Circulating Tumor Cells in Breast Cancer Patients: A Balancing Act between Stemness, EMT Features and DNA Damage Responses
Abstract
Simple Summary
Abstract
1. DNA Repair Defects Play Key Roles during the Development of Breast Cancer
2. Dynamic Changes in DNA Damage Responses Are Intricately Linked with EMT and Stemness Features during Breast Cancer Progression
2.1. EMT and DNA Repair
2.1.1. Signaling from the Cellular Surface
2.1.2. Crosstalk between Nuclear DNA Damage Response Components and EMT
2.2. Stemness and DNA Repair
3. Dynamic Changes in the Regulatory Network of Stemness, EMT Features and DNA Damage Responses of CTCs
3.1. Plasticity Causes Stress in CTCs
3.1.1. Interplay between Stemness, EMT and DNA Damage Response Pathways in CTCs
3.1.2. Sources of DNA Damage in CTCs
3.2. Evidence for CTC-Specific DNA Damage Responses and Their Manifestation at the Genomic Level in Breast Cancer Patients
3.2.1. Accumulation of Genomic Instabilities in CTCs from Breast Cancer Patients
3.2.2. CTC-Specific DNA Damage Responses of Breast Cancer Patients
4. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tayoun, T.; Oulhen, M.; Aberlenc, A.; Farace, F.; Pawlikowska, P. Tumor Evolution and Therapeutic Choice Seen through a Prism of Circulating Tumor Cell Genomic Instability. Cells 2021, 10, 337. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Ni, X.; Guo, H.; Su, Z.; Ba, Y.; Tong, Z.; Guo, Z.; Yao, X.; Chen, X.; Yin, J.; et al. Single-Cell Sequencing Deciphers a Convergent Evolution of Copy Number Alterations from Primary to Circulating Tumor Cells. Genome Res. 2017, 27, 1312–1322. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA Double-Strand Break Repair-Pathway Choice in Somatic Mammalian Cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Tutt, A.; Ashworth, A. Hallmarks of “BRCAness” in Sporadic Cancers. Nat. Rev. Cancer 2004, 4, 814–819. [Google Scholar] [CrossRef]
- Sotiriou, S.K.; Kamileri, I.; Lugli, N.; Evangelou, K.; Da-Ré, C.; Huber, F.; Padayachy, L.; Tardy, S.; Nicati, N.L.; Barriot, S.; et al. Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol. Cell 2016, 64, 1127–1134. [Google Scholar] [CrossRef]
- Deniz, M.; Kaufmann, J.; Stahl, A.; Gundelach, T.; Janni, W.; Hoffmann, I.; Keimling, M.; Hampp, S.; Ihle, M.; Wiesmüller, L. In Vitro Model for DNA Double-Strand Break Repair Analysis in Breast Cancer Reveals Cell Type-Specific Associations with Age and Prognosis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016, 30, 3786–3799. [Google Scholar] [CrossRef] [PubMed]
- Keimling, M.; Kaur, J.; Bagadi, S.A.R.; Kreienberg, R.; Wiesmüller, L.; Ralhan, R. A Sensitive Test for the Detection of Specific DSB Repair Defects in Primary Cells from Breast Cancer Specimens. Int. J. Cancer 2008, 123, 730–736. [Google Scholar] [CrossRef]
- Deniz, M.; Holzmann, K.; Wiesmüller, L. Functional Analysis—Make or Break for Cancer Predictability. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2013, 743–744, 132–141. [Google Scholar] [CrossRef]
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and Reprogramming: Origins of Cancer Stem Cells. EMBO Rep. 2014, 15, 244. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xiang, D.; Liu, B.; He, A.; Randle, H.J.; Zhang, K.X.; Dongre, A.; Sachs, N.; Clark, A.P.; Tao, L.; et al. Inadequate DNA Damage Repair Promotes Mammary Transdifferentiation Leading to BRCA1 Breast Cancer. Cell 2019, 178, 135. [Google Scholar] [CrossRef]
- Gorodetska, I.; Kozeretska, I.; Dubrovska, A. BRCA Genes: The Role in Genome Stability, Cancer Stemness and Therapy Resistance. J. Cancer 2019, 10, 2109–2127. [Google Scholar] [CrossRef] [PubMed]
- Chopra, N.; Tovey, H.; Pearson, A.; Cutts, R.; Toms, C.; Proszek, P.; Hubank, M.; Dowsett, M.; Dodson, A.; Daley, F.; et al. Homologous Recombination DNA Repair Deficiency and PARP Inhibition Activity in Primary Triple Negative Breast Cancer. Nat. Commun. 2020, 11, 2662. [Google Scholar] [CrossRef] [PubMed]
- Schlacher, K.; Christ, N.; Siaud, N.; Egashira, A.; Wu, H.; Jasin, M. Double-Strand Break Repair-Independent Role for BRCA2 in Blocking Stalled Replication Fork Degradation by MRE11. Cell 2011, 145, 529–542. [Google Scholar] [CrossRef]
- Schlacher, K.; Wu, H.; Jasin, M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer Cell 2012, 22, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; O’Shaughnessy, J.; Untch, M.; Sikov, W.M.; Rugo, H.S.; McKee, M.D.; Huober, J.; Golshan, M.; von Minckwitz, G.; Maag, D.; et al. Addition of the PARP Inhibitor Veliparib plus Carboplatin or Carboplatin Alone to Standard Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer (BrighTNess): A Randomised, Phase 3 Trial. Lancet Oncol. 2018, 19, 497–509. [Google Scholar] [CrossRef]
- Stork, C.T.; Bocek, M.; Crossley, M.P.; Sollier, J.; Sanz, L.A.; Chédin, F.; Swigut, T.; Cimprich, K.A. Co-Transcriptional R-Loops Are the Main Cause of Estrogen-Induced DNA Damage. eLife 2016, 5, e17548. [Google Scholar] [CrossRef]
- Chiang, H.C.; Zhang, X.; Li, J.; Zhao, X.; Chen, J.; Wang, H.T.H.; Jatoi, I.; Brenner, A.; Hu, Y.; Li, R. BRCA1-Associated R-Loop Affects Transcription and Differentiation in Breast Luminal Epithelial Cells. Nucleic Acids Res. 2019, 47, 5086–5099. [Google Scholar] [CrossRef]
- Hatchi, E.; Skourti-Stathaki, K.; Ventz, S.; Pinello, L.; Yen, A.; Kamieniarz-Gdula, K.; Dimitrov, S.; Pathania, S.; McKinney, K.M.; Eaton, M.L.; et al. BRCA1 Recruitment to Transcriptional Pause Sites Is Required for R-Loop-Driven DNA Damage Repair. Mol. Cell 2015, 57, 636–647. [Google Scholar] [CrossRef]
- Alluri, P.; Newman, L.A. Basal-like and Triple-Negative Breast Cancers: Searching for Positives among Many Negatives. Surg. Oncol. Clin. N. Am. 2014, 23, 567–577. [Google Scholar] [CrossRef]
- Yang, J.; Weinberg, R.A. Epithelial-Mesenchymal Transition: At the Crossroads of Development and Tumor Metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef]
- Couch, F.J.; Hart, S.N.; Sharma, P.; Toland, A.E.; Wang, X.; Miron, P.; Olson, J.E.; Godwin, A.K.; Pankratz, V.S.; Olswold, C.; et al. Inherited Mutations in 17 Breast Cancer Susceptibility Genes among a Large Triple-Negative Breast Cancer Cohort Unselected for Family History of Breast Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Angulo, A.M.; Timms, K.M.; Liu, S.; Chen, H.; Litton, J.K.; Potter, J.; Lanchbury, J.S.; Stemke-Hale, K.; Hennessy, B.T.; Arun, B.K.; et al. Incidence and Outcome of BRCA Mutations in Unselected Patients with Triple Receptor-Negative Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, E.; Ganzinelli, M.; Andreis, D.; Bertoni, R.; Giardini, R.; Fox, S.B.; Broggini, M.; Bottini, A.; Zanoni, V.; Bazzola, L.; et al. Triple Negative Breast Cancers Have a Reduced Expression of DNA Repair Genes. PLoS ONE 2013, 8, e66243. [Google Scholar] [CrossRef] [PubMed]
- Linn, S.C.; van ’t Veer, L.J. Clinical Relevance of the Triple-Negative Breast Cancer Concept: Genetic Basis and Clinical Utility of the Concept. Eur. J. Cancer 2009, 45, 11–26. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Caparica, R.; Lambertini, M.; de Azambuja, E. How I Treat Metastatic Triple-Negative Breast Cancer. ESMO Open 2019, 4, e000504. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef]
- Mittendorf, E.A.; Zhang, H.; Barrios, C.H.; Saji, S.; Jung, K.H.; Hegg, R.; Koehler, A.; Sohn, J.; Iwata, H.; Telli, M.L.; et al. Neoadjuvant Atezolizumab in Combination with Sequential Nab-Paclitaxel and Anthracycline-Based Chemotherapy versus Placebo and Chemotherapy in Patients with Early-Stage Triple-Negative Breast Cancer (IMpassion031): A Randomised, Double-Blind, Phase 3 Trial. Lancet 2020, 396, 1090–1100. [Google Scholar] [CrossRef]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Gonçalves, A.; Lee, K.-H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Robson, M.E.; Tung, N.; Conte, P.; Im, S.A.; Senkus, E.; Xu, B.; Masuda, N.; Delaloge, S.; Li, W.; Armstrong, A.; et al. OlympiAD Final Overall Survival and Tolerability Results: Olaparib versus Chemotherapy Treatment of Physician’s Choice in Patients with a Germline BRCA Mutation and HER2-Negative Metastatic Breast Cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 558–566. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Metcalfe, K.; Hanna, W.; Lynch, H.T.; Ghadirian, P.; Tung, N.; Olopade, O.; Weber, B.; McLennan, J.; Olivotto, I.A.; et al. Disruption of the Expected Positive Correlation between Breast Tumor Size and Lymph Node Status in BRCA1-Related Breast Carcinoma. Cancer 2003, 98, 1569–1577. [Google Scholar] [CrossRef]
- Luo, M.; Brooks, M.; Wicha, M. Epithelial-Mesenchymal Plasticity of Breast Cancer Stem Cells: Implications for Metastasis and Therapeutic Resistance. Curr. Pharm. Des. 2015, 21, 1301–1310. [Google Scholar] [CrossRef]
- Drápela, S.; Bouchal, J.; Jolly, M.K.; Culig, Z.; Souček, K. ZEB1: A Critical Regulator of Cell Plasticity, DNA Damage Response, and Therapy Resistance. Front. Mol. Biosci. 2020, 7, 36. [Google Scholar] [CrossRef]
- Singh, R.; Shankar, B.S.; Sainis, K.B. TGF-Β1-ROS-ATM-CREB Signaling Axis in Macrophage Mediated Migration of Human Breast Cancer MCF7 Cells. Cell. Signal. 2014, 26, 1604–1615. [Google Scholar] [CrossRef]
- BeLow, M.; Osipo, C. Notch Signaling in Breast Cancer: A Role in Drug Resistance. Cells 2020, 9, 2204. [Google Scholar] [CrossRef]
- Dubrovska, A.; Kanamoto, T.; Lomnytska, M.; Heldin, C.H.; Volodko, N.; Souchelnytskyi, S. TGFbeta1/Smad3 Counteracts BRCA1-Dependent Repair of DNA Damage. Oncogene 2005, 24, 2289–2297. [Google Scholar] [CrossRef]
- Ewan, K.B.; Henshall-Powell, R.L.; Ravani, S.A.; Pajares, M.J.; Arteaga, C.; Warters, R.; Akhurst, R.J.; Barcellos-Hoff, M.H. Transforming Growth Factor-Β1 Mediates Cellular Response to DNA Damage in Situ. Cancer Res. 2002, 62, 5627–5631. [Google Scholar]
- Wang, Y.; Yu, Y.; Tsuyada, A.; Ren, X.; Wu, X.; Stubblefield, K.; Rankin-Gee, E.K.; Wang, S.E. Transforming Growth Factor-β Regulates the Sphere-Initiating Stem Cell-like Feature in Breast Cancer through MiRNA-181 and ATM. Oncogene 2011, 30, 1470–1480. [Google Scholar] [CrossRef]
- Rodriguez-Vargas, J.M.; Nguekeu-Zebaze, L.; Dantzer, F. PARP3 Comes to Light as a Prime Target in Cancer Therapy. Cell Cycle 2019, 18, 1295–1301. [Google Scholar] [CrossRef]
- Prodhomme, M.K.; Péricart, S.; Pommier, R.M.; Morel, A.P.; Brunac, A.C.; Franchet, C.; Moyret-Lalle, C.; Brousset, P.; Puisieux, A.; Hoffmann, J.S.; et al. Opposite Roles for ZEB1 and TMEJ in the Regulation of Breast Cancer Genome Stability. Front. Cell Dev. Biol. 2021, 9, 2245. [Google Scholar] [CrossRef]
- Qiang, L.; Shah, P.; Barcellos-Hoff, M.H.; He, Y.Y. TGF-β Signaling Links E-Cadherin Loss to Suppression of Nucleotide Excision Repair. Oncogene 2016, 35, 3293–3302. [Google Scholar] [CrossRef]
- Xu, Y.; Lee, D.K.; Feng, Z.; Xu, Y.; Bu, W.; Li, Y.; Liao, L.; Xu, J. Breast Tumor Cell-Specific Knockout of Twist1 Inhibits Cancer Cell Plasticity, Dissemination, and Lung Metastasis in Mice. Proc. Natl. Acad. Sci. USA 2017, 114, 11494–11499. [Google Scholar] [CrossRef]
- Xu, R.; Won, J.Y.; Kim, C.H.; Kim, D.E.; Yim, H. Roles of the Phosphorylation of Transcriptional Factors in Epithelial-Mesenchymal Transition. J. Oncol. 2019, 2019, 5810465. [Google Scholar] [CrossRef]
- Stemmler, M.P.; Eccles, R.L.; Brabletz, S.; Brabletz, T. Non-Redundant Functions of EMT Transcription Factors. Nat. Cell Biol. 2019, 21, 102–112. [Google Scholar] [CrossRef]
- Prodhomme, M.K.; Pommier, R.M.; Franchet, C.; Fauvet, F.; Bergoglio, V.; Brousset, P.; Morel, A.P.; Brunac, A.C.; Devouassoux-Shisheboran, M.; Petrilli, V.; et al. EMT Transcription Factor ZEB1 Represses the Mutagenic POLθ-Mediated End-Joining Pathway in Breast Cancers. Cancer Res. 2021, 81, 1595–1606. [Google Scholar] [CrossRef]
- Wang, H.; Bierie, B.; Li, A.G.; Pathania, S.; Toomire, K.; Dimitrov, S.D.; Liu, B.; Gelman, R.; Giobbie-Hurder, A.; Feunteun, J.; et al. BRCA1/FANCD2/BRG1-Driven DNA Repair Stabilizes the Differentiation State of Human Mammary Epithelial Cells. Mol. Cell 2016, 63, 277–292. [Google Scholar] [CrossRef]
- Buckley, N.E.; Nic An Tsaoir, C.B.; Blayney, J.K.; Oram, L.C.; Crawford, N.T.; D’Costa, Z.C.; Quinn, J.E.; Kennedy, R.D.; Harkin, D.P.; Mullan, P.B. BRCA1 Is a Key Regulator of Breast Differentiation through Activation of Notch Signalling with Implications for Anti-Endocrine Treatment of Breast Cancers. Nucleic Acids Res. 2013, 41, 8601–8614. [Google Scholar] [CrossRef]
- Wu, Z.Q.; Li, X.Y.; Hu, C.Y.; Ford, M.; Kleer, C.G.; Weiss, S.J. Canonical Wnt Signaling Regulates Slug Activity and Links Epithelial-Mesenchymal Transition with Epigenetic Breast Cancer 1, Early Onset (BRCA1) Repression. Proc. Natl. Acad. Sci. USA 2012, 109, 16654–16659. [Google Scholar] [CrossRef]
- Park, S.Y.; Korm, S.; Chung, H.J.; Choi, S.J.; Jang, J.J.; Cho, S.; Lim, Y.T.; Kim, H.; Lee, J.Y. RAP80 Regulates Epithelial-Mesenchymal Transition Related with Metastasis and Malignancy of Cancer. Cancer Sci. 2016, 107, 267–273. [Google Scholar] [CrossRef]
- Jin, G.; Mao, X.; Qiao, Z.; Chen, B.; Jin, F. RAP80 Expression in Breast Cancer and Its Relationship with Apoptosis in Breast Cancer Cells. OncoTargets Ther. 2019, 12, 625. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Kim, H.S.; Li, X.Y.; Lee, I.; Choi, H.S.; Kang, S.E.; Cha, S.Y.; Ryu, J.K.; Yoon, D.; Fearon, E.R.; et al. A P53/MiRNA-34 Axis Regulates Snail1-Dependent Cancer Cell Epithelial-Mesenchymal Transition. J. Cell Biol. 2011, 195, 417–433. [Google Scholar] [CrossRef]
- Fu, R.; Han, C.F.; Ni, T.; Di, L.; Liu, L.J.; Lv, W.C.; Bi, Y.R.; Jiang, N.; He, Y.; Li, H.M.; et al. A ZEB1/P53 Signaling Axis in Stromal Fibroblasts Promotes Mammary Epithelial Tumours. Nat. Commun. 2019, 10, 3210. [Google Scholar] [CrossRef] [PubMed]
- Powell, E.; Piwnica-Worms, D.; Piwnica-Worms, H. Contribution of P53 to Metastasis. Cancer Discov. 2014, 4, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Chao, C.H.; Xia, W.; Yang, J.Y.; Xiong, Y.; Li, C.W.; Yu, W.H.; Rehman, S.K.; Hsu, J.L.; Lee, H.H.; et al. P53 Regulates Epithelial-Mesenchymal Transition and Stem Cell Properties through Modulating MiRNAs. Nat. Cell Biol. 2011, 13, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Ni, T.; Li, X.Y.; Lu, N.; An, T.; Liu, Z.P.; Fu, R.; Lv, W.C.; Zhang, Y.W.; Xu, X.J.; Grant Rowe, R.; et al. Snail1-Dependent P53 Repression Regulates Expansion and Activity of Tumour-Initiating Cells in Breast Cancer. Nat. Cell Biol. 2016, 18, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Weyemi, U.; Redon, C.E.; Sethi, T.K.; Burrell, A.S.; Jailwala, P.; Kasoji, M.; Abrams, N.; Merchant, A.; Bonner, W.M. Twist1 and Slug Mediate H2AX-Regulated Epithelial-Mesenchymal Transition in Breast Cells. Cell Cycle 2016, 15, 2398–2404. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kuang, J.; Min, L.; Liu, C.; Chen, S.; Gao, C.; Ma, J.; Wu, X.; Li, W.; Wu, L.; Zhu, L. RNF8 Promotes Epithelial-Mesenchymal Transition in Lung Cancer Cells via Stabilization of Slug. Mol. Cancer Res. MCR 2020, 18, 1638–1649. [Google Scholar] [CrossRef]
- Zhang, P.; Wei, Y.; Wang, L.I.; Debeb, B.G.; Yuan, Y.; Zhang, J.; Yuan, J.; Wang, M.; Chen, D.; Sun, Y.; et al. ATM-Mediated Stabilization of ZEB1 Promotes DNA Damage Response and Radioresistance through CHK1. Nat. Cell Biol. 2014, 16, 864–875. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Z.; Zhang, Q.; Zhang, Q.; Sun, P.; Xiang, R.; Ren, G.; Yang, S. ZEB1 Confers Chemotherapeutic Resistance to Breast Cancer by Activating ATM. Cell Death Dis. 2018, 9, 57. [Google Scholar] [CrossRef]
- Kumar, M.; Jaiswal, R.K.; Yadava, P.K.; Singh, R.P. An Assessment of Poly (ADP-Ribose) Polymerase-1 Role in Normal and Cancer Cells. BioFactors 2020, 46, 894–905. [Google Scholar] [CrossRef]
- Sterneck, E.; Poria, D.K.; Balamurugan, K. Slug and E-Cadherin: Stealth Accomplices? Front. Mol. Biosci. 2020, 7, 138. [Google Scholar] [CrossRef]
- Patteson, A.E.; Vahabikashi, A.; Pogoda, K.; Adam, S.A.; Mandal, K.; Kittisopikul, M.; Sivagurunathan, S.; Goldman, A.; Goldman, R.D.; Janmey, P.A. Vimentin Protects Cells against Nuclear Rupture and DNA Damage during Migration. J. Cell Biol. 2019, 218, 4079–4092. [Google Scholar] [CrossRef] [PubMed]
- Fleisch, M.C.; Maxwell, C.A.; Barcellos-Hoff, M.H. The Pleiotropic Roles of Transforming Growth Factor Beta in Homeostasis and Carcinogenesis of Endocrine Organs. Endocr.-Relat. Cancer 2006, 13, 379–400. [Google Scholar] [CrossRef]
- Meng, J.; Chen, S.; Han, J.X.; Qian, B.; Wang, X.R.; Zhong, W.L.; Qin, Y.; Zhang, H.; Gao, W.F.; Lei, Y.Y.; et al. Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma. Cancer Res. 2018, 78, 4150–4162. [Google Scholar] [CrossRef] [PubMed]
- Alpaugh, R.K.; Zhou, L.L.; Dicker, D.T.; Matthew, E.; El-Deiry, W.S. Circulating Tumor Cells: Silent Predictors of Metastasis. F1000Research 2017, 6, 28868131. [Google Scholar] [CrossRef]
- Bachelder, R.E.; Yoon, S.O.; Franci, C.; García De Herreros, A.; Mercurio, A.M. Glycogen Synthase Kinase-3 Is an Endogenous Inhibitor of Snail Transcription: Implications for the Epithelial-Mesenchymal Transition. J. Cell Biol. 2005, 168, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Schindlbeck, C.; Andergassen, U.; Jueckstock, J.; Rack, B.; Janni, W.; Jeschke, U. Disseminated and Circulating Tumor Cells in Bone Marrow and Blood of Breast Cancer Patients: Properties, Enrichment, and Potential Targets. J. Cancer Res. Clin. Oncol. 2016, 142, 1883–1895. [Google Scholar] [CrossRef]
- Bukholm, I.K.; Nesland, J.M.; Børresen-Dale, A.L. Re-Expression of E-Cadherin, Alpha-Catenin and Beta-Catenin, but Not of Gamma-Catenin, in Metastatic Tissue from Breast Cancer Patients [Seecomments]. J. Pathol. 2000, 190, 15–19. [Google Scholar] [CrossRef]
- Lee, K.; Krempely, K.; Roberts, M.E.; Anderson, M.J.; Carneiro, F.; Chao, E.; Dixon, K.; Figueiredo, J.; Ghosh, R.; Huntsman, D.; et al. Specifications of the ACMG/AMP Variant Curation Guidelines for the Analysis of Germline CDH1 Sequence Variants. Hum. Mutat. 2018, 39, 1553–1568. [Google Scholar] [CrossRef]
- Hansford, S.; Kaurah, P.; Li-Chang, H.; Woo, M.; Senz, J.; Pinheiro, H.; Schrader, K.A.; Schaeffer, D.F.; Shumansky, K.; Zogopoulos, G.; et al. Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA Oncol. 2015, 1, 23–32. [Google Scholar] [CrossRef]
- Berx, G.; Cleton-Jansen, A.M.; Nollet, F.; de Leeuw, W.J.F.; van de Vijver, M.J.; Cornelisse, C.; van Roy, F. E-Cadherin Is a Tumour/Invasion Suppressor Gene Mutated in Human Lobular Breast Cancers. EMBO J. 1995, 14, 6107–6115. [Google Scholar] [CrossRef]
- Vincent-Salomon, A.; Thiery, J.P. Host Microenvironment in Breast Cancer Development: Epithelial-Mesenchymal Transition in Breast Cancer Development. Breast Cancer Res. BCR 2003, 5, 101–106. [Google Scholar] [CrossRef]
- Bryan, S.; Witzel, I.; Borgmann, K.; Oliveira-Ferrer, L. Molecular Mechanisms Associated with Brain Metastases in HER2-Positive and Triple Negative Breast Cancers. Cancers 2021, 13, 4137. [Google Scholar] [CrossRef]
- MA, O.; RM, N.; HA, L.; NE, H. The ErbB Signaling Network: Receptor Heterodimerization in Development and Cancer. EMBO J. 2000, 19, 3159–3167. [Google Scholar] [CrossRef]
- Ingthorsson, S.; Andersen, K.; Hilmarsdottir, B.; Maelandsmo, G.M.; Magnusson, M.K.; Gudjonsson, T. HER2 Induced EMT and Tumorigenicity in Breast Epithelial Progenitor Cells Is Inhibited by Coexpression of EGFR. Oncogene 2016, 35, 4244–4255. [Google Scholar] [CrossRef]
- Lian, X.; Zhu, C.; Lin, H.; Gao, Z.; Li, G.; Zhang, N.; Cao, B.; Kang, Y. Radiosensitization of HER2-Positive Esophageal Cancer Cells by Pyrotinib. Biosci. Rep. 2020, 40, BSR20194167. [Google Scholar] [CrossRef]
- Balaji, K.; Vijayaraghavan, S.; Diao, L.; Tong, P.; Fan, Y.; Carey, J.P.W.; Bui, T.N.; Warner, S.; Heymach, J.V.; Hunt, K.K.; et al. AXL Inhibition Suppresses the DNA Damage Response and Sensitizes Cells to PARP Inhibition in Multiple Cancers. Mol. Cancer Res. MCR 2017, 15, 45–58. [Google Scholar] [CrossRef]
- Harradine, K.A.; Akhurst, R.J. Mutations of TGFbeta Signaling Molecules in Human Disease. Ann. Med. 2006, 38, 403–414. [Google Scholar] [CrossRef]
- Siegel, P.M.; Massagué, J. Cytostatic and Apoptotic Actions of TGF-Beta in Homeostasis and Cancer. Nat. Rev. Cancer 2003, 3, 807–820. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J. MicroRNA-Mediated Breast Cancer Metastasis: From Primary Site to Distant Organs. Oncogene 2012, 31, 2499–2511. [Google Scholar] [CrossRef]
- Tarapore, P.; Fukasawa, K. Loss of P53 and Centrosome Hyperamplification. Oncogene 2002, 21, 6234–6240. [Google Scholar] [CrossRef]
- Luongo, F.; Colonna, F.; Calapà, F.; Vitale, S.; Fiori, M.E.; de Maria, R. PTEN Tumor-Suppressor: The Dam of Stemness in Cancer. Cancers 2019, 11, 1076. [Google Scholar] [CrossRef]
- Easton, D.F.; Pharoah, P.D.P.; Antoniou, A.C.; Tischkowitz, M.; Tavtigian, S.V.; Nathanson, K.L.; Devilee, P.; Meindl, A.; Couch, F.J.; Southey, M.; et al. Gene-Panel Sequencing and the Prediction of Breast-Cancer Risk. N. Engl. J. Med. 2015, 372, 2243–2257. [Google Scholar] [CrossRef]
- Sinha, A.; Saleh, A.; Endersby, R.; Yuan, S.H.; Chokshi, C.R.; Brown, K.R.; Kuzio, B.; Kauppinen, T.; Singh, S.K.; Baker, S.J.; et al. RAD51-Mediated DNA Homologous Recombination Is Independent of PTEN Mutational Status. Cancers 2020, 12, 3178. [Google Scholar] [CrossRef]
- Hunt, C.R.; Gupta, A.; Horikoshi, N.; Pandita, T.K. Does PTEN Loss Impair DNA Double-Strand Break Repair by Homologous Recombination? Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 920–922. [Google Scholar] [CrossRef]
- He, J.; Zhang, Z.; Ouyang, M.; Yang, F.; Hao, H.; Lamb, K.L.; Yang, J.; Yin, Y.; Shen, W.H. PTEN Regulates EG5 to Control Spindle Architecture and Chromosome Congression during Mitosis. Nat. Commun. 2016, 7, 12355. [Google Scholar] [CrossRef]
- Shen, W.H.; Balajee, A.S.; Wang, J.; Wu, H.; Eng, C.; Pandolfi, P.P.; Yin, Y. Essential Role for Nuclear PTEN in Maintaining Chromosomal Integrity. Cell 2007, 128, 157–170. [Google Scholar] [CrossRef]
- Yun, M.H.; Hiom, K. CtIP-BRCA1 Modulates the Choice of DNA Double-Strand-Break Repair Pathway throughout the Cell Cycle. Nature 2009, 459, 460–463. [Google Scholar] [CrossRef]
- Zhao, W.; Steinfeld, J.B.; Liang, F.; Chen, X.; Maranon, D.G.; Jian Ma, C.; Kwon, Y.; Rao, T.; Wang, W.; Sheng, C.; et al. BRCA1-BARD1 Promotes RAD51-Mediated Homologous DNA Pairing. Nature 2017, 550, 360–365. [Google Scholar] [CrossRef]
- Sengodan, S.K.; Sreelatha, K.H.; Nadhan, R.; Srinivas, P. Regulation of Epithelial to Mesenchymal Transition by BRCA1 in Breast Cancer. Crit. Rev. Oncol./Hematol. 2018, 123, 74–82. [Google Scholar] [CrossRef]
- Chen, W.C.; Lai, Y.A.; Lin, Y.C.; Ma, J.W.; Huang, L.F.; Yang, N.S.; Ho, C.T.; Kuo, S.C.; Way, T. der Curcumin Suppresses Doxorubicin-Induced Epithelial-Mesenchymal Transition via the Inhibition of TGF-β and PI3K/AKT Signaling Pathways in Triple-Negative Breast Cancer Cells. J. Agric. Food Chem. 2013, 61, 11817–11824. [Google Scholar] [CrossRef]
- Anandi, L.; Chakravarty, V.; Ashiq, K.A.; Bodakuntla, S.; Lahiri, M. DNA-Dependent Protein Kinase Plays a Central Role in Transformation of Breast Epithelial Cells Following Alkylation Damage. J. Cell Sci. 2017, 130, 3749–3763. [Google Scholar] [CrossRef]
- Yoshida, T.; Ozawa, Y.; Kimura, T.; Sato, Y.; Kuznetsov, G.; Xu, S.; Uesugi, M.; Agoulnik, S.; Taylor, N.; Funahashi, Y.; et al. Eribulin Mesilate Suppresses Experimental Metastasis of Breast Cancer Cells by Reversing Phenotype from Epithelial-Mesenchymal Transition (EMT) to Mesenchymal-Epithelial Transition (MET) States. Br. J. Cancer 2014, 110, 1497–1505. [Google Scholar] [CrossRef]
- Haynes, B.; Saadat, N.; Myung, B.; Shekhar, M.P.V. Crosstalk between Translesion Synthesis, Fanconi Anemia Network, and Homologous Recombination Repair Pathways in Interstrand DNA Crosslink Repair and Development of Chemoresistance. Mutat. Res. Rev. Mutat. Res. 2015, 763, 258–266. [Google Scholar] [CrossRef]
- Bilardi, R.A.; Kimura, K.I.; Phillips, D.R.; Cutts, S.M. Processing of Anthracycline-DNA Adducts via DNA Replication and Interstrand Crosslink Repair Pathways. Biochem. Pharmacol. 2012, 83, 1241–1250. [Google Scholar] [CrossRef]
- Kaina, B.; Fritz, G.; Ochs, K.; Haas, S.; Grombacher, T.; Dosch, J.; Christmann, M.; Lund, P.; Gregel, M.C.; Becker, K. Transgenic Systems in Studies on Genotoxicity of Alkylating Agents: Critical Lesions, Thresholds and Defense Mechanisms. Mutat. Res. 1998, 405, 179–191. [Google Scholar] [CrossRef]
- Wu, W.; Koike, A.; Takeshita, T.; Ohta, T. The Ubiquitin E3 Ligase Activity of BRCA1 and Its Biological Functions. Cell Div. 2008, 3, 1. [Google Scholar] [CrossRef]
- Weyemi, U.; Redon, C.E.; Choudhuri, R.; Aziz, T.; Maeda, D.; Boufraqech, M.; Parekh, P.R.; Sethi, T.K.; Kasoji, M.; Abrams, N.; et al. The Histone Variant H2A.X Is a Regulator of the Epithelial-Mesenchymal Transition. Nat. Commun. 2016, 7, 10711. [Google Scholar] [CrossRef]
- Ihle, M.; Biber, S.; Schroeder, I.S.; Blattner, C.; Deniz, M.; Damia, G.; Gottifredi, V.; Wiesmuller, L. Impact of the Interplay between Stemness Features, P53 and Pol Iota on Replication Pathway Choices. Nucleic Acids Res. 2021, 49, 7457–7475. [Google Scholar] [CrossRef]
- Gottifredi, V.; Wiesmüller, L. The Tip of an Iceberg: Replication-Associated Functions of the Tumor Suppressor P53. Cancers 2018, 10, 250. [Google Scholar] [CrossRef]
- Hampp, S.; Kiessling, T.; Buechle, K.; Mansilla, S.F.; Thomale, J.; Rall, M.; Ahn, J.; Pospiech, H.; Gottifredi, V.; Wiesmüller, L. DNA Damage Tolerance Pathway Involving DNA Polymerase ι and the Tumor Suppressor P53 Regulates DNA Replication Fork Progression. Proc. Natl. Acad. Sci. USA 2016, 113, E4311–E4319. [Google Scholar] [CrossRef]
- Gatz, S.A.; Wiesmüller, L. P53 in Recombination and Repair. Cell Death Differ. 2006, 13, 1003–1016. [Google Scholar] [CrossRef]
- Bertrand, P.; Saintigny, Y.; Lopez, B.S. P53′s Double Life: Transactivation-Independent Repression of Homologous Recombination. Trends Genet. TIG 2004, 20, 235–243. [Google Scholar] [CrossRef]
- Parfenyev, S.; Singh, A.; Fedorova, O.; Daks, A.; Kulshreshtha, R.; Barlev, N. cA Interplay between P53 and Non-Coding RNAs in the Regulation of EMT in Breast Cancer. Cell Death Dis. 2021, 12, 17. [Google Scholar] [CrossRef]
- Kaller, M.; Hermeking, H. Interplay Between Transcription Factors and MicroRNAs Regulating Epithelial-Mesenchymal Transitions in Colorectal Cancer. Adv. Exp. Med. Biol. 2016, 937, 71–92. [Google Scholar] [CrossRef]
- Kao, C.-N.; Moi, S.-H.; Hou, M.-F.; Luo, C.-W.; Chen, F.-M.; Pan, M.-R. RNF8–CDH1 Co-Expression Predicts Clinical Benefit of Chemoradiotherapy in Triple-Negative Breast Cancer. J. Pers. Med. 2021, 11, 655. [Google Scholar] [CrossRef]
- Kuang, J.; Li, L.; Guo, L.; Su, Y.; Wang, Y.; Xu, Y.; Wang, X.; Meng, S.; Lei, L.; Xu, L.; et al. RNF8 Promotes Epithelial-Mesenchymal Transition of Breast Cancer Cells. J. Exp. Clin. Cancer Res. 2016, 35, 88. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kong, G. Roles and Epigenetic Regulation of Epithelial-Mesenchymal Transition and Its Transcription Factors in Cancer Initiation and Progression. Cell. Mol. Life Sci. CMLS 2016, 73, 4643–4660. [Google Scholar] [CrossRef]
- Miao, K.; Lei, J.H.; Valecha, M.V.; Zhang, A.; Xu, J.; Wang, L.; Lyu, X.; Chen, S.; Miao, Z.; Zhang, X.; et al. NOTCH1 Activation Compensates BRCA1 Deficiency and Promotes Triple-Negative Breast Cancer Formation. Nat. Commun. 2020, 11, 3256. [Google Scholar] [CrossRef]
- Schochter, F.; Werner, K.; Köstler, C.; Faul, A.; Tzschaschel, M.; Alberter, B.; Müller, V.; Neubauer, H.; Fehm, T.; Friedl, T.W.P.; et al. 53BP1 Accumulation in Circulating Tumor Cells Identifies Chemotherapy-Responsive Metastatic Breast Cancer Patients. Cancers 2020, 12, 930. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive Molecular Portraits of Human Breast Tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- McMullin, R.P.; Wittner, B.S.; Yang, C.; Denton-Schneider, B.R.; Hicks, D.; Singavarapu, R.; Moulis, S.; Lee, J.; Akbari, M.R.; Narod, S.A.; et al. A BRCA1 Deficient-like Signature Is Enriched in Breast Cancer Brain Metastases and Predicts DNA Damage-Induced Poly (ADP-Ribose) Polymerase Inhibitor Sensitivity. Breast Cancer Res. BCR 2014, 16, R25. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.Q.; Huo, H.Y.; Ao, S.; Liu, T.; Yang, L.; Fei, Z.Y.; Zhang, Z.Q.; Ding, L.; Cui, Q.H.; Lin, J.; et al. TGF-β1-induced Epithelial-mesenchymal Transition Increases Fatty Acid Oxidation and OXPHOS Activity via the P-AMPK Pathway in Breast Cancer Cells. Oncol. Rep. 2020, 44, 1206–1215. [Google Scholar] [CrossRef]
- Agnoletto, C.; Corrà, F.; Minotti, L.; Baldassari, F.; Crudele, F.; Cook, W.J.J.; di Leva, G.; D’Adamo, A.P.; Gasparini, P.; Volinia, S. Heterogeneity in Circulating Tumor Cells: The Relevance of the Stem-Cell Subset. Cancers 2019, 11, 483. [Google Scholar] [CrossRef]
- Nilendu, P.; Kumar, A.; Kumar, A.; Pal, J.K.; Sharma, N.K. Breast Cancer Stem Cells as Last Soldiers Eluding Therapeutic Burn: A Hard Nut to Crack. Int. J. Cancer 2018, 142, 7–17. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Z.; Na, L.; Dong, D.; Wang, W.; Zhao, C. FZD5 Contributes to TNBC Proliferation, DNA Damage Repair and Stemness. Cell Death Dis. 2020, 11, 1060. [Google Scholar] [CrossRef]
- Maugeri-Saccà, M.; Bartucci, M.; Maria, R. de DNA Damage Repair Pathways in Cancer Stem Cells. Mol. Cancer Ther. 2012, 11, 1627–1636. [Google Scholar] [CrossRef]
- Pavlopoulou, A.; Oktay, Y.; Vougas, K.; Louka, M.; Vorgias, C.E.; Georgakilas, A.G. Determinants of Resistance to Chemotherapy and Ionizing Radiation in Breast Cancer Stem Cells. Cancer Lett. 2016, 380, 485–493. [Google Scholar] [CrossRef]
- Zhang, M.; Behbod, F.; Atkinson, R.L.; Landis, M.D.; Kittrell, F.; Edwards, D.; Medina, D.; Tsimelzon, A.; Hilsenbeck, S.; Green, J.E.; et al. Identification of Tumor-Initiating Cells in a P53-Null Mouse Model of Breast Cancer. Cancer Res. 2008, 68, 4674–4682. [Google Scholar] [CrossRef]
- Al-Assar, O.; Mantoni, T.; Lunardi, S.; Kingham, G.; Helleday, T.; Brunner, T.B. Breast Cancer Stem-like Cells Show Dominant Homologous Recombination Due to a Larger S-G2 Fraction. Cancer Biol. Ther. 2011, 11, 1028–1035. [Google Scholar] [CrossRef][Green Version]
- Wang, Q.; Zhang, H.; Kajino, K.; Greene, M.I. BRCA1 Binds C-Myc and Inhibits Its Transcriptional and Transforming Activity in Cells. Oncogene 1998, 17, 1939–1948. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt Pathways in Cancer Stem Cells: Clinical Update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Liu, S.; Ginestier, C.; Charafe-Jauffret, E.; Foco, H.; Kleer, C.G.; Merajver, S.D.; Dontu, G.; Wicha, M.S. BRCA1 Regulates Human Mammary Stem/Progenitor Cell Fate. Proc. Natl. Acad. Sci. USA 2008, 105, 1680–1685. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, M.; Strappazzon, F.; Arisi, I.; Brandi, R.; D’Onofrio, M.; Sambucci, M.; Manic, G.; Vitale, I.; Barilà, D.; Stagni, V. ATM Kinase Sustains Breast Cancer Stem-like Cells by Promoting ATG4C Expression and Autophagy. Oncotarget 2017, 8, 21692–21709. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Paulson, A.; Charafe-Jauffret, E.; Ginestier, C.; Brown, M.; Dutcher, J.; Clouthier, S.G.; Wicha, M.S. Regulation of Mammary Stem/Progenitor Cells by PTEN/Akt/Beta-Catenin Signaling. PLoS Biol. 2009, 7, e1000121. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.P.; Deng, J.; Xia, W.; Xu, J.; Li, Y.M.; Gunduz, M.; Hung, M.C. Dual Regulation of Snail by GSK-3beta-Mediated Phosphorylation in Control of Epithelial-Mesenchymal Transition. Nat. Cell Biol. 2004, 6, 931–940. [Google Scholar] [CrossRef]
- Plo, I.; Laulier, C.; Gauthier, L.; Lebrun, F.; Calvo, F.; Lopez, B.S. AKT1 Inhibits Homologous Recombination by Inducing Cytoplasmic Retention of BRCA1 and RAD51. Cancer Res. 2008, 68, 9404–9412. [Google Scholar] [CrossRef]
- Yi, Y.W.; Hong, W.; Kang, H.J.; Kim, H.J.; Zhao, W.; Wang, A.; Seong, Y.S.; Bae, I. Inhibition of the PI3K/AKT Pathway Potentiates Cytotoxicity of EGFR Kinase Inhibitors in Triple-Negative Breast Cancer Cells. J. Cell. Mol. Med. 2013, 17, 648–656. [Google Scholar] [CrossRef]
- Ye, X.; Tam, W.L.; Shibue, T.; Kaygusuz, Y.; Reinhardt, F.; Ng Eaton, E.; Weinberg, R.A. Distinct EMT Programs Control Normal Mammary Stem Cells and Tumour-Initiating Cells. Nature 2015, 525, 256–260. [Google Scholar] [CrossRef]
- Storci, G.; Sansone, P.; Mari, S.; D’Uva, G.; Tavolari, S.; Guarnieri, T.; Taffurelli, M.; Ceccarelli, C.; Santini, D.; Chieco, P.; et al. TNFalpha Up-Regulates SLUG via the NF-KappaB/HIF1alpha Axis, Which Imparts Breast Cancer Cells with a Stem Cell-like Phenotype. J. Cell. Physiol. 2010, 225, 682–691. [Google Scholar] [CrossRef]
- Chimge, N.O.; Baniwal, S.K.; Little, G.H.; Chen, Y.B.; Kahn, M.; Tripathy, D.; Borok, Z.; Frenkel, B. Regulation of Breast Cancer Metastasis by Runx2 and Estrogen Signaling: The Role of SNAI2. Breast Cancer Res. BCR 2011, 13, R127. [Google Scholar] [CrossRef] [PubMed]
- Oliveras-Ferraros, C.; Vall-Llovera, A.M.; Vazquez-Martin, A.; Salip, D.C.; Queralt, B.; Cufí, S.; Martin-Castillo, B.; Bosch-Barrera, J.; Brunet, J.; de Llorens, R.; et al. Transcriptional Upregulation of HER2 Expression in the Absence of HER2 Gene Amplification Results in Cetuximab Resistance That Is Reversed by Trastuzumab Treatment. Oncol. Rep. 2012, 27, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Morel, A.P.; Ginestier, C.; Pommier, R.M.; Cabaud, O.; Ruiz, E.; Wicinski, J.; Devouassoux-Shisheboran, M.; Combaret, V.; Finetti, P.; Chassot, C.; et al. A Stemness-Related ZEB1-MSRB3 Axis Governs Cellular Pliancy and Breast Cancer Genome Stability. Nat. Med. 2017, 23, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Fougner, C.; Bergholtz, H.; Norum, J.H.; Sørlie, T. Re-Definition of Claudin-Low as a Breast Cancer Phenotype. Nat. Commun. 2020, 11, 1787. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Shang, L.; Brooks, M.D.; Jiagge, E.; Zhu, Y.; Buschhaus, J.M.; Conley, S.; Fath, M.A.; Davis, A.; Gheordunescu, E.; et al. Targeting Breast Cancer Stem Cell State Equilibrium through Modulation of Redox Signaling. Cell Metab. 2018, 28, 69–86.e6. [Google Scholar] [CrossRef]
- Thery, L.; Meddis, A.; Cabel, L.; Proudhon, C.; Latouche, A.; Pierga, J.-Y.; Bidard, F.-C. Circulating Tumor Cells in Early Breast Cancer. JNCI Cancer Spectr. 2019, 3, pkz026. [Google Scholar] [CrossRef]
- Rack, B.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; Hepp, P.; Zwingers, T.; Friedl, T.W.P.; Lorenz, R.; Tesch, H.; Fasching, P.A.; et al. Circulating Tumor Cells Predict Survival in Early Average-to-High Risk Breast Cancer Patients. J. Natl. Cancer Inst. 2014, 106, dju066. [Google Scholar] [CrossRef]
- Polzer, B.; Medoro, G.; Pasch, S.; Fontana, F.; Zorzino, L.; Pestka, A.; Andergassen, U.; Meier-Stiegen, F.; Czyz, Z.T.; Alberter, B.; et al. Molecular Profiling of Single Circulating Tumor Cells with Diagnostic Intention. EMBO Mol. Med. 2014, 6, 1371–1386. [Google Scholar] [CrossRef]
- Heitzer, E.; Auer, M.; Gasch, C.; Pichler, M.; Ulz, P.; Hoffmann, E.M.; Lax, S.; Waldispuehl-Geigl, J.; Mauermann, O.; Lackner, C.; et al. Complex Tumor Genomes Inferred from Single Circulating Tumor Cells by Array-CGH and next-Generation Sequencing. Cancer Res. 2013, 73, 2965–2975. [Google Scholar] [CrossRef]
- Pantel, K.; Res, C.A.P. Functional Studies on Viable Circulating Tumor Cells. Clin. Chem. 2016, 62, 328–334. [Google Scholar] [CrossRef]
- Ladan, M.M.; van Gent, D.C.; Jager, A. Homologous Recombination Deficiency Testing for BRCA-Like Tumors: The Road to Clinical Validation. Cancers 2021, 13, 1004. [Google Scholar] [CrossRef] [PubMed]
- Varga, D.; Deniz, M.; Schwentner, L.; Wiesmüller, L. Ovarian Cancer: In Search of Better Marker Systems Based on DNA Repair Defects. Int. J. Mol. Sci. 2013, 14, 640–673. [Google Scholar] [CrossRef] [PubMed]
- Naipal, K.A.T.; Verkaik, N.S.; Ameziane, N.; van Deurzen, C.H.M.; Brugge, P.T.; Meijers, M.; Sieuwerts, A.M.; Martens, J.W.; O’Connor, M.J.; Vrieling, H.; et al. Functional Ex Vivo Assay to Select Homologous Recombination-Deficient Breast Tumors for PARP Inhibitor Treatment. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 4816–4826. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Pfister, T.D.; Parchment, R.E.; Kummar, S.; Rubinstein, L.; Evrard, Y.A.; Gutierrez, M.E.; Murgo, A.J.; Tomaszewski, J.E.; Doroshow, J.H.; et al. Monitoring Drug-Induced GammaH2AX as a Pharmacodynamic Biomarker in Individual Circulating Tumor Cells. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Nagelkerke, A.; van Kuijk, S.J.A.; Sweep, F.C.G.J.; Nagtegaal, I.D.; Hoogerbrugge, N.; Martens, J.W.M.; Timmermans, M.A.; van Laarhoven, H.W.M.; Bussink, J.; Span, P.N. Constitutive Expression of γ-H2AX Has Prognostic Relevance in Triple Negative Breast Cancer. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2011, 101, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Kummar, S.; Ji, J.; Morgan, R.; Lenz, H.J.; Puhalla, S.L.; Belani, C.P.; Gandara, D.R.; Allen, D.; Kiesel, B.; Beumer, J.H.; et al. A Phase I Study of Veliparib in Combination with Metronomic Cyclophosphamide in Adults with Refractory Solid Tumors and Lymphomas. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 1726–1734. [Google Scholar] [CrossRef]
- Nel, I.; Gauler, T.C.; Eberhardt, W.E.; Nickel, A.C.; Schuler, M.; Thomale, J.; Hoffmann, A.C. Formation and Repair Kinetics of Pt-(GpG) DNA Adducts in Extracted Circulating Tumour Cells and Response to Platinum Treatment. Br. J. Cancer 2013, 109, 1223–1229. [Google Scholar] [CrossRef]
- Gong, C.; Liu, B.; Yao, Y.; Qu, S.; Luo, W.; Tan, W.; Liu, Q.; Yao, H.; Zou, L.; Su, F.; et al. Potentiated DNA Damage Response in Circulating Breast Tumor Cells Confers Resistance to Chemotherapy. J. Biol. Chem. 2015, 290, 14811–14825. [Google Scholar] [CrossRef]
- Castroviejo-Bermejo, M.; Cruz, C.; Llop-Guevara, A.; Gutiérrez-Enríquez, S.; Ducy, M.; Ibrahim, Y.H.; Gris-Oliver, A.; Pellegrino, B.; Bruna, A.; Guzmán, M.; et al. A RAD51 Assay Feasible in Routine Tumor Samples Calls PARP Inhibitor Response beyond BRCA Mutation. EMBO Mol. Med. 2018, 10, e9172. [Google Scholar] [CrossRef]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bäuerle, T.; Wallwiener, M.; et al. Identification of a Population of Blood Circulating Tumor Cells from Breast Cancer Patients That Initiates Metastasis in a Xenograft Assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Dubash, T.D.; Edd, J.F.; Jewett, M.K.; Garre, S.G.; Karabacak, N.M.; Rabe, D.C.; Mutlu, B.R.; Walsh, J.R.; Kapur, R.; et al. Ultrahigh-Throughput Magnetic Sorting of Large Blood Volumes for Epitope-Agnostic Isolation of Circulating Tumor Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 16839–16847. [Google Scholar] [CrossRef] [PubMed]
- Andree, K.C.; Mentink, A.; Zeune, L.L.; Terstappen, L.W.M.M.; Stoecklein, N.H.; Neves, R.P.; Driemel, C.; Lampignano, R.; Yang, L.; Neubauer, H.; et al. Toward a Real Liquid Biopsy in Metastatic Breast and Prostate Cancer: Diagnostic LeukApheresis Increases CTC Yields in a European Prospective Multicenter Study (CTCTrap). Int. J. Cancer 2018, 143, 2584–2591. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Aceto, N.; Bersani, F.; Madden, M.W.; Donaldson, M.C.; Desai, R.; Zhu, H.; Comaills, V.; Zheng, Z.; et al. Cancer Therapy. Ex Vivo Culture of Circulating Breast Tumor Cells for Individualized Testing of Drug Susceptibility. Science 2014, 345, 216–220. [Google Scholar] [CrossRef]
- Koch, C.; Kuske, A.; Joosse, S.A.; Yigit, G.; Sflomos, G.; Thaler, S.; Smit, D.J.; Werner, S.; Borgmann, K.; Gärtner, S.; et al. Characterization of Circulating Breast Cancer Cells with Tumorigenic and Metastatic Capacity. EMBO Mol. Med. 2020, 12, e11908. [Google Scholar] [CrossRef]
- Bulfoni, M.; Turetta, M.; del Ben, F.; di Loreto, C.; Beltrami, A.P.; Cesselli, D. Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack. Int. J. Mol. Sci. 2016, 17, 1775. [Google Scholar] [CrossRef]
- Lim, Y.C.; Wiegmans, A.P. Tracking Metastatic Breast Cancer: The Future of Biology in Biosensors. Med. Oncol. 2016, 33, 36. [Google Scholar] [CrossRef]
- Riebensahm, C.; Joosse, S.A.; Mohme, M.; Hanssen, A.; Matschke, J.; Goy, Y.; Witzel, I.; Lamszus, K.; Kropidlowski, J.; Petersen, C.; et al. Clonality of Circulating Tumor Cells in Breast Cancer Brain Metastasis Patients. Breast Cancer Res. BCR 2019, 21, 101. [Google Scholar] [CrossRef]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The Identification and Characterization of Breast Cancer CTCs Competent for Brain Metastasis. Sci. Transl. Med. 2013, 5, 180ra48. [Google Scholar] [CrossRef]
- Cortés-Hernández, L.E.; Eslami-S, Z.; Alix-Panabières, C. Circulating Tumor Cell as the Functional Aspect of Liquid Biopsy to Understand the Metastatic Cascade in Solid Cancer. Mol. Asp. Med. 2020, 72, 100816. [Google Scholar] [CrossRef]
- Paoletti, C.; Cani, A.K.; Larios, J.M.; Hovelson, D.H.; Aung, K.; Darga, E.P.; Cannell, E.M.; Baratta, P.J.; Liu, C.J.; Chu, D.; et al. Comprehensive Mutation and Copy Number Profiling in Archived Circulating Breast Cancer Tumor Cells Documents Heterogeneous Resistance Mechanisms. Cancer Res. 2018, 78, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Lorentzen, A.; Becker, P.F.; Kosla, J.; Saini, M.; Weidele, K.; Ronchi, P.; Klein, C.; Wolf, M.J.; Geist, F.; Seubert, B.; et al. Single Cell Polarity in Liquid Phase Facilitates Tumour Metastasis. Nat. Commun. 2018, 9, 887. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Boareto, M.; Huang, B.; Jia, D.; Lu, M.; Onuchic, J.N.; Levine, H.; Ben-Jacob, E. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol. 2015, 5, 155. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.X.; Chauhan, V.P.; Posada, J.; Ng, M.R.; Wu, M.W.; Adstamongkonkul, P.; Huang, P.; Lindeman, N.; Langer, R.; Jain, R.K. Blocking CXCR4 Alleviates Desmoplasia, Increases T-Lymphocyte Infiltration, and Improves Immunotherapy in Metastatic Breast Cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 4558–4566. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Zou, J.M.; Luo, C.; Shu, Y.; Luo, J.; Qin, J.; Wang, Y.; Li, D.; Wang, S.S.; Chi, G.; et al. Circulating Tumor Cells Promote the Metastatic Colonization of Disseminated Carcinoma Cells by Inducing Systemic Inflammation. Oncotarget 2017, 8, 28418–28430. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Raghuram, G.V.; Mittra, I. Is Inflammation a Direct Response to DsDNA Breaks? Mutat. Res. 2018, 808, 48–52. [Google Scholar] [CrossRef]
- Zheng, Y.; Miyamoto, D.T.; Wittner, B.S.; Sullivan, J.P.; Aceto, N.; Jordan, N.V.; Yu, M.; Karabacak, N.M.; Comaills, V.; Morris, R.; et al. Expression of β-Globin by Cancer Cells Promotes Cell Survival during Blood-Borne Dissemination. Nat. Commun. 2017, 8, 14344. [Google Scholar] [CrossRef]
- Amantini, C.; Morelli, M.B.; Nabissi, M.; Piva, F.; Marinelli, O.; Maggi, F.; Bianchi, F.; Bittoni, A.; Berardi, R.; Giampieri, R.; et al. Expression Profiling of Circulating Tumor Cells in Pancreatic Ductal Adenocarcinoma Patients: Biomarkers Predicting Overall Survival. Front. Oncol. 2019, 9, 874. [Google Scholar] [CrossRef]
- Kröger, C.; Afeyan, A.; Mraz, J.; Eaton, E.N.; Reinhardt, F.; Khodor, Y.L.; Thiru, P.; Bierie, B.; Ye, X.; Burge, C.B.; et al. Acquisition of a Hybrid E/M State Is Essential for Tumorigenicity of Basal Breast Cancer Cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7353–7362. [Google Scholar] [CrossRef]
- Kumar, D.; Gupta, D.; Shankar, S.; Srivastava, R.K. Biomolecular Characterization of Exosomes Released from Cancer Stem Cells: Possible Implications for Biomarker and Treatment of Cancer. Oncotarget 2015, 6, 3280–3291. [Google Scholar] [CrossRef]
- Savelieva, O.E.; Tashireva, L.A.; Kaigorodova, E.V.; Buzenkova, A.V.; Mukhamedzhanov, R.K.; Grigoryeva, E.S.; Zavyalova, M.V.; Tarabanovskaya, N.A.; Cherdyntseva, N.V.; Perelmuter, V.M. Heterogeneity of Stemlike Circulating Tumor Cells in Invasive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 2780. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, M.A.; Stoupis, G.; Theodoropoulos, P.A.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Circulating Tumor Cells with Stemness and Epithelial-to-Mesenchymal Transition Features Are Chemoresistant and Predictive of Poor Outcome in Metastatic Breast Cancer. Mol. Cancer Ther. 2019, 18, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Schuster, E.; Taftaf, R.; Reduzzi, C.; Albert, M.K.; Romero-Calvo, I.; Liu, H. Better Together: Circulating Tumor Cell Clustering in Metastatic Cancer. Trends Cancer 2021, 7, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- Gkountela, S.; Castro-Giner, F.; Szczerba, B.M.; Vetter, M.; Landin, J.; Scherrer, R.; Krol, I.; Scheidmann, M.C.; Beisel, C.; Stirnimann, C.U.; et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019, 176, 98–112.e14. [Google Scholar] [CrossRef]
- Zhao, L.; Wu, X.; Zheng, J.; Dong, D. DNA Methylome Profiling of Circulating Tumor Cells in Lung Cancer at Single Base-Pair Resolution. Oncogene 2021, 40, 1884–1895. [Google Scholar] [CrossRef]
- Gorski, J.J.; James, C.R.; Quinn, J.E.; Stewart, G.E.; Staunton, K.C.; Buckley, N.E.; McDyer, F.A.; Kennedy, R.D.; Wilson, R.H.; Mullan, P.B.; et al. BRCA1 Transcriptionally Regulates Genes Associated with the Basal-like Phenotype in Breast Cancer. Breast Cancer Res. Treat. 2010, 122, 721–731. [Google Scholar] [CrossRef]
- Pieraccioli, M.; Nicolai, S.; Antonov, A.; Somers, J.; Malewicz, M.; Melino, G.; Raschella, G. ZNF281 Contributes to the DNA Damage Response by Controlling the Expression of XRCC2 and XRCC4. Oncogene 2016, 35, 2592–2601. [Google Scholar] [CrossRef]
- Gorges, T.M.; Kuske, A.; Röck, K.; Mauermann, O.; Müller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016, 62, 1504–1515. [Google Scholar] [CrossRef]
- Mani, C.; Tripathi, K.; Omy, T.R.; Reedy, M.; Manne, U.; Palle, K. GLI1-Targeting Drugs Induce Replication Stress and Homologous Recombination Deficiency and Synergize with PARP-Targeted Therapies in Triple Negative Breast Cancer Cells. Biochim. Et Biophys. Acta. Mol. Basis Dis. 2022, 1868, 166300. [Google Scholar] [CrossRef]
- Zhao, W.; Shan, B.; He, D.; Cheng, Y.; Li, B.; Zhang, C.; Duan, C. Recent Progress in Characterizing Long Noncoding RNAs in Cancer Drug Resistance. J. Cancer 2019, 10, 6693–6702. [Google Scholar] [CrossRef]
- Lv, J.; Liu, Y.; Cheng, F.; Li, J.; Zhou, Y.; Zhang, T.; Zhou, N.; Li, C.; Wang, Z.; Ma, L.; et al. Cell Softness Regulates Tumorigenicity and Stemness of Cancer Cells. EMBO J. 2021, 40, e106123. [Google Scholar] [CrossRef] [PubMed]
- Ketema, M.; Kreft, M.; Secades, P.; Janssen, H.; Sonnenberg, A. Nesprin-3 Connects Plectin and Vimentin to the Nuclear Envelope of Sertoli Cells but Is Not Required for Sertoli Cell Function in Spermatogenesis. Mol. Biol. Cell 2013, 24, 2454–2466. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.D.; Liu, P.Z.; Kandula, V.; Chen, H.; Almassalha, L.M.; Herman, C.; Backman, V.; O’Halloran, T.; Adam, S.A.; Goldman, R.D.; et al. Physicochemical Mechanotransduction Alters Nuclear Shape and Mechanics via Heterochromatin Formation. Mol. Biol. Cell 2019, 30, 2320–2330. [Google Scholar] [CrossRef] [PubMed]
- Schafer, Z.T.; Grassian, A.R.; Song, L.; Jiang, Z.; Gerhart-Hines, Z.; Irie, H.Y.; Gao, S.; Puigserver, P.; Brugge, J.S. Antioxidant and Oncogene Rescue of Metabolic Defects Caused by Loss of Matrix Attachment. Nature 2009, 461, 109–113. [Google Scholar] [CrossRef]
- Yoshida, G.J. Emerging Roles of Myc in Stem Cell Biology and Novel Tumor Therapies. J. Exp. Clin. Cancer Res. CR 2018, 37, 173. [Google Scholar] [CrossRef]
- Lebok, P.; Schütt, K.; Kluth, M.; Witzel, I.; Wölber, L.; Paluchowski, P.; Terracciano, L.; Wilke, C.; Heilenkötter, U.; Müller, V.; et al. High Mitochondrial Content Is Associated with Breast Cancer Aggressiveness. Mol. Clin. Oncol. 2021, 15, 2049–9450. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Cayrefourcq, L.; Mazard, T.; Maudelonde, T.; Assenat, E.; Assou, S. Molecular Portrait of Metastasis-Competent Circulating Tumor Cells in Colon Cancer Reveals the Crucial Role of Genes Regulating Energy Metabolism and DNA Repair. Clin. Chem. 2017, 63, 700–713. [Google Scholar] [CrossRef]
- Sen, N.; Satija, Y.K.; Das, S. P53 and Metabolism: Old Player in a New Game. Transcription 2012, 3, 119–123. [Google Scholar] [CrossRef]
- Tonzi, P.; Huang, T.T. Role of Y-Family Translesion DNA Polymerases in Replication Stress: Implications for New Cancer Therapeutic Targets. DNA Repair 2019, 78, 20–26. [Google Scholar] [CrossRef]
- Ksiakiewicz, M.; Markiewicz, A.; Zaczek, A.J. Epithelial-Mesenchymal Transition: A Hallmark in Metastasis Formation Linking Circulating Tumor Cells and Cancer Stem Cells. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2012, 79, 195–208. [Google Scholar] [CrossRef]
- Nadal, R.; Salido, M.; Nonell, L.; Rodríguez-Rivera, M.; Puigdecanet, E.; Garcia-Puche, J.L.; Macià, M.; Corominas, J.M.; Serrano, M.J.; Lorente, J.A.; et al. Combined Analysis of Copy Number Alterations by Single-Nucleotide Polymorphism Array and MYC Status in Non-Metastatic Breast Cancer Patients: Comparison According to the Circulating Tumor Cell Status. Tumour Biol. J. Int. Soc. Oncol. Biol. Med. 2015, 36, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Pusch, O.; Bernaschek, G.; Eilers, M.; Hengstschläger, M. Activation of C-Myc Uncouples DNA Replication from Activation of G1-Cyclin-Dependent Kinases. Oncogene 1997, 15, 649–656. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Herold, S.; Herkert, B.; Eilers, M. Facilitating Replication under Stress: An Oncogenic Function of MYC? Nat. Rev. Cancer 2009, 9, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Baluapuri, A.; Wolf, E.; Eilers, M. Target Gene-Independent Functions of MYC Oncoproteins. Nat. Rev. Mol. Cell Biol. 2020, 21, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Gross, K.M.; Zhou, W.; Breindel, J.L.; Ouyang, J.; Jin, D.X.; Sokol, E.S.; Gupta, P.B.; Huber, K.; Zou, L.; Kuperwasser, C.M. Loss of Slug Compromises DNA Damage Repair and Accelerates Stem Cell Aging in Mammary Epithelium. Cell Rep. 2019, 28, 394–407.e6. [Google Scholar] [CrossRef]
- Feng, L.; Wei, W.; Heng, Z.; Yantao, H.; Chunbo, W. Knockdown of REV7 Inhibits Breast Cancer Cell Migration and Invasion. Oncol. Res. 2016, 24, 315–325. [Google Scholar] [CrossRef]
- Shilkin, E.S.; Boldinova, E.O.; Stolyarenko, A.D.; Goncharova, R.I.; Chuprov-Netochin, R.N.; Smal, M.P.; Makarova, A.V. Translesion DNA Synthesis and Reinitiation of DNA Synthesis in Chemotherapy Resistance. Biochem. Biokhimiia 2020, 85, 869–882. [Google Scholar] [CrossRef]
- Clairmont, C.S.; D’Andrea, A.D. REV7 Directs DNA Repair Pathway Choice. Trends Cell Biol. 2021, 31, 965–978. [Google Scholar] [CrossRef]
- Zou, S.; Xu, Y.; Chen, X.; He, C.; Gao, A.; Zhou, J.; Chen, Y. DNA Polymerase Iota (Pol ι) Promotes the Migration and Invasion of Breast Cancer Cell via EGFR-ERK-Mediated Epithelial to Mesenchymal Transition. Cancer Biomark. Sect. A Dis. Markers 2019, 24, 363–370. [Google Scholar] [CrossRef]
- Ireno, I.C.; Wiehe, R.S.; Stahl, A.I.; Hampp, S.; Aydin, S.; Troester, M.A.; Selivanova, G.; Wiesmüller, L. Modulation of the Poly (ADP-Ribose) Polymerase Inhibitor Response and DNA Recombination in Breast Cancer Cells by Drugs Affecting Endogenous Wild-Type P53. Carcinogenesis 2014, 35, 2273–2282. [Google Scholar] [CrossRef]
- Ciriello, G.; Miller, M.L.; Aksoy, B.A.; Senbabaoglu, Y.; Schultz, N.; Sander, C. Emerging Landscape of Oncogenic Signatures across Human Cancers. Nat. Genet. 2013, 45, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Hastings, P.J.; Lupski, J.R.; Rosenberg, S.M.; Ira, G. Mechanisms of Change in Gene Copy Number. Nat. Rev. Genet. 2009, 10, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Tripathy, D.; Shete, S.; Ashfaq, R.; Haley, B.; Perkins, S.; Beitsch, P.; Khan, A.; Euhus, D.; Osborne, C.; et al. HER-2 Gene Amplification Can Be Acquired as Breast Cancer Progresses. Proc. Natl. Acad. Sci. USA 2004, 101, 9393–9398. [Google Scholar] [CrossRef]
- Silvestri, M.; Reduzzi, C.; Feliciello, G.; Vismara, M.; Schamberger, T.; Köstler, C.; Motta, R.; Calza, S.; Ferraris, C.; Vingiani, A.; et al. Detection of Genomically Aberrant Cells within Circulating Tumor Microemboli (CTMs) Isolated from Early-Stage Breast Cancer Patients. Cancers 2021, 13, 1409. [Google Scholar] [CrossRef] [PubMed]
- Kasimir-Bauer, S.; Bittner, A.K.; König, L.; Reiter, K.; Keller, T.; Kimmig, R.; Hoffmann, O. Does Primary Neoadjuvant Systemic Therapy Eradicate Minimal Residual Disease? Analysis of Disseminated and Circulating Tumor Cells before and after Therapy. Breast Cancer Res. BCR 2016, 18, 20. [Google Scholar] [CrossRef]
- Liu, M.; Casimiro, M.C.; Wang, C.; Shirley, L.A.; Jiao, X.; Katiyar, S.; Ju, X.; Li, Z.; Yu, Z.; Zhou, J.; et al. P21CIP1 Attenuates Ras- and c-Myc-Dependent Breast Tumor Epithelial Mesenchymal Transition and Cancer Stem Cell-like Gene Expression in Vivo. Proc. Natl. Acad. Sci. USA 2009, 106, 19035–19039. [Google Scholar] [CrossRef] [PubMed]
- Schardt, J.A.; Meyer, M.; Hartmann, C.H.; Schubert, F.; Schmidt-Kittler, O.; Fuhrmann, C.; Polzer, B.; Petronio, M.; Eils, R.; Klein, C.A. Genomic Analysis of Single Cytokeratin-Positive Cells from Bone Marrow Reveals Early Mutational Events in Breast Cancer. Cancer Cell 2005, 8, 227–239. [Google Scholar] [CrossRef]
- Munzone, E.; Nolé, F.; Goldhirsch, A.; Botteri, E.; Esposito, A.; Zorzino, L.; Curigliano, G.; Minchella, I.; Adamoli, L.; Cassatella, M.C.; et al. Changes of HER2 Status in Circulating Tumor Cells Compared with the Primary Tumor during Treatment for Advanced Breast Cancer. Clin. Breast Cancer 2010, 10, 392–397. [Google Scholar] [CrossRef]
- Jackson, A.L.; Loeb, L.A. The Mutation Rate and Cancer. Genetics 1998, 148, 1483–1490. [Google Scholar] [CrossRef]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef]
- Lindahl, T.; Barnes, D.E. Repair of Endogenous DNA Damage. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 127–133. [Google Scholar] [CrossRef] [PubMed]
- van Loon, B.; Markkanen, E.; Hübscher, U. Oxygen as a Friend and Enemy: How to Combat the Mutational Potential of 8-Oxo-Guanine. DNA Repair 2010, 9, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Song, I.; Jo, A.; Shin, J.H.; Cho, H.; Eoff, R.L.; Guengerich, F.P.; Choi, J.Y. Biochemical Analysis of Six Genetic Variants of Error-Prone Human DNA Polymerase ι Involved in Translesion DNA Synthesis. Chem. Res. Toxicol. 2014, 27, 1837–1852. [Google Scholar] [CrossRef]
- Yang, Y.; Li, X.; Hao, L.; Jiang, D.; Wu, B.; He, T.; Tang, Y. The Diagnostic Value of DNA Repair Gene in Breast Cancer Metastasis. Sci. Rep. 2020, 10, 19626. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.D.; Zheng, S.; Xiu, J.; Zhou, S.; Khasraw, M.; Brastianos, P.K.; Kesari, S.; Hu, J.; Rudnick, J.; Salacz, M.E.; et al. Profiles of Brain Metastases: Prioritization of Therapeutic Targets. Int. J. Cancer 2018, 143, 3019–3026. [Google Scholar] [CrossRef] [PubMed]
- Woditschka, S.; Evans, L.; Duchnowska, R.; Reed, L.T.; Palmieri, D.; Qian, Y.; Badve, S.; Sledge, G.; Gril, B.; Aladjem, M.I.; et al. DNA Double-Strand Break Repair Genes and Oxidative Damage in Brain Metastasis of Breast Cancer. J. Natl. Cancer Inst. 2014, 106, dju145. [Google Scholar] [CrossRef]
- Friboulet, L.; Postel-Vinay, S.; Sourisseau, T.; Adam, J.; Stoclin, A.; Ponsonnailles, F.; Dorvault, N.; Commo, F.; Saulnier, P.; Salome-Desmoulez, S.; et al. ERCC1 Function in Nuclear Excision and Interstrand Crosslink Repair Pathways Is Mediated Exclusively by the ERCC1-202 Isoform. Cell Cycle 2013, 12, 3298–3306. [Google Scholar] [CrossRef]
- Friboulet, L.; Olaussen, K.A.; Pignon, J.-P.; Shepherd, F.A.; Tsao, M.-S.; Graziano, S.; Kratzke, R.; Douillard, J.-Y.; Seymour, L.; Pirker, R.; et al. ERCC1 Isoform Expression and DNA Repair in Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2013, 368, 1101–1110. [Google Scholar] [CrossRef]
- Bouwman, P.; Aly, A.; Escandell, J.M.; Pieterse, M.; Bartkova, J.; van der Gulden, H.; Hiddingh, S.; Thanasoula, M.; Kulkarni, A.; Yang, Q.; et al. 53BP1 Loss Rescues BRCA1 Deficiency and Is Associated with Triple-Negative and BRCA-Mutated Breast Cancers. Nat. Struct. Mol. Biol. 2010, 17, 688–695. [Google Scholar] [CrossRef]
- Bunting, S.F.; Callén, E.; Wong, N.; Chen, H.T.; Polato, F.; Gunn, A.; Bothmer, A.; Feldhahn, N.; Fernandez-Capetillo, O.; Cao, L.; et al. 53BP1 Inhibits Homologous Recombination in Brca1-Deficient Cells by Blocking Resection of DNA Breaks. Cell 2010, 141, 243–254. [Google Scholar] [CrossRef]
- Lottersberger, F.; Karssemeijer, R.A.; Dimitrova, N.; de Lange, T. 53BP1 and the LINC Complex Promote Microtubule-Dependent DSB Mobility and DNA Repair. Cell 2015, 163, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Gao, R.; Sei, E.; Brandt, R.; Hartman, J.; Hatschek, T.; Crosetto, N.; Foukakis, T.; Navin, N.E. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018, 173, 879–893.e13. [Google Scholar] [CrossRef] [PubMed]
- Eldridge, C.B.; Allen, F.J.; Crisp, A.; Grandy, R.A.; Vallier, L.; Sale, J.E. A P53-Dependent Checkpoint Induced upon DNA Damage Alters Cell Fate during HiPSC Differentiation. Stem Cell Rep. 2020, 15, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Vanderstichele, A.; Busschaert, P.; Olbrecht, S.; Lambrechts, D.; Vergote, I. Genomic Signatures as Predictive Biomarkers of Homologous Recombination Deficiency in Ovarian Cancer. Eur. J. Cancer 2017, 86, 5–14. [Google Scholar] [CrossRef]
- Cleary, J.M.; Aguirre, A.J.; Shapiro, G.I.; D’Andrea, A.D. Biomarker-Guided Development of DNA Repair Inhibitors. Mol. Cell 2020, 78, 1070–1085. [Google Scholar] [CrossRef]
- Malka, M.M.; Eberle, J.; Niedermayer, K.; Zlotos, D.P.; Wiesmüller, L. Dual PARP and RAD51 Inhibitory Drug Conjugates Show Synergistic and Selective Effects on Breast Cancer Cells. Biomolecules 2021, 11, 981. [Google Scholar] [CrossRef]
- Chang, X.; Sun, D.; Shi, D.; Wang, G.; Chen, Y.; Zhang, K.; Tan, H.; Liu, J.; Liu, B.; Ouyang, L. Design, Synthesis, and Biological Evaluation of Quinazolin-4(3 H)-One Derivatives Co-Targeting Poly(ADP-Ribose) Polymerase-1 and Bromodomain Containing Protein 4 for Breast Cancer Therapy. Acta Pharm. Sin. B 2021, 11, 156–180. [Google Scholar] [CrossRef]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem Cell and Epithelial-Mesenchymal Transition Markers Are Frequently Overexpressed in Circulating Tumor Cells of Metastatic Breast Cancer Patients. Breast Cancer Res. BCR 2009, 11, R46. [Google Scholar] [CrossRef]
- Carey, J.P.W.; Karakas, C.; Bui, T.; Chen, X.; Vijayaraghavan, S.; Zhao, Y.; Wang, J.; Mikule, K.; Litton, J.K.; Hunt, K.K.; et al. Synthetic Lethality of PARP Inhibitors in Combination with MYC Blockade Is Independent of BRCA Status in Triple-Negative Breast Cancer. Cancer Res. 2018, 78, 742–757. [Google Scholar] [CrossRef]
- Sato, H.; Niimi, A.; Yasuhara, T.; Permata, T.B.M.; Hagiwara, Y.; Isono, M.; Nuryadi, E.; Sekine, R.; Oike, T.; Kakoti, S.; et al. DNA Double-Strand Break Repair Pathway Regulates PD-L1 Expression in Cancer Cells. Nat. Commun. 2017, 8, 1751. [Google Scholar] [CrossRef]
- Tan, Z.; Yue, C.; Ji, S.; Zhao, C.; Jia, R.; Zhang, Y.; Liu, R.; Li, D.; Yu, Q.; Li, P.; et al. Assessment of PD-L1 Expression on Circulating Tumor Cells for Predicting Clinical Outcomes in Patients with Cancer Receiving PD-1/PD-L1 Blockade Therapies. Oncologist 2021, 26, e2227–e2238. [Google Scholar] [CrossRef]
- Berti, M.; Cortez, D.; Lopes, M. The Plasticity of DNA Replication Forks in Response to Clinically Relevant Genotoxic Stress. Nat. Rev. Mol. Cell Biol. 2020, 21, 633–651. [Google Scholar] [CrossRef]
- Patel, S.M.; Dash, R.C.; Hadden, M.K. Translesion Synthesis Inhibitors as a New Class of Cancer Chemotherapeutics. Expert Opin. Investig. Drugs 2021, 30, 13–24. [Google Scholar] [CrossRef]
- Yang, J.; Chen, Z.; Liu, Y.; Hickey, R.J.; Malkas, L.H. Altered DNA polymerase iota expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenenesis. Cancer Res. 2004, 64, 5597–5607. [Google Scholar] [CrossRef] [PubMed]
- Vassel, F.M.; Bian, K.; Walker, G.C.; Hemann, M.T. Rev7 Loss Alters Cisplatin Response and Increases Drug Efficacy in Chemotherapy-Resistant Lung Cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 28922–28924. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Ross Chapman, J.; Brandsma, I.; Yuan, J.; Mistrik, M.; Bouwman, P.; Bartkova, J.; Gogola, E.; Warmerdam, D.; Barazas, M.; et al. REV7 Counteracts DNA Double-Strand Break Resection and Affects PARP Inhibition. Nature 2015, 521, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Tomicic, M.T.; Thust, R.; Sobol, R.W.; Kaina, B. DNA Polymerase β Mediates Protection of Mammalian Cells against Ganciclovir-Induced Cytotoxicity and DNA Breakage. Cancer Res. 2001, 61, 7399–7403. [Google Scholar] [PubMed]
- Srivastava, A.K.; Han, C.; Zhao, R.; Cui, T.; Dai, Y.; Mao, C.; Zhao, W.; Zhang, X.; Yu, J.; Wang, Q.E. Enhanced Expression of DNA Polymerase Eta Contributes to Cisplatin Resistance of Ovarian Cancer Stem Cells. Proc. Natl. Acad. Sci. USA 2015, 112, 4411–4416. [Google Scholar] [CrossRef]
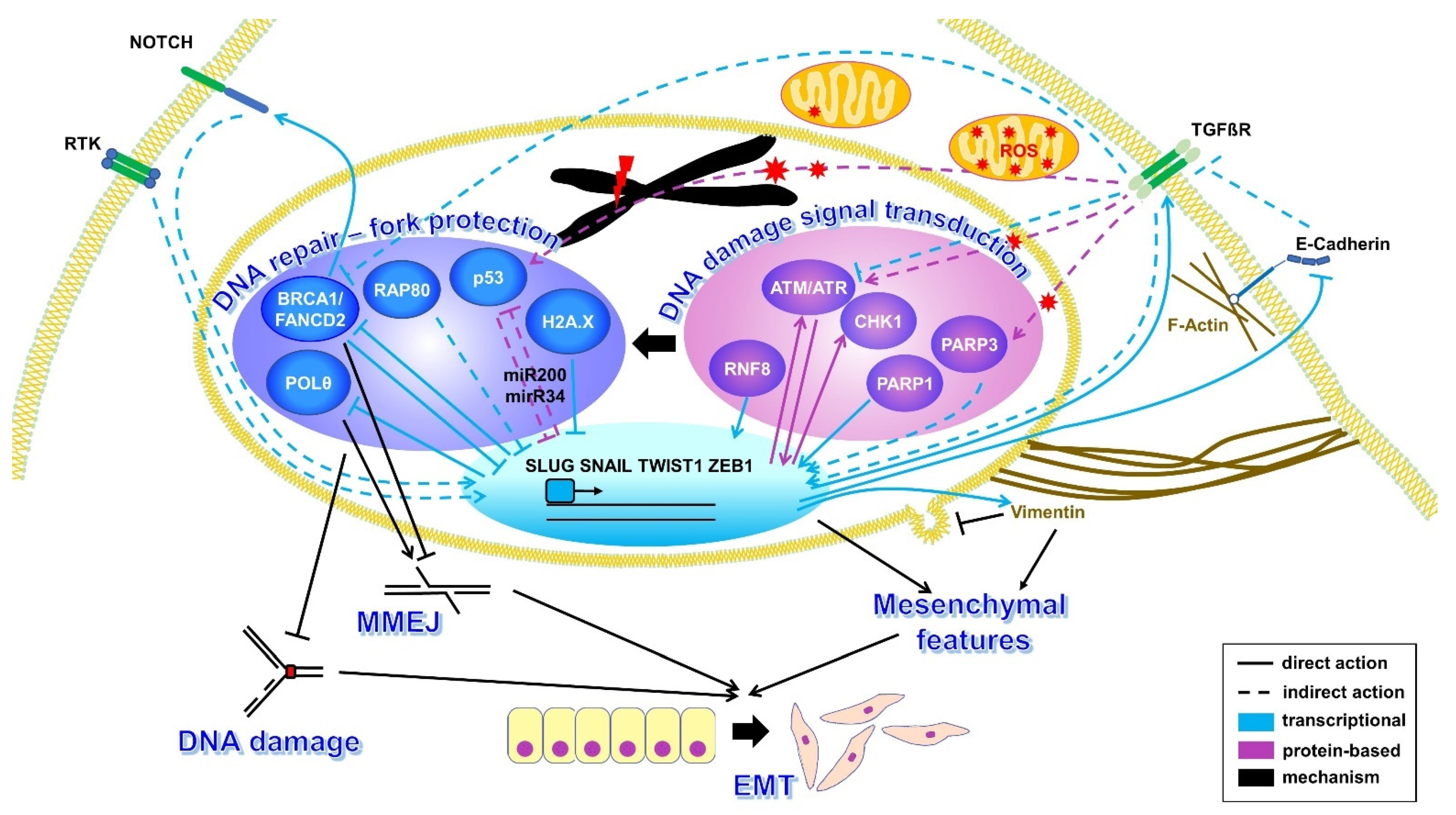
| Effector/Effector Group | Target/Group of Targets | References |
|---|---|---|
| RTK | SLUG/SNAIL/TWIST1/ZEB1 | [34,35] |
| NOTCH | SLUG/SNAIL/TWIST1/ZEB1 | [36] |
| TGFß | BRCA1/FANCD2 | [37] |
| TGFß | p53 | [38] |
| TGFß | ATM/ATR | [35,39] |
| TGFß | PARP3 | [40] |
| TGFß | SLUG/SNAIL/TWIST1/ZEB1 | [41] |
| SLUG/SNAIL/TWIST1/ZEB1 | TGFß | [34] |
| E-Cadherin | TGFß | [42] |
| SLUG/SNAIL/TWIST1/ZEB1 | E-Cadherin | [34] |
| Vimentin | TGFß | [42] |
| SLUG/SNAIL/TWIST1/ZEB1 | Vimentin | [43,44,45] |
| SLUG/SNAIL/TWIST1/ZEB1 | polymerase θ | [46] |
| BRCA1, FANCD2 | SLUG, SNAIL, TWIST1, ZEB1 | [47] |
| BRCA1, FANCD2 | NOTCH | [48] |
| SLUG/SNAIL/TWIST1/ZEB1 | BRCA1/FANCD2 | [49] |
| RAP80 | SLUG/SNAIL/TWIST1/ZEB1 | [50,51] |
| p53 | SLUG/SNAIL/TWIST1/ZEB1 | [52,53,54] |
| SLUG, SNAIL, TWIST1, ZEB1 | p53 | [34,55,56] |
| H2A.X | SLUG/SNAIL/TWIST1/ZEB1 | [57] |
| RNF8 | SLUG/SNAIL/TWIST1/ZEB1 | [58] |
| ATM/ATR | SLUG/SNAIL/TWIST1/ZEB1 | [34,41,59,60] |
| SLUG/SNAIL/TWIST1/ZEB1 | ATM/ATR | [45,59,60] |
| SLUG/SNAIL/TWIST1/ZEB1 | CHK1 | [34,59] |
| PARP1 | SLUG/SNAIL/TWIST1/ZEB1 | [61] |
| PARP3 | SLUG/SNAIL/TWIST1/ZEB1 | [40] |
| Effect in CTCs | Observations | References |
|---|---|---|
| CNAs in primary BC correlate with CTC numbers | Copy number alterations (CNAs) in BC specimen of CTC-positive cases. | [191] |
| CNAs rise with invasiveness | CNAs differ between CTCs from individual patients but not between CTCs from same patient. CNA numbers increase from patients with ductal carcinoma in situ (DCIS) to patients with invasive ductal carcinoma. | [2] |
| Clonality of CTCs in MBC patients | NGS reveals high genomic clonality in CTCs from BC patients with brain metastases. | [158] |
| HER2amplification acquired | Fluorescence in situ hybridization (FISH)-based detection of HER2 amplification provides evidence for acquisition in 37.5% of BC patients during progression and/or treatment. | [203] |
| Microevolution of genomic rearrangements | Genomic disparity between primary BC and single CTCs detected by NGS; driver mutation-specific rise of CNAs. | [139,204] |
| ESR1mutations acquired | Activating ESR1 mutations in CTCs from MBC patients after endocrine therapy; 85% concordance between key mutations and CNAs in CTCs and metastases. | [161] |
| Increased oxidative stress | Intracellular ROS is elevated and counterbalanced by endogenous antioxidants in CTCs but not primary BC or MBC, which prevents apoptosis and permits metastasis. | [149,167] |
| Potentiated DNA repair confers chemoresistance | γH2AX-marked basal DNA damage is elevated in CTCs versus attached BC cells and partially activates DNA damage responses. Comet assay- and γH2AX-marked DNA damage induced by cytostatics (Epirubicin, Cisplatin) is repaired faster in CTCs vs. attached BC cells irrespective of BC stemness. | [149] |
| γH2AX monitors response to DNA damaging drugs | γH2AX signals accumulate in CTCs from BC and other patients after combined cyclophosphamide and PARP inhibitor treatment (Phase I). | [145,147] |
| ERCC1expression before and after chemotherapy | ERCC1 mRNA expression analyzed by multiplex RT-PCR of separated CTCs shows expression in 60–70% of patients before and after neoadjuvant therapy. | [205] |
| CNAs coupled with DNA repair gene alterations in CTC line | ER+ CTC line from MBC patient with wide spectrum of CNAs carries pathogenic TP53 mutation and predicted deleterious change in ATM. | [155] |
| 53BP1 associates with chemotherapy response | 53BP1 accumulates in CTCs from MBC patients with hormone receptor-positive metastases and in Eribulin-responsive patients. | [111] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heitmeir, B.; Deniz, M.; Janni, W.; Rack, B.; Schochter, F.; Wiesmüller, L. Circulating Tumor Cells in Breast Cancer Patients: A Balancing Act between Stemness, EMT Features and DNA Damage Responses. Cancers 2022, 14, 997. https://doi.org/10.3390/cancers14040997
Heitmeir B, Deniz M, Janni W, Rack B, Schochter F, Wiesmüller L. Circulating Tumor Cells in Breast Cancer Patients: A Balancing Act between Stemness, EMT Features and DNA Damage Responses. Cancers. 2022; 14(4):997. https://doi.org/10.3390/cancers14040997
Chicago/Turabian StyleHeitmeir, Benedikt, Miriam Deniz, Wolfgang Janni, Brigitte Rack, Fabienne Schochter, and Lisa Wiesmüller. 2022. "Circulating Tumor Cells in Breast Cancer Patients: A Balancing Act between Stemness, EMT Features and DNA Damage Responses" Cancers 14, no. 4: 997. https://doi.org/10.3390/cancers14040997
APA StyleHeitmeir, B., Deniz, M., Janni, W., Rack, B., Schochter, F., & Wiesmüller, L. (2022). Circulating Tumor Cells in Breast Cancer Patients: A Balancing Act between Stemness, EMT Features and DNA Damage Responses. Cancers, 14(4), 997. https://doi.org/10.3390/cancers14040997







