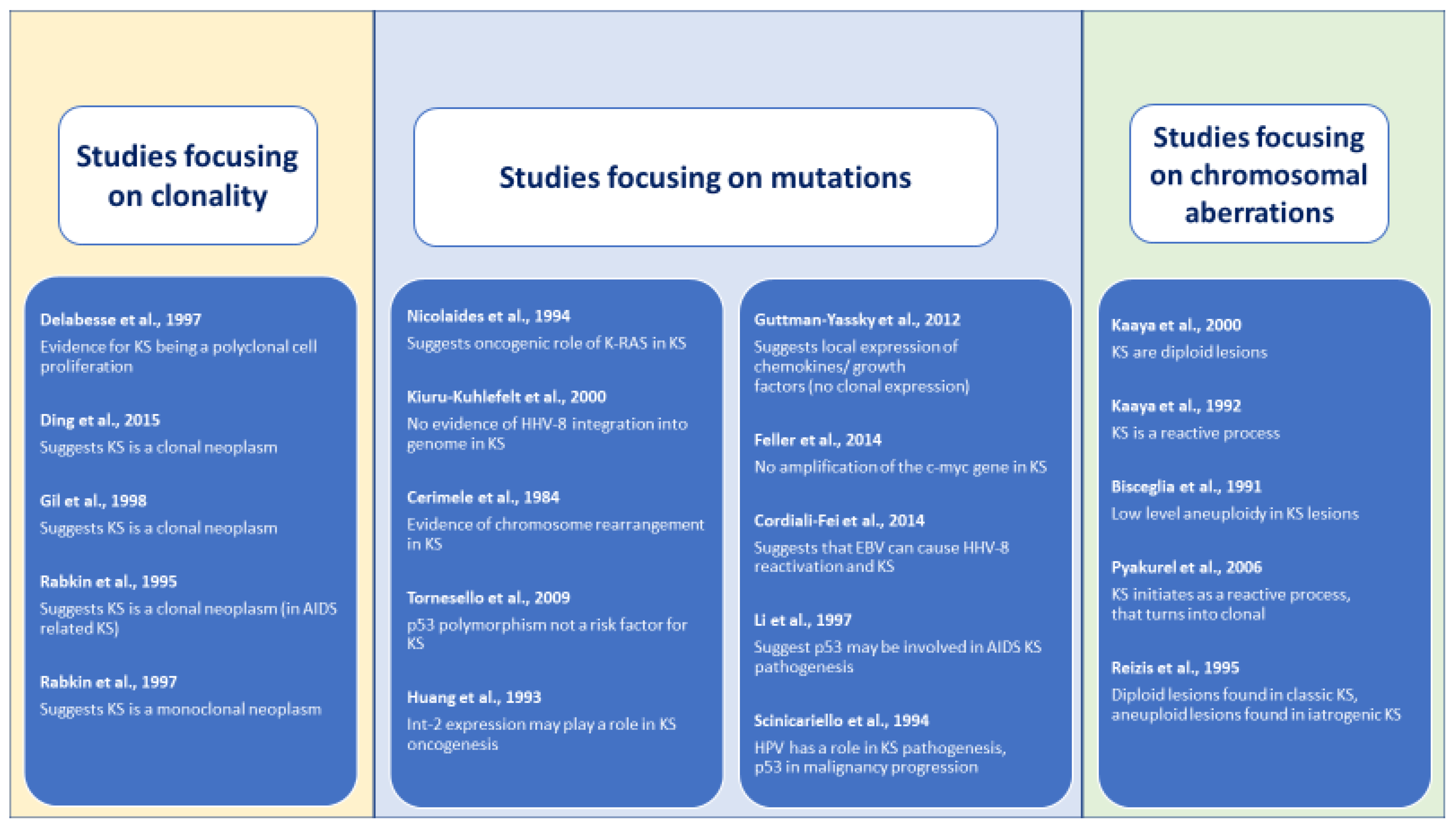
- Clinical and morphological description of chromosomal aberrations studies: 58 (27%) of the 214 cases were HIV associated, 147 (69%) classic and 9 (4%) iatrogenic KS presentations. Only 3 studies [36,37,39] reported on morphological stages of these cases summing 3 (2%) plaque, 37 (17%) nodular, 23 (11%) lesion samples and 151 (70%) cases where it was not reported.Outcomes assessing the clonal nature of KS provided by the 20 included studies (Table 2, Table 3 and Table 4) resulted very heterogeneous (Figure 2). For a more comprehensive synthesis of the results, we decided to report retrieved outcomes grouped into the mentioned three categories of studied genetic alterations. However, differences in study aims, outcome definitions, applied methods and reported outcome measures limited seriously our possibility to pool and/or compare the published data. This heterogeneity in reported data was also the reason for ruling out a quantitative synthesis of the retrieved results, and the performance of a meta-analysis was excluded.
- Determining clonality: outcomes on clonality in KS samples were mostly determined by monoclonal patterns of gene inactivation or methylation and obtained mixed results. Four case series [24,25,26,27] obtained results suggesting that KS is a clonal neoplasm, while one study obtained results compatible with the description of a polyclonal cell [23]. All studies presented a high risk of bias as assessed by the adapted Quadas-2 tool due to bias inherent to their study design and insufficient reporting.
- Detecting mutations: Ten studies aimed to detect mutations in KS lesions, focusing three on p53 mutations [21,34,35] and seven on other different single mutations (IL-6, c-myc, LEC and BEC gene signatures, FGF3, HLA, FGF4 and KRAS). Each study applied different laboratory technics, determined different outcomes and reported diverse findings. Of these ten studies, three [21,29,32] reported negative results for the investigated mutation, and the other seven obtained outcomes suggested a possible relationship. Of the three studies [21,34,35] focusing on a possible role of p53 mutation in the KS oncogenesis, two [29,35] obtained results that point to a possible implication, and one case control study [21] with a large sample and well-performed statistical analysis failed in obtaining evidence of a possible association. These studies presented a high risk of bias, including the prospective case series [22] and the case control study [21] due to difficulties in assessing the internal and external validity of the studies based on the reported data.
- Demonstrating chromosomal aberrations: Finally, of the five selected studies [36,37,38,39,40] that investigated chromosomal aberrations, two studies [36,38] reported diploidy Reizis et al. [38] (a type of pattern for iatrogenic forms of KS) and aneuploid patterns for the classic form of KS. Another study [40] detected low levels of DNA aneuploidy, and the other two [37,41] reported results compatible with a hyperplastic reactive cell proliferation. Again, a high risk of bias was detected for all included studies due to limitations inherent to the study design and through a lack of detailed reporting of methods. Table 2, Table 3 and Table 4 summarize our findings.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Declarations
References
- Curtiss, P.; Strazzulla, L.C.; Friedman-Kien, A.E. An Update on Kaposi’s Sarcoma: Epidemiology, Pathogenesis and Treatment. Dermatol. Ther. 2016, 6, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Shiels, R.A. A history of Kaposi’s sarcoma. J. R. Soc. Med. 1986, 79, 532–534. [Google Scholar] [CrossRef] [PubMed]
- Vangipuram, R.; Tyring, S.K. AIDS-Associated Malignancies. Cancer Treat. Res. 2019, 177, 1–21. [Google Scholar] [PubMed]
- Vangipuram, R.; Tyring, S.K. Epidemiology of Kaposi sarcoma: Review and description of the nonepidemic variant. Int. J. Dermatol. 2019, 58, 538–542. [Google Scholar] [CrossRef]
- Mariggio, G.; Koch, S.; Schulz, T.F. Kaposi sarcoma herpesvirus pathogenesis. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2017, 372, 20160275. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Etta, E.M.; Alayande, D.P.; Mavhandu-Ramarumo, L.G.; Gachara, G.; Bessong, P.O. HHV-8 Seroprevalence and Genotype Distribution in Africa, 1998(-)2017: A Systematic Review. Viruses 2018, 10, 458. [Google Scholar] [CrossRef]
- Phipps, W.; Ssewankambo, F.; Nguyen, H.; Saracino, M.; Wald, A.; Corey, L.; Orem, J.; Kambugu, A.; Casper, C. Gender differences in clinical presentation and outcomes of epidemic Kaposi sarcoma in Uganda. PLoS ONE 2010, 5, e13936. [Google Scholar] [CrossRef]
- Willis, R.A. Tumor seminar. Tex. State J. Med. 1950, 46, 611–638. [Google Scholar]
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Parsons, B.L. Multiclonal tumor origin: Evidence and implications. Mutat. Res. Rev. Mutat. Res. 2018, 777, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Erickson-Johnson, M.R.; Chou, M.M.; Evers, B.R.; Roth, C.W.; Seys, A.R.; Jin, L.; Ye, Y.; Lau, A.W.; Wang, X.; Oliveira, A.M. Nodular fasciitis: A novel model of transient neoplasia induced by MYH9-USP6 gene fusion. Lab. Investig. A J. Tech. Methods Pathology. 2011, 91, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Callan, M.F.; Steven, N.; Krausa, P.; Wilson, J.D.; Moss, P.A.; Gillespie, G.M.; Bell, J.I.; Rickinson, A.B.; McMichael, A.J. Large clonal expansions of CD8+ T cells in acute infectious mononucleosis. Nat. Med. 1996, 2, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.R.; Jackson, C.; Lewis, J.E.; Taylor, G.S.; Thomas, O.G.; Stagg, H.R. Predictors of Epstein-Barr virus serostatus and implications for vaccine policy: A systematic review of the literature. J. Glob. Health. 2020, 10, 010404. [Google Scholar] [CrossRef] [PubMed]
- Knudson, A.G., Jr. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef]
- Martincorena, I.; Raine, K.M.; Gerstung, M.; Dawson, K.J.; Haase, K.; Van Loo, P.; Davies, H.; Stratton, M.R.; Campbell, P.J. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 2017, 171, 1029–1041.e21. [Google Scholar] [CrossRef]
- Martincorena, I. Somatic mutation and clonal expansions in human tissues. Genome Med. 2019, 11, 35. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Biryahwaho, B.; Downing, R.; Hatzakis, A.; Alessi, E.; Cusini, M.; Ruocco, V.; Katongole-Mbidde, E.; Buonaguro, L.; Buonaguro, F.M. TP53 codon 72 polymorphism in classic, endemic and epidemic Kaposi’s sarcoma in African and Caucasian patients. Oncology 2009, 77, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Cerimele, D.; Contu, L.; Scappaticci, S.; Cottoni, F. Kaposi’s sarcoma in Sardinia: An epidemiologic and genetic investigation. Ann. N. Y. Acad. Sci. 1984, 437, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Delabesse, E.; Oksenhendler, E.; Lebbe, C.; Verola, O.; Varet, B.; Turhan, A.G. Molecular analysis of clonality in Kaposi’s sarcoma. J. Clin. Pathol. 1997, 50, 664–668. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yuan, D.; XiuJuan, W.; Yan, Z.; JunQin, L.; Fang, X.; Shirong, Y.; Xiaojing, K.; Yanyan, F.; Weidong, W.; Dong, L.; et al. Use of X-Chromosome Inactivation Pattern to Analyze the Clonality of 14 Female Cases of Kaposi Sarcoma. Med. Sci. Monit. Basic Res. 2015, 21, 116–122. [Google Scholar]
- Gill, P.S.; Tsai, Y.C.; Rao, A.P.; Spruck, C.H.; Zheng, T.; Harrington, W.A.; Cheung, T.; Nathwani, B.; Jones, P.A. Evidence for multiclonality in multicentric Kaposi’s sarcoma. Proc. Natl. Acad. Sci. USA 1998, 95, 8257–8261. [Google Scholar] [CrossRef]
- Rabkin, C.S.; Biggar, R.J.; Coleman, A.; Musaba, E.; Chibwe, G.; Janz, S. Clonality of aids-related kaposis-sarcoma. Aids Res. Hum. Retrovir. 1995, 11, S74. [Google Scholar]
- Rabkin, C.S.; Janz, S.; Lash, A.; Coleman, A.E.; Musaba, E.; Liotta, L.; Biggar, R.J.; Zhuang, Z. Monoclonal origin of multicentric Kaposi’s sarcoma lesions. N. Engl. J. Med. 1997, 336, 988–993. [Google Scholar] [CrossRef]
- Nicolaides, A.; Huang, Y.Q.; Li, J.J.; Zhang, W.G.; Friedman-Kien, A.E. Gene amplification and multiple mutations of the K-ras oncogene in Kaposi’s sarcoma. Anticancer Res. 1994, 14, 921–926. [Google Scholar]
- Kiuru-Kuhlefelt, S.; Sarlomo-Rikala, M.; Larramendy, M.L.; Söderlund, M.; Hedman, K.; Miettinen, M.; Knuutila, S. FGF4 and INT2 oncogenes are amplified and expressed in Kaposi’s sarcoma. Mod. Pathol. 2000, 13, 433–437. [Google Scholar] [CrossRef]
- Huang, Y.Q.; Li, J.J.; Moscatelli, D.; Basilico, C.; Nicolaides, A.; Zhang, W.G.; Poiesz, B.J.; Friedman-Kien, A.E. Expression of int-2 oncogene in Kaposi’s sarcoma lesions. J. Clin. Investig. 1993, 91, 1191–1197. [Google Scholar] [CrossRef]
- Ensoli, B.; Sturzl, M.; Monini, P. Cytokine-mediated growth promotion of Kaposi’s sarcoma and primary effusion lymphoma. Semin. Cancer Biol. 2000, 10, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Feller, K.; Yang, S.; Tung, N.; Lee, J.; Mahalingam, M. c-myc in Kaposi’s sarcoma: Analyses by fluorescent in situ hybridization and immunohistochemistry. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Cordiali-Fei, P.; Latini, A.; Trento, E.; Zampatti, S.; Ferraresi, V.; Cota, C.; Volpi, S.; D’agosto, G.; Bordignon, V.; Giardina, E.; et al. Familial Kaposi’s Sarcoma in HHV8 infected subjects presenting the G-174C allele of the IL-6 promoter: A possible role for EBV? Eur. J. Dermatol. 2014, 24, 503–504. [Google Scholar] [CrossRef]
- Li, J.J.; Huang, Y.Q.; Cockerell, C.J.; Zhang, W.G.; Nicolaides, A.; Friedman-Kien, A.E. Expression and mutation of the tumor suppressor gene p53 in AIDS-associated Kaposi’s sarcoma. Am. J. Dermatopathol. 1997, 19, 373–378. [Google Scholar] [CrossRef]
- Scinicariello, F.; Dolan, M.J.; Nedelcu, I.; Tyring, S.K.; Hilliard, J.K. Occurrence of human papillomavirus and p53 gene mutations in Kaposi’s sarcoma. Virology 1994, 203, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Kaaya, E.; Castanos-Velez, E.; Heiden, T.; Ekman, M.; Catrina, A.I.; Kitinya, J.; Andersson, L.; Biberfeld, P. Proliferation and apoptosis in the evolution of endemic and acquired immunodeficiency syndrome-related Kaposi’s sarcoma. Med. Oncol. 2000, 17, 325–332. [Google Scholar] [CrossRef]
- Kaaya, E.E.; Parravicini, C.; Sundelin, B.; Mgaya, E.; Kitinya, J.; Lema, L.; Luande, J.; Biberfeld, P. Spindle cell ploidy and proliferation in endemic and epidemic African Kaposi’s sarcoma. Eur. J. Cancer 1992, 28a, 1890–1894. [Google Scholar] [CrossRef]
- Reizis, Z.; Trattner, A.; Katzenelson, V.; David, M.; Rotem, A.; Nativ, O.; Mor, Y. Flow cytometric DNA analysis of classic and steroid-induced Kaposi’s sarcoma. Br. J. Dermatol. 1995, 132, 548–550. [Google Scholar] [CrossRef]
- Pyakurel, P.; Montag, U.; Castaños-Vélez, E.; Kaaya, E.; Christensson, B.; Tönnies, H.; Biberfeld, P.; Heiden, T. CGH of microdissected Kaposi’s sarcoma lesions reveals recurrent loss of chromosome Y in early and additional chromosomal changes in late tumour stages. AIDS 2006, 20, 1805–1812. [Google Scholar] [CrossRef]
- Bisceglia, M.; Bosman, C.; Quirke, P. A histologic and flow cytometric study of Kaposi’s sarcoma. Cancer 1992, 69, 793–798. [Google Scholar] [CrossRef]
- Pyakurel, P.; Pak, F.; Mwakigonja, A.R.; Kaaya, E.; Heiden, T.; Biberfeld, P. Lymphatic and vascular origin of Kaposi’s sarcoma spindle cells during tumor development. Int. J. Cancer 2006, 119, 1262–1267. [Google Scholar] [CrossRef]
- Murad, M.H.; Asi, N.; Alsawas, M.; Alahdab, F. New evidence pyramid. Evid. Based Med. 2016, 21, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Burns, P.B.; Rohrich, R.J.; Chung, K.C. The levels of evidence and their role in evidence-based medicine. Plast Reconstr. Surg. 2011, 128, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Goncales, J.P.; Júnior, J.V.; Lopes, T.R.; Tozetto-Mendoza, T.R.; de Farias Guimarães, D.; de Morais, V.M.; Coêlho, M.R. Association of polymorphisms in NFκB1 promoter and NFκBIA gene with the development of antibodies against HHV-8 in HIV-infected individuals. Virology 2019, 535, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Sbihi, Z.; Dossier, A.; Boutboul, D.; Galicier, L.; Parizot, C.; Emarre, A.; Hoareau, B.; Dupin, N.; Marcelin, A.-G.; Oudin, A.; et al. iNKT and memory B-cell alterations in HHV-8 multicentric Castleman disease. Blood 2017, 129, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.C.; Dickson, M.A.; Sadjadi, M.; Gessain, A.; Abel, L.; Jouanguy, E.; Casanova, J.-L. Kaposi Sarcoma of Childhood: Inborn or Acquired Immunodeficiency to Oncogenic HHV-8. Pediatric Blood Cancer 2016, 63, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Barbarov, I.; Koren-Michowitz, M.; Schiby, G.; Portnoy, O.; Livingstone, D.; Segal, G. Fulminant HHV-8 associated Castleman’s disease in a non-HIV, Kaposi sarcoma patient with borderline hemophagocytic syndrome. IMAJ 2015, 17, 253–255. [Google Scholar]
- Koreishi, A.F.; Saenz, A.J.; Arcila, M.E.; Hedvat, C.; Fleming, S.; Teruya-Feldstein, J. Synchronous Follicular Lymphoma, Kaposi Sarcoma, and Castleman’s Disease in a HIV-Negative Patient With EBV and HHV-8 Coinfection. Int. J. Surg. Pathol. 2011, 19, 685–691. [Google Scholar] [CrossRef]
- Jain, V.; Plaisance-Bonstaff, K.; Sangani, R.; Lanier, C.; Dolce, A.; Hu, J.; Brulois, K.; Haecker, I.; Turner, P.; Renne, R.; et al. A Toolbox for Herpesvirus miRNA Research: Construction of a Complete Set of KSHV miRNA Deletion Mutants. Viruses 2016, 8, 54. [Google Scholar] [CrossRef]
- Katano, H.; Sato, Y.; Sata, T. Expression of p53 and human herpesvirus-8 (HHV-8)-encoded latency-associated nuclear antigen with inhibition of apoptosis in HHV-8-associated malignancies. Cancer 2001, 92, 3076–3084. [Google Scholar] [CrossRef]
- Krause, C.J.; Popp, O.; Thirunarayanan, N.; Dittmar, G.; Lipp, M.; Muller, G. MicroRNA-34a promotes genomic instability by a broad suppression of genome maintenance mechanisms downstream of the oncogene KSHV-vGPCR. Oncotarget 2016, 7, 10414–10432. [Google Scholar] [CrossRef]
- Mui, U.N.; Haley, C.T.; Tyring, S.K. Viral Oncology: Molecular Biology and Pathogenesis. J. Clin. Med. 2017, 6, 111. [Google Scholar] [CrossRef] [PubMed]
- Egashira, S.; Jinnin, M.; Harada, M.; Masuguchi, S.; Fukushima, S.; Ihn, H. Exome sequence analysis of Kaposiform hemangioendothelioma: Identification of putative driver mutations. An. Bras. De Dermatol. 2016, 91, 748–753. [Google Scholar] [CrossRef]
- Naipauer, J.; Salyakina, D.; Journo, G.; Rosario, S.; Williams, S.; Abba, M.; Shamay, M.; Mesri, E.A. High-throughput sequencing analysis of a “hit and run” cell and animal model of KSHV tumorigenesis. PLoS Pathog. 2020, 16, e1008589. [Google Scholar] [CrossRef] [PubMed]
- Noel, J.C.; De Thier, F.; Simonart, T.; Andre, J.; Hermans, P.; Van Vooren, J.P.; Heenen, M. p53 protein overexpression is a common but late event in the pathogenesis of iatrogenic and AIDS-related Kaposi’s sarcoma. Arch. Dermatol. Res. 1997, 289, 660–661. [Google Scholar] [CrossRef] [PubMed]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.-C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef] [PubMed]
- Juhasz, A.; Remenyik, E.; Szarka, K.; Veress, G.; Hunyadi, J.; Gergely, L. Consistent polymerase chain reaction-single-strand conformation polymorphism pattern of human herpesvirus-8 in the course of classical Kaposi’s sarcoma assumes its clonal origin. J. Med. Virol. 1998, 54, 300–304. [Google Scholar] [CrossRef]
- Duprez, R.; Lacoste, V.; Brière, J.; Couppie, P.; Frances, C.; Sainte-Marie, D.; Kassa-Kelembho, E.; Lando, M.-J.; Oyono, J.-L.E.; Nkegoum, B.; et al. Evidence for a multiclonal origin of multicentric advanced lesions of Kaposi sarcoma. J. Natl. Cancer Inst. 2007, 99, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Judde, J.G.; Lacoste, V.; Briere, J.; Kassa-Kelembho, E.; Clyti, E.; Couppié, P.; Buchrieser, C.; Tulliez, M.; Morvan, J.; Gessain, A. Monoclonality or oligoclonality of human herpesvirus 8 terminal repeat sequences in Kaposi’s sarcoma and other diseases. J. Natl. Cancer Inst. 2000, 92, 729–736. [Google Scholar] [CrossRef]
- Brooks, J.J. Kaposi’s sarcoma: A reversible hyperplasia. Lancet 1986, 2, 1309–1311. [Google Scholar] [CrossRef]
- Lunardi-Iskandar, Y.; Gill, P.; Lam, V.H.; Zeman, R.A.; Michaels, F.; Mann, D.L.; Reitz, M.S., Jr.; Kaplan, M.; Berneman, Z.N.; Carter, D.; et al. Isolation and characterization of an immortal neoplastic cell line (KS Y-1) from AIDS-associated Kaposi’s sarcoma. J. Natl. Cancer Inst. 1995, 87, 974–981. [Google Scholar] [CrossRef] [PubMed]
| Search term | Keywords | MeSH | Records |
|---|---|---|---|
| Concept 1 | |||
| Kaposi sarcoma | Kaposi * AND sarcoma * | Sarcoma, Kaposi | 1988 |
| Concept 2 | |||
| Clonality | Clonal * | Clonal Evolution | 2012 |
| Mutation | Monoclonal * | Mutation | 1964 |
| Oligoclonal * | Polymorphism, Genetic | 2005 (1968) | |
| Polyclonal * | Clone Cells | 1968 (1964) | |
| Mutant * | Cell Proliferation | 2005 | |
| Reactive * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Indave Ruiz, B.I.; Armon, S.; Watanabe, R.; Uttley, L.; White, V.A.; Lazar, A.J.; Cree, I.A. Clonality, Mutation and Kaposi Sarcoma: A Systematic Review. Cancers 2022, 14, 1201. https://doi.org/10.3390/cancers14051201
Indave Ruiz BI, Armon S, Watanabe R, Uttley L, White VA, Lazar AJ, Cree IA. Clonality, Mutation and Kaposi Sarcoma: A Systematic Review. Cancers. 2022; 14(5):1201. https://doi.org/10.3390/cancers14051201
Chicago/Turabian StyleIndave Ruiz, Blanca Iciar, Subasri Armon, Reiko Watanabe, Lesley Uttley, Valerie A. White, Alexander J. Lazar, and Ian A. Cree. 2022. "Clonality, Mutation and Kaposi Sarcoma: A Systematic Review" Cancers 14, no. 5: 1201. https://doi.org/10.3390/cancers14051201
APA StyleIndave Ruiz, B. I., Armon, S., Watanabe, R., Uttley, L., White, V. A., Lazar, A. J., & Cree, I. A. (2022). Clonality, Mutation and Kaposi Sarcoma: A Systematic Review. Cancers, 14(5), 1201. https://doi.org/10.3390/cancers14051201






