Computational Analyses of YY1 and Its Target RKIP Reveal Their Diagnostic and Prognostic Roles in Lung Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chromatin Binding Prediction and Chromatin-Immunoprecipitation Sequencing (ChIP-Seq) Data Analysis
2.2. Dataset Repositories
2.3. Statistical Analyses
3. Results
3.1. The RKIP Gene Expression Is Directly Repressed by the YY1 Transcription Factor
3.2. TCGA Lung Cancer Datasets Analyses Disclose Diagnostic and Prognostic Roles for Both YY1 and RKIP
3.3. Lung Cancer GEO Dataset Analyses Confirm Both the Diagnostic and Prognostic Functions of YY1 and RKIP
3.4. NSCLC Mixed Dataset Analyses Suggest That YY1 and RKIP Expression Assessments Have a Robust PreDIctive Power
3.5. Single-Cell RNA-Seq Lung Cancer Dataset Analyses Reveal That Both YY1 and RKIP Gene Expressions Are Cell Type-Dependent
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Pikor, L.A.; Ramnarine, V.R.; Lam, S.; Lam, W.L. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer 2013, 82, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Bade, B.C.; Dela Cruz, C.S. Lung Cancer 2020. Clin. Chest Med. 2020, 41, 1–24. [Google Scholar] [CrossRef]
- Rapisarda, V.; Salemi, R.; Marconi, A.; Loreto, C.; Graziano, A.C.; Cardile, V.; Basile, M.S.; Candido, S.; Falzone, L.; Spandidos, D.A.; et al. Fluoro-edenite induces fibulin-3 overexpression in non-malignant human mesothelial cells. Oncol. Lett. 2016, 12, 3363–3367. [Google Scholar] [CrossRef] [PubMed][Green Version]
- National Lung Screening Trial Research Team. Lung Cancer Incidence and Mortality with Extended Follow-up in the National Lung Screening Trial. J. Thorac. Oncol. 2019, 14, 1732–1742. [Google Scholar] [CrossRef]
- Pastorino, U.; Silva, M.; Sestini, S.; Sabia, F.; Boeri, M.; Cantarutti, A.; Sverzellati, N.; Sozzi, G.; Corrao, G.; Marchianò, A. Prolonged lung cancer screening reduced 10-year mortality in the MILD trial: New confirmation of lung cancer screening efficacy. Ann. Oncol. 2019, 30, 1162–1169. [Google Scholar] [CrossRef]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.-W.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef]
- Indini, A.; Rijavec, E.; Bareggi, C.; Grossi, F. Novel treatment strategies for early-stage lung cancer: The oncologist’s perspective. J. Thorac. Dis. 2020, 12, 3390–3398. [Google Scholar] [CrossRef]
- Alexander, M.; Kim, S.Y.; Cheng, H. Update 2020: Management of Non-Small Cell Lung Cancer. Lung 2020, 198, 897–907. [Google Scholar] [CrossRef]
- Middleton, G.; Fletcher, P.; Popat, S.; Savage, J.; Summers, Y.; Greystoke, A.; Gilligan, D.; Cave, J.; O’Rourke, N.; Brewster, A.; et al. The National Lung Matrix Trial of personalized therapy in lung cancer. Nature 2020, 583, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Mustachio, L.; Roszik, J. Current Targeted Therapies for the Fight against Non-Small Cell Lung Cancer. Pharmaceuticals 2020, 13, 374. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Reckamp, K.L. Immunotherapy in Advanced Non-Small Cell Lung Cancer. Semin. Respir. Crit. Care Med. 2020, 41, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Qiao, M.; Zhou, C. The cutting-edge progress of immune-checkpoint blockade in lung cancer. Cell. Mol. Immunol. 2021, 18, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Xiong, S.; Li, C.; Liang, H.; Zhao, Y.; Li, J.; Shi, J.; Ou, L.; Chen, Z.; Liang, P.; et al. An annual review of the remarkable advances in lung cancer clinical research in 2019. J. Thorac. Dis. 2020, 12, 1056–1069. [Google Scholar] [CrossRef] [PubMed]
- Christofi, T.; Baritaki, S.; Falzone, L.; Libra, M.; Zaravinos, A. Current Perspectives in Cancer Immunotherapy. Cancers 2019, 11, 1472. [Google Scholar] [CrossRef]
- Tafe, L.J. Non–Small Cell Lung Cancer as a Precision Oncology Paradigm: Emerging Targets and Tumor Mutational Burden (TMB). Adv. Anat. Pathol. 2020, 27, 3–10. [Google Scholar] [CrossRef]
- Chen, R.; Manochakian, R.; James, L.; Azzouqa, A.-G.; Shi, H.; Zhang, Y.; Zhao, Y.; Zhou, K.; Lou, Y. Emerging therapeutic agents for advanced non-small cell lung cancer. J. Hematol. Oncol. 2020, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Aramini, B.; Masciale, V.; Banchelli, F.; D’Amico, R.; Dominici, M.; Husnain Haider, K. Precision Medicine in Lung Cancer: Challenges and Opportunities in Diagnostic and Therapeutic Purposes. In Lung Cancer—Modern Multidisciplinary Management; IntechOpen: London, UK, 2021. [Google Scholar]
- Ostrin, E.J.; Sidransky, D.; Spira, A.; Hanash, S.M. Biomarkers for Lung Cancer Screening and Detection. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2411–2415. [Google Scholar] [CrossRef] [PubMed]
- Kan, C.F.K.; Unis, G.D.; Li, L.Z.; Gunn, S.; Li, L.; Soyer, H.P.; Stark, M.S. Circulating Biomarkers for Early Stage Non-Small Cell Lung Carcinoma Detection: Supplementation to Low-Dose Computed Tomography. Front. Oncol. 2021, 11, 555331. [Google Scholar] [CrossRef]
- Seijo, L.M.; Peled, N.; Ajona, D.; Boeri, M.; Field, J.K.; Sozzi, G.; Pio, R.; Zulueta, J.J.; Spira, A.; Massion, P.P.; et al. Biomarkers in Lung Cancer Screening: Achievements, Promises, and Challenges. J. Thorac. Oncol. 2019, 14, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Akopyan, G.; Garban, H.; Bonavida, B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene 2006, 25, 1125–1142. [Google Scholar] [CrossRef] [PubMed]
- Meliala, I.T.S.; Hosea, R.; Kasim, V.; Wu, S. The biological implications of Yin Yang 1 in the hallmarks of cancer. Theranostics 2020, 4, 4183–4200. [Google Scholar] [CrossRef] [PubMed]
- Sarvagalla, S.; Kolapalli, S.P.; Vallabhapurapu, S. The Two Sides of YY1 in Cancer: A Friend and a Foe. Front. Oncol. 2019, 9, 1230. [Google Scholar] [CrossRef] [PubMed]
- Vivarelli, S.; Falzone, L.; Candido, S.; Bonavida, B.; Libra, M. YY1 Silencing Induces 5-Fluorouracil-Resistance and BCL2L15 Downregulation in Colorectal Cancer Cells: Diagnostic and Prognostic Relevance. Int. J. Mol. Sci. 2021, 22, 8481. [Google Scholar] [CrossRef]
- Vivarelli, S.; Falzone, L.; Ligresti, G.; Candido, S.; Garozzo, A.; Magro, G.G.; Bonavida, B.; Libra, M. Role of the Transcription Factor Yin Yang 1 and Its Selectively Identified Target Survivin in High-Grade B-Cells non-Hodgkin Lymphomas: Potential Diagnostic and Therapeutic Targets. Int. J. Mol. Sci. 2020, 21, 6446. [Google Scholar] [CrossRef]
- Huang, T.; Wang, G.; Yang, L.; Peng, B.; Wen, Y.; Ding, G.; Wang, Z. Transcription Factor YY1 Modulates Lung Cancer Progression by Activating lncRNA-PVT1. DNA Cell Biol. 2017, 36, 947–958. [Google Scholar] [CrossRef]
- Landry-Truchon, K.; Houde, N.; Lhuillier, M.; Charron, L.; Hadchouel, A.; Delacourt, C.; Foulkes, W.D.; Galmiche-Rolland, L.; Jeannotte, L. Deletion of Yy1 in mouse lung epithelium unveils molecular mechanisms governing pleuropulmonary blastoma pathogenesis. Dis. Models Mech. 2020, 13, dmm045989. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, H.; Zhang, J.; Shan, L.; Yibureyimu, B.; Nurlan, A.; Aerxiding, P.; Luo, Q. MiR-142-5p Suppresses Lung Cancer Cell Metastasis by Targeting Yin Yang 1 to Regulate Epithelial–Mesenchymal Transition. Cell. Reprogram. 2020, 22, 328–336. [Google Scholar] [CrossRef]
- Xu, P.; Xiao, H.; Yang, Q.; Hu, R.; Jiang, L.; Bi, R.; Jiang, X.; Wang, L.; Mei, J.; Ding, F.; et al. The USP21/YY1/SNHG16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer. Exp. Mol. Med. 2020, 52, 41–55. [Google Scholar] [CrossRef]
- Jiang, W.; Zhang, C.; Kang, Y.; Yu, X.; Pang, P.; Li, G.; Feng, Y. MRPL42 is activated by YY1 to promote lung adenocarcinoma progression. J. Cancer 2021, 12, 2403–2411. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-C.; Kuo, I.-Y.; Wu, L.-T.; Kuan, W.-H.; Liao, S.-Y.; Jen, J.; Yang, Y.-E.; Tang, C.-W.; Chen, Y.-R.; Wang, Y.-C. Dysregulated Kras/YY1/ZNF322A/Shh transcriptional axis enhances neo-angiogenesis to promote lung cancer progression. Theranostics 2020, 10, 10001–10015. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhou, W.; Zhang, Y. Transcription factor YY1 enhances the stemness of lung cancer cells by stabilizing hypoxia factor HIF-1α under a hypoxic microenvironment. Environ. Toxicol. 2021, 36, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Bonavida, B. RKIP: A Pivotal Gene Product in the Pathogenesis of Cancer. Cancers 2021, 13, 2488. [Google Scholar] [CrossRef]
- Lamiman, K.; Keller, J.M.; Mizokami, A.; Zhang, J.; Keller, E.T. Survey of Raf Kinase Inhibitor Protein (RKIP) in Multiple Cancer Types. Crit. Rev. Oncog. 2014, 19, 455–468. [Google Scholar] [CrossRef]
- Xie, S.-Y.; Li, G.; Han, C.; Yu, Y.-Y.; Li, N. RKIP reduction enhances radioresistance by activating the Shh signaling pathway in non-small-cell lung cancer. Onco Targets Ther. 2017, 10, 5605–5619. [Google Scholar] [CrossRef]
- Wang, A.; Duan, G.; Zhao, C.; Gao, Y.; Liu, X.; Wang, Z.; Li, W.; Wang, K.; Wang, W. Reduced RKIP expression levels are associated with frequent non-small cell lung cancer metastasis and STAT3 phosphorylation and activation. Oncol. Lett. 2017, 13, 3039–3045. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, X.; Wu, T.; Li, G.; Shi, Y. Clinical significance of RKIP mRNA expression in non-small cell lung cancer. Tumor Biol. 2014, 35, 4377–4380. [Google Scholar] [CrossRef]
- Yan, H. Reduction of Raf Kinase Inhibitor Protein Expression is Associated with Lymph Node Metastasis in Resectable Non-small Cell Lung Cancer. Open Respir. Med. J. 2012, 6, 135–138. [Google Scholar] [CrossRef]
- Huerta-Yepez, S.; Yoon, N.K.; Hernandez-Cueto, A.; Mah, V.; Rivera-Pazos, C.M.; Chatterjee, D.; Vega, M.I.; Maresh, E.L.; Horvath, S.; Chia, D.; et al. Expression of phosphorylated raf kinase inhibitor protein (pRKIP) is a predictor of lung cancer survival. BMC Cancer 2011, 11, 259. [Google Scholar] [CrossRef]
- Zhu, C.; Wang, Q.; Xie, J.; Shi, J.; Zhou, X.; Li, D.; Xiong, F.; Zhang, L. Expression and Significance of RKIP and E-cadherin in Lung Squamous Cell Carcinoma. Pathol. Oncol. Res. 2013, 19, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Raquel-Cunha, A.; Cardoso-Carneiro, D.; Reis, R.M.; Martinho, O. Current Status of Raf Kinase Inhibitor Protein (RKIP) in Lung Cancer: Behind RTK Signaling. Cells 2019, 8, 442. [Google Scholar] [CrossRef]
- Bonavida, B. RKIP-Mediated Chemo-Immunosensitization of Resistant Cancer Cells via Disruption of the NF-κB/Snail/YY1/RKIP Resistance-Driver Loop. Crit. Rev. Oncog. 2014, 19, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Stormo, G.D. Modeling the specificity of protein-DNA interactions. Quant. Biol. 2013, 1, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Navarro Gonzalez, J.; Zweig, A.S.; Speir, M.L.; Schmelter, D.; Rosenbloom, K.R.; Raney, B.J.; Powell, C.C.; Nassar, L.R.; Maulding, N.D.; Lee, C.M.; et al. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021, 49, D1046–D1057. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Koster, J. R2: Genomics Analysis and Visualization Platform. Available online: http://r2.amc.nl (accessed on 11 October 2021).
- Bild, A.H.; Yao, G.; Chang, J.T.; Wang, Q.; Potti, A.; Chasse, D.; Joshi, M.-B.; Harpole, D.; Lancaster, J.M.; Berchuck, A.; et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006, 439, 353–357. [Google Scholar] [CrossRef]
- Tarca, A.L.; Lauria, M.; Unger, M.; Bilal, E.; Boue, S.; Kumar Dey, K.; Hoeng, J.; Koeppl, H.; Martin, F.; Meyer, P.; et al. Strengths and limitations of microarray-based phenotype prediction: Lessons learned from the IMPROVER Diagnostic Signature Challenge. Bioinformatics 2013, 29, 2892–2899. [Google Scholar] [CrossRef]
- Landi, M.T.; Dracheva, T.; Rotunno, M.; Figueroa, J.D.; Liu, H.; Dasgupta, A.; Mann, F.E.; Fukuoka, J.; Hames, M.; Bergen, A.W.; et al. Gene Expression Signature of Cigarette Smoking and Its Role in Lung Adenocarcinoma Development and Survival. PLoS ONE 2008, 3, e1651. [Google Scholar] [CrossRef]
- Hou, J.; Aerts, J.; den Hamer, B.; van IJcken, W.; den Bakker, M.; Riegman, P.; van der Leest, C.; van der Spek, P.; Foekens, J.A.; Hoogsteden, H.C.; et al. Gene Expression-Based Classification of Non-Small Cell Lung Carcinomas and Survival Prediction. PLoS ONE 2010, 5, e10312. [Google Scholar] [CrossRef]
- Sanchez-Palencia, A.; Gomez-Morales, M.; Gomez-Capilla, J.A.; Pedraza, V.; Boyero, L.; Rosell, R.; Fárez-Vidal, M.E. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int. J. Cancer 2011, 129, 355–364. [Google Scholar] [CrossRef]
- Chen, F.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019, 10, 5679. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- EMBL-EBI. Single Cell Expression Atlas. Available online: https://www.ebi.ac.uk/gxa/sc/home (accessed on 24 September 2020).
- KOBIC. User-Friendly Interface Tool to Explore Cell Atlas (URECA). Available online: http://ureca-singlecell.kr/ (accessed on 24 September 2020).
- Single Cell Portal Broad Institute. Available online: https://singlecell.broadinstitute.org/single_cell?order=popular (accessed on 28 June 2021).
- Kim, N.; Kim, H.K.; Lee, K.; Hong, Y.; Cho, J.H.; Choi, J.W.; Lee, J.-I.; Suh, Y.-L.; Ku, B.M.; Eum, H.H.; et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020, 11, 2285. [Google Scholar] [CrossRef]
- Lambrechts, D.; Wauters, E.; Boeckx, B.; Aibar, S.; Nittner, D.; Burton, O.; Bassez, A.; Decaluwé, H.; Pircher, A.; Van den Eynde, K.; et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018, 24, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Goveia, J.; Rohlenova, K.; Taverna, F.; Treps, L.; Conradi, L.-C.; Pircher, A.; Geldhof, V.; de Rooij, L.P.M.H.; Kalucka, J.; Sokol, L.; et al. An Integrated Gene Expression Landscape Profiling Approach to Identify Lung Tumor Endothelial Cell Heterogeneity and Angiogenic Candidates. Cancer Cell 2020, 37, 21–36.e13. [Google Scholar] [CrossRef] [PubMed]
- Kinker, G.S.; Greenwald, A.C.; Tal, R.; Orlova, Z.; Cuoco, M.S.; McFarland, J.M.; Warren, A.; Rodman, C.; Roth, J.A.; Bender, S.A.; et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 2020, 52, 1208–1218. [Google Scholar] [CrossRef]
- Marjanovic, N.D.; Hofree, M.; Chan, J.E.; Canner, D.; Wu, K.; Trakala, M.; Hartmann, G.G.; Smith, O.C.; Kim, J.Y.; Evans, K.V.; et al. Emergence of a High-Plasticity Cell State during Lung Cancer Evolution. Cancer Cell 2020, 38, 229–246.e13. [Google Scholar] [CrossRef]
- Montoro, D.T.; Haber, A.L.; Biton, M.; Vinarsky, V.; Lin, B.; Birket, S.E.; Yuan, F.; Chen, S.; Leung, H.M.; Villoria, J.; et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018, 560, 319–324. [Google Scholar] [CrossRef]
- Pennell, N.A.; Arcila, M.E.; Gandara, D.R.; West, H. Biomarker Testing for Patients With Advanced Non–Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am. Soc. Clin. Oncol. Educ. B. 2019, 39, 531–542. [Google Scholar] [CrossRef]
- Fang, R.; Zhu, Y.; Khadka, V.S.; Zhang, F.; Jiang, B.; Deng, Y. The Evaluation of Serum Biomarkers for Non-small Cell Lung Cancer (NSCLC) Diagnosis. Front. Physiol. 2018, 9, 1710. [Google Scholar] [CrossRef] [PubMed]
- Calvayrac, O.; Pradines, A.; Pons, E.; Mazières, J.; Guibert, N. Molecular biomarkers for lung adenocarcinoma. Eur. Respir. J. 2017, 49, 1601734. [Google Scholar] [CrossRef] [PubMed]
- Wottrich, S.; Kaufhold, S.; Chrysos, E.; Zoras, O.; Baritaki, S.; Bonavida, B. Inverse correlation between the metastasis suppressor RKIP and the metastasis inducer YY1: Contrasting roles in the regulation of chemo/immuno-resistance in cancer. Drug Resist. Updates 2017, 30, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Bonavida, B.; Baritaki, S. The Novel Role of Yin Yang 1 in the Regulation of Epithelial to Mesenchymal Transition in Cancer Via the Dysregulated NF-κB/Snail/YY1/RKIP/PTEN Circuitry. Crit. Rev. Oncog. 2011, 16, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Yeung, K.C.; Rose, D.W.; Dhillon, A.S.; Yaros, D.; Gustafsson, M.; Chatterjee, D.; McFerran, B.; Wyche, J.; Kolch, W.; Sedivy, J.M. Raf Kinase Inhibitor Protein Interacts with NF-κB-Inducing Kinase and TAK1 and Inhibits NF-κB Activation. Mol. Cell. Biol. 2001, 21, 7207–7217. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.B.; Majumder, P.; Cooper, J.C.; Yoon, H.; Wade, P.A.; Boss, J.M. Yin Yang 1 Regulates the Expression of Snail through a Distal Enhancer. Mol. Cancer Res. 2009, 7, 221–229. [Google Scholar] [CrossRef]
- Zaravinos, A.; Bonavida, B.; Chatzaki, E.; Baritaki, S. RKIP: A Key Regulator in Tumor Metastasis Initiation and Resistance to Apoptosis: Therapeutic Targeting and Impact. Cancers 2018, 10, 287. [Google Scholar] [CrossRef]
- Beach, S.; Tang, H.; Park, S.; Dhillon, A.S.; Keller, E.T.; Kolch, W.; Yeung, K.C. Snail is a repressor of RKIP transcription in metastatic prostate cancer cells. Oncogene 2008, 27, 2243–2248. [Google Scholar] [CrossRef]
- Shvartsur, A.; Givechian, K.B.; Garban, H.; Bonavida, B. Overexpression of RKIP and its cross-talk with several regulatory gene products in multiple myeloma. J. Exp. Clin. Cancer Res. 2017, 36, 62. [Google Scholar] [CrossRef]
- Notarbartolo, M.; Giannitrapani, L.; Vivona, N.; Poma, P.; Labbozzetta, M.; Florena, A.M.; Porcasi, R.; Muggeo, V.M.R.; Sandonato, L.; Cervello, M.; et al. Frequent Alteration of the Yin Yang 1/Raf-1 Kinase Inhibitory Protein Ratio in Hepatocellular Carcinoma. Omi. J. Integr. Biol. 2011, 15, 267–272. [Google Scholar] [CrossRef]
- HPA. The Human Protein Atlas YY1 Pathology Lung Cancer. Available online: https://www.proteinatlas.org/ENSG00000100811-YY1/pathology/lung+cancer (accessed on 3 January 2022).
- HPA. The Human Protein Atlas PEBP1 Pathology Lung Cancer. Available online: https://www.proteinatlas.org/ENSG00000089220-PEBP1/pathology/lung+cancer (accessed on 3 January 2022).
- Özenver, N.; Efferth, T. Therapeutic targeting of SNAIL, RKIP, and YY1 in tumor metastasis and drug resistance. In Prognostic and Therapeutic Applications of RKIP in Cancer; Elsevier: Amsterdam, The Netherlands, 2020; pp. 357–387. [Google Scholar]



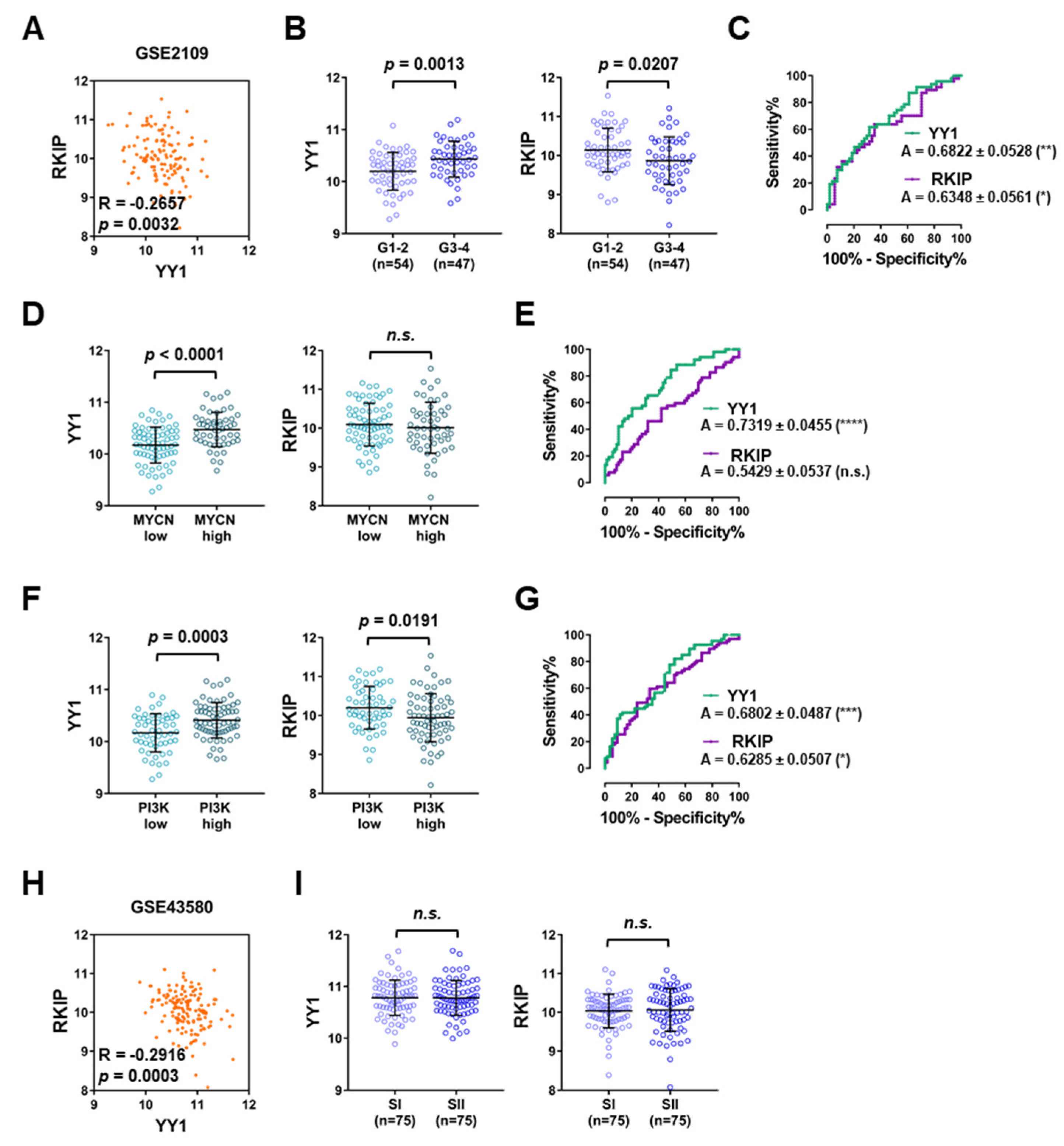
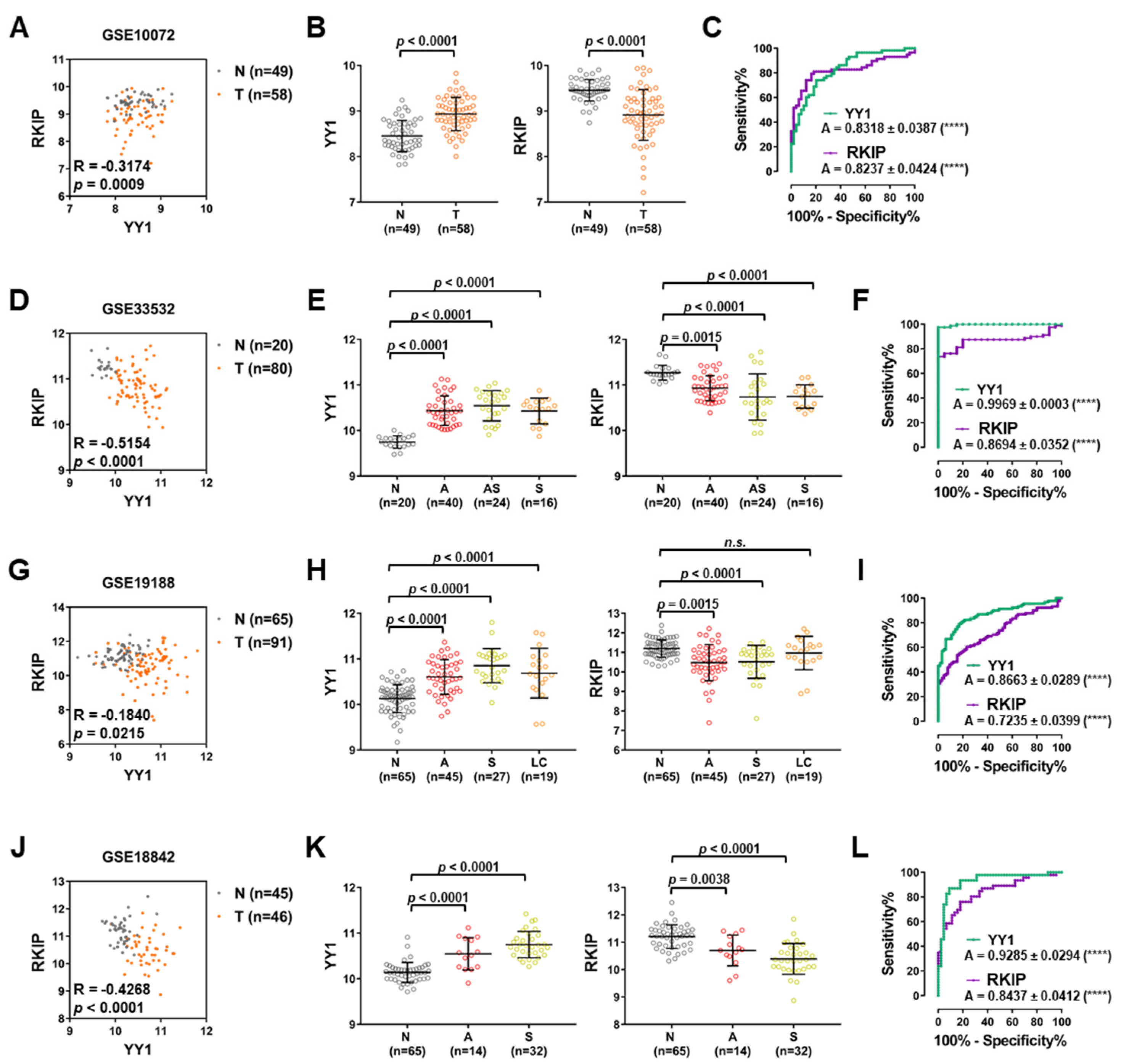


| GEO ID | Contributors | Platform | Tumor Samples | Matching Non-Tumor Samples | Pearson Correlation | p-Value | Reference |
|---|---|---|---|---|---|---|---|
| GSE3141 | Bild AH et al. | Affymetrix HG-U133 2.0 | 114 | 0 | −0.2054 | 0.0283 | [49] |
| GSE2109 | n.a. | Affymetrix HG-U133 2.0 | 121 | 0 | −0.2657 | 0.0032 | n.a. |
| GSE43580 | Peitsch MC et al. | Affymetrix HG-U133 2.0 | 150 | 0 | −0.2916 | 0.0003 | [50] |
| GSE10072 | Jen J et al. | Affymetrix HG-U133A | 58 | 49 | −0.3174 | 0.0009 | [51] |
| GSE33532 | Muley T et al. | Affymetrix HG-U133 2.0 | 80 | 20 | −0.5154 | <0.0001 | n.a. |
| GSE19188 | Hou J et al. | Affymetrix HG-U133 2.0 | 91 | 65 | −0.1840 | 0.0215 | [52] |
| GSE18842 | Farez-Vidal ME et al. | Affymetrix HG-U133 2.0 | 46 | 45 | −0.4268 | <0.0001 | [53] |
| Study ID | Technology | Number of Cells | Analysis Portal | Reference |
|---|---|---|---|---|
| SCP542 | Droplet-based scRNA-seq (H sapiens) | 53,513 | Single cell Portal (Broad Institute) | [62] |
| GSE152607 | Illumina NextSeq 500 (M musculus) | 3891 | Single cell Portal (Broad Institute) | [63] |
| GSE103354 | Illumina NextSeq 500 (M musculus) | 7193 | Single cell Portal (Broad Institute) | [64] |
| GSE131907 | Illumina HiSeq 2500 (H sapiens) | 45,149; 42,995 | URECA (Kobic Center) | [59] |
| E-MTAB-6653 | Droplet-based scRNA-seq (H sapiens) | 33,208 | Single cell expression atlas (EMBL-EBI) | [60] |
| E-MTAB-6308 | Droplet-based scRNA-seq (H sapiens) | 56,771 | Single cell expression atlas (EMBL-EBI) | [61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vivarelli, S.; Falzone, L.; Grillo, C.M.; Bonavida, B.; Crimi, C.; La Mantia, I.; Libra, M. Computational Analyses of YY1 and Its Target RKIP Reveal Their Diagnostic and Prognostic Roles in Lung Cancer. Cancers 2022, 14, 922. https://doi.org/10.3390/cancers14040922
Vivarelli S, Falzone L, Grillo CM, Bonavida B, Crimi C, La Mantia I, Libra M. Computational Analyses of YY1 and Its Target RKIP Reveal Their Diagnostic and Prognostic Roles in Lung Cancer. Cancers. 2022; 14(4):922. https://doi.org/10.3390/cancers14040922
Chicago/Turabian StyleVivarelli, Silvia, Luca Falzone, Caterina Maria Grillo, Benjamin Bonavida, Claudia Crimi, Ignazio La Mantia, and Massimo Libra. 2022. "Computational Analyses of YY1 and Its Target RKIP Reveal Their Diagnostic and Prognostic Roles in Lung Cancer" Cancers 14, no. 4: 922. https://doi.org/10.3390/cancers14040922
APA StyleVivarelli, S., Falzone, L., Grillo, C. M., Bonavida, B., Crimi, C., La Mantia, I., & Libra, M. (2022). Computational Analyses of YY1 and Its Target RKIP Reveal Their Diagnostic and Prognostic Roles in Lung Cancer. Cancers, 14(4), 922. https://doi.org/10.3390/cancers14040922












