A Negative Regulatory Role for RKIP in Breast Cancer Immune Response
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Cell Lines and Reagents
2.2. Microarray
2.3. Analysis of Tumor Datasets
2.4. Quantitative Real-Time Reverse Transcriptase–Polymerase Chain Reaction (qRT-PCR)
3. Results
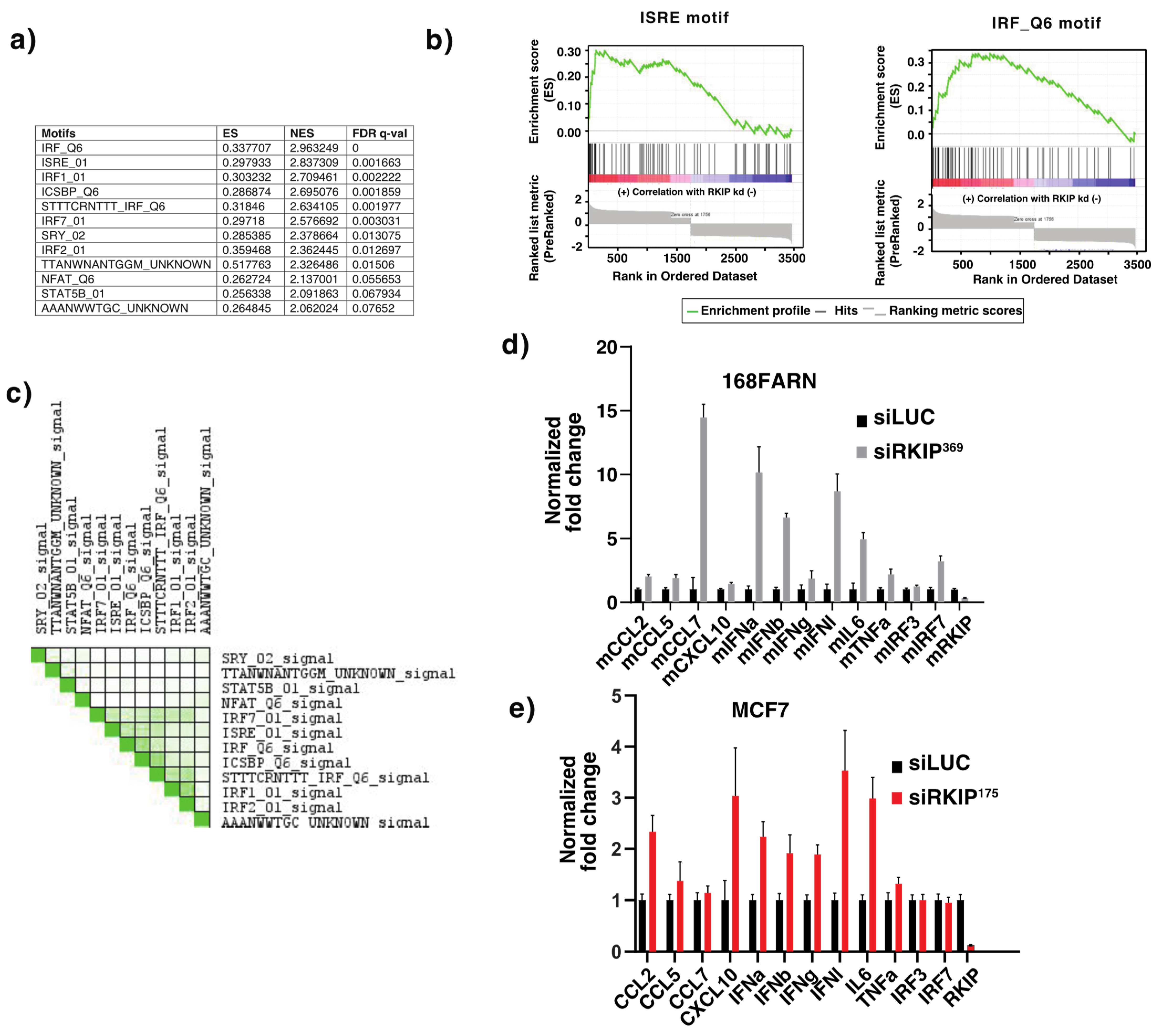
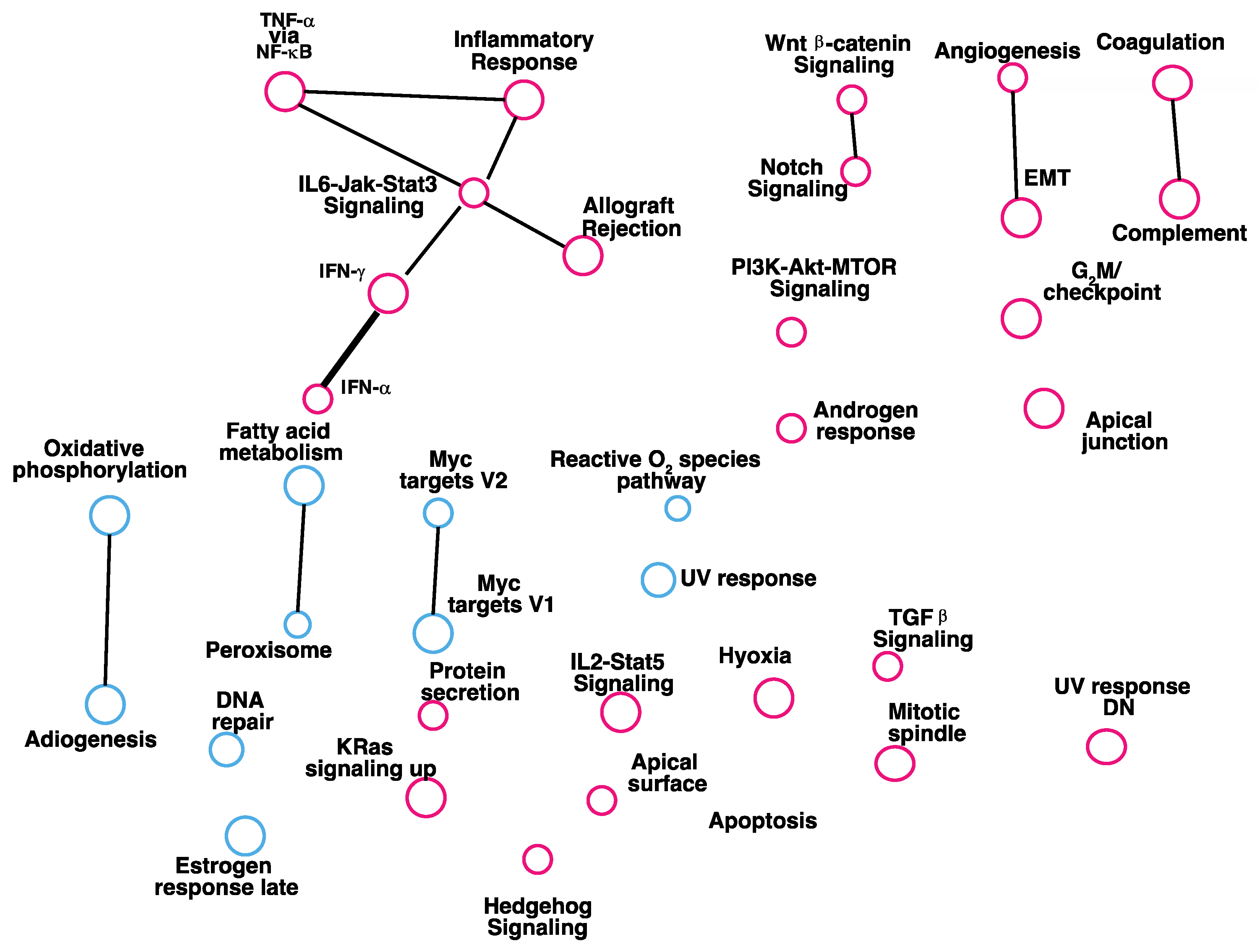
3.1. Gene Set Enrichment Analysis of Genes Affected by Reduced Expression of RKIP
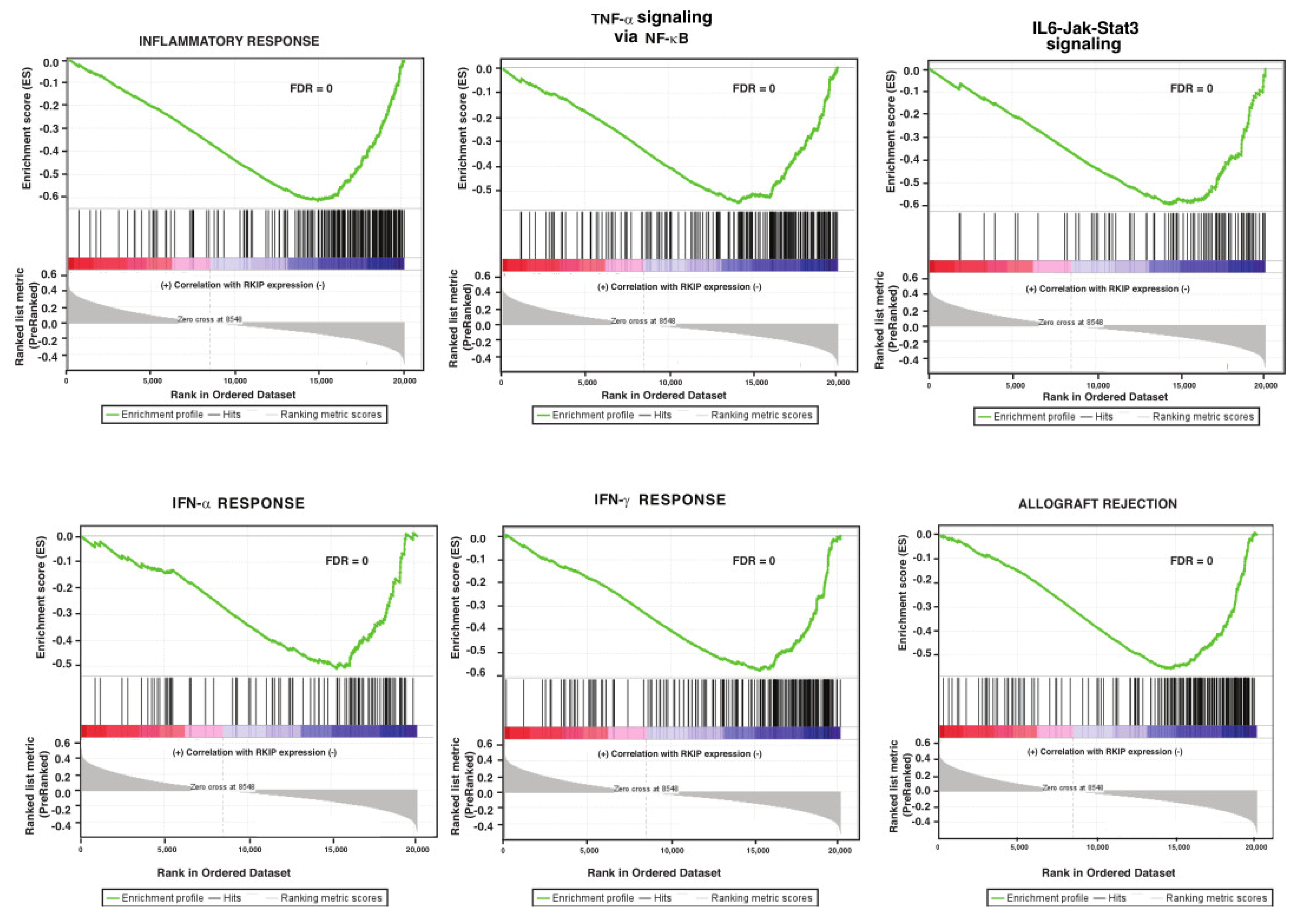
3.2. Gene Set Enrichment Analysis of Genes Correlate with Expression of RKIP in Human Breast Tumors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yeung, K.; Seitz, T.; Li, S.; Janosch, P.; McFerran, B.; Kaiser, C.; Fee, F.; Katsanakis, K.D.; Rose, D.W.; Mischak, H.; et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 1999, 401, 173–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolch, W. Meaningful relationships: The regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000, 351 Pt 2, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, H.J.; Weber, M.J. Mitogen-activated protein kinases: Specific messages from ubiquitous messengers. Mol. Cell Biol. 1999, 19, 2435–2444. [Google Scholar] [CrossRef] [Green Version]
- Schoentgen, F.; Jolles, P. From structure to function: Possible biological roles of a new widespread protein family binding hydrophobic ligands and displaying a nucleotide binding site. FEBS Lett. 1995, 369, 22–26. [Google Scholar] [CrossRef] [Green Version]
- Beshir, A.B.; Ren, G.; Magpusao, A.N.; Barone, L.M.; Yeung, K.C.; Fenteany, G. Raf kinase inhibitor protein suppresses nuclear factor-kappaB-dependent cancer cell invasion through negative regulation of matrix metalloproteinase expression. Cancer Lett. 2010, 299, 137–149. [Google Scholar] [CrossRef] [Green Version]
- Escara-Wilke, J.; Keller, J.M.; Ignatoski, K.M.; Dai, J.; Shelley, G.; Mizokami, A.; Zhang, J.; Yeung, M.L.; Yeung, K.C.; Keller, E.T. Raf kinase inhibitor protein (RKIP) deficiency decreases latency of tumorigenesis and increases metastasis in a murine genetic model of prostate cancer. Prostate 2015, 75, 292–302. [Google Scholar] [CrossRef] [Green Version]
- Dangi-Garimella, S.; Yun, J.; Eves, E.M.; Newman, M.; Erkeland, S.J.; Hammond, S.M.; Minn, A.J.; Rosner, M.R. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009, 28, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Yun, J.; Frankenberger, C.A.; Kuo, W.L.; Boelens, M.C.; Eves, E.M.; Cheng, N.; Liang, H.; Li, W.H.; Ishwaran, H.; Minn, A.J.; et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011, 30, 4500–4514. [Google Scholar] [CrossRef]
- Datar, I.; Feng, J.; Qiu, X.; Lewandowski, J.; Yeung, M.; Ren, G.; Aras, S.; Al-Mulla, F.; Cui, H.; Trumbly, R.; et al. RKIP Inhibits Local Breast Cancer Invasion by Antagonizing the Transcriptional Activation of MMP13. PLoS ONE 2015, 10, e0134494. [Google Scholar] [CrossRef]
- Granovsky, A.E.; Clark, M.C.; McElheny, D.; Heil, G.; Hong, J.; Liu, X.; Kim, Y.; Joachimiak, G.; Joachimiak, A.; Koide, S.; et al. Raf kinase inhibitory protein function is regulated via a flexible pocket and novel phosphorylation-dependent mechanism. Mol. Cell Biol. 2009, 29, 1306–1320. [Google Scholar] [CrossRef] [Green Version]
- Wenzel, S.E.; Tyurina, Y.Y.; Zhao, J.; St Croix, C.M.; Dar, H.H.; Mao, G.; Tyurin, V.A.; Anthonymuthu, T.S.; Kapralov, A.A.; Amoscato, A.A.; et al. PEBP1 Wardens Ferroptosis by Enabling Lipoxygenase Generation of Lipid Death Signals. Cell 2017, 171, 628–641.e26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenz, K.; Lohse, M.J.; Quitterer, U. Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature 2003, 426, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Al-Mulla, F.; Bitar, M.S.; Feng, J.; Park, S.; Yeung, K.C. A new model for raf kinase inhibitory protein induced chemotherapeutic resistance. PLoS ONE 2012, 7, e29532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Mulla, F.; Bitar, M.S.; Al-Maghrebi, M.; Behbehani, A.I.; Al-Ali, W.; Rath, O.; Doyle, B.; Tan, K.Y.; Pitt, A.; Kolch, W. Raf kinase inhibitor protein RKIP enhances signaling by glycogen synthase kinase-3beta. Cancer Res. 2011, 71, 1334–1343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, D.; Sabo, E.; Tavares, R.; Resnick, M.B. Inverse association between Raf Kinase Inhibitory Protein and signal transducers and activators of transcription 3 expression in gastric adenocarcinoma patients: Implications for clinical outcome. Clin. Cancer Res. 2008, 14, 2994–3001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, D.; Sabo, E.; Resnick, M.; Yeung, K.; Chin, Y. The RKIP and STAT3 Axis in Cancer Chemotherapy: Opposites Attract. In Sensitization of Cancer Cells for Chemo/Immuno/Radio-Therapy; Bonavida, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 159–174. [Google Scholar]
- Al-Mulla, F.; Bitar, M.S.; Taqi, Z.; Yeung, K.C. RKIP: Much more than Raf kinase inhibitory protein. J. Cell Physiol. 2013, 228, 1688–1702. [Google Scholar] [CrossRef] [PubMed]
- Eves, E.M.; Shapiro, P.; Naik, K.; Klein, U.R.; Trakul, N.; Rosner, M.R. Raf kinase inhibitory protein regulates aurora B kinase and the spindle checkpoint. Mol. Cell 2006, 23, 561–574. [Google Scholar] [CrossRef] [Green Version]
- Gu, M.; Liu, Z.; Lai, R.; Liu, S.; Lin, W.; Ouyang, C.; Ye, S.; Huang, H.; Wang, X. RKIP and TBK1 form a positive feedback loop to promote type I interferon production in innate immunity. EMBO J. 2016, 35, 2553–2565. [Google Scholar] [CrossRef]
- Lai, R.; Gu, M.; Jiang, W.; Lin, W.; Xu, P.; Liu, Z.; Huang, H.; An, H.; Wang, X. Raf Kinase Inhibitor Protein Preferentially Promotes TLR3-Triggered Signaling and Inflammation. J. Immunol. 2017, 198, 4086–4095. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Su, F.; Gautam, R.; Wang, N.; Zhang, Y.; Wang, X. Raf kinase inhibitor protein negatively regulates FcepsilonRI-mediated mast cell activation and allergic response. Proc. Natl. Acad. Sci. USA 2018, 115, E9859–E9868. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Ma, C.; Su, F.; Jiang, Y.; Lai, R.; Zhang, T.; Sun, K.; Fan, L.; Cai, Z.; Li, Z.; et al. Raf kinase inhibitor protein mediates intestinal epithelial cell apoptosis and promotes IBDs in humans and mice. Gut 2017, 66, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Liu, H.; Shou, J.; Jiang, Y.; Yu, H.; Wang, X. The inhibitor effect of RKIP on inflammasome activation and inflammasome-dependent diseases. Cell. Mol. Immunol. 2021, 18, 992–1004. [Google Scholar] [CrossRef] [PubMed]
- Schmid, E.; Neef, S.; Berlin, C.; Tomasovic, A.; Kahlert, K.; Nordbeck, P.; Deiss, K.; Denzinger, S.; Herrmann, S.; Wettwer, E.; et al. Cardiac RKIP induces a beneficial beta-adrenoceptor-dependent positive inotropy. Nat. Med. 2015, 21, 1298–1306. [Google Scholar] [CrossRef]
- Antoun, G.; Bouchard-Cannon, P.; Cheng, H.Y. Regulation of MAPK/ERK signaling and photic entrainment of the suprachiasmatic nucleus circadian clock by Raf kinase inhibitor protein. J. Neurosci. 2012, 32, 4867–4877. [Google Scholar] [CrossRef] [PubMed]
- Zamparo, I.; Francia, S.; Franchi, S.A.; Redolfi, N.; Costanzi, E.; Kerstens, A.; Fukutani, Y.; Battistutta, R.; Polverino de Laureto, P.; Munck, S.; et al. Axonal Odorant Receptors Mediate Axon Targeting. Cell Rep. 2019, 29, 4334–4348.e7. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Netea, M.G.; Schlitzer, A.; Placek, K.; Joosten, L.A.B.; Schultze, J.L. Innate and Adaptive Immune Memory: An Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe 2019, 25, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Bonjardim, C.A.; Ferreira, P.C.; Kroon, E.G. Interferons: Signaling, antiviral and viral evasion. Immunol. Lett. 2009, 122, 1–11. [Google Scholar] [CrossRef]
- Newton, K.; Dixit, V.M. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 2012, 4, a006049. [Google Scholar] [CrossRef]
- Wright, K.T.; Vella, A.T. RKIP contributes to IFN-gamma synthesis by CD8+ T cells after serial TCR triggering in systemic inflammatory response syndrome. J. Immunol. 2013, 191, 708–716. [Google Scholar] [CrossRef] [Green Version]
- Nan, J.; Wang, Y.; Yang, J.; Stark, G.R. IRF9 and unphosphorylated STAT2 cooperate with NF-kappaB to drive IL6 expression. Proc. Natl. Acad. Sci. USA 2018, 115, 3906–3911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Zaravinos, A.; Bonavida, B.; Chatzaki, E.; Baritaki, S. RKIP: A key regulator in tumor metastasis initiation and resistance to apoptosis: Therapeutic targeting and impact. Cancers 2018, 10, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabriela-Freitas, M.; Pinheiro, J.; Raquel-Cunha, A.; Cardoso-Cameiro, D.; Martino, O. RKIP as an inflammatory and immune system modulator: Implication in cancer. Biomolecules 2019, 9, 769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, K.B.; Cousens, L.P.; Doughty, L.A.; Pien, G.C.; Durbin, J.E.; Biron, C.A. Interferon alpha/beta-mediated inhibition and promotion of interferon gamma: STAT1 resolves a paradox. Nat. Immunol. 2000, 1, 70–76. [Google Scholar] [CrossRef]
- Young, H.A.; Hardy, K.J. Role of interferon-gamma in immune cell regulation. J. Leukoc. Biol. 1995, 58, 373–381. [Google Scholar] [CrossRef]
- Boehm, J.S.; Zhao, J.J.; Yao, J.; Kim, S.Y.; Firestein, R.; Dunn, I.F.; Sjostrom, S.K.; Garraway, L.A.; Weremowicz, S.; Richardson, A.L.; et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 2007, 129, 1065–1079. [Google Scholar] [CrossRef] [Green Version]
- Dunn, G.P.; Bruce, A.T.; Sheehan, K.C.; Shankaran, V.; Uppaluri, R.; Bui, J.D.; Diamond, M.S.; Koebel, C.M.; Arthur, C.; White, J.M.; et al. A critical function for type I interferons in cancer immunoediting. Nat. Immunol. 2005, 6, 722–729. [Google Scholar] [CrossRef]
- Dunn, G.P.; Koebel, C.M.; Schreiber, R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006, 6, 836–848. [Google Scholar] [CrossRef]
- Kim, M.K.; Kim, H.W.; Jang, M.; Oh, S.S.; Yong, S.J.; Jeong, Y.; Jung, S.H.; Choi, J.W. LOX family and ZFPM2 as novel diagnostic biomarkers for malignant pleural mesothelioma. Biomark. Res. 2020, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Wischhusen, J.; Waschbisch, A.; Wiendl, H. Immune-refractory cancers and their little helpers—An extended role for immunetolerogenic MHC molecules HLA-G and HLA-E? Semin. Cancer Biol. 2007, 17, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Ogony, J.; Choi, H.J.; Lui, A.; Cristofanilli, M.; Lewis-Wambi, J. Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner. Breast Cancer Res. 2016, 18, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buess, M.; Nuyten, D.S.; Hastie, T.; Nielsen, T.; Pesich, R.; Brown, P.O. Characterization of heterotypic interaction effects in vitro to deconvolute global gene expression profiles in cancer. Genome Biol. 2007, 8, R191. [Google Scholar] [CrossRef] [PubMed] [Green Version]

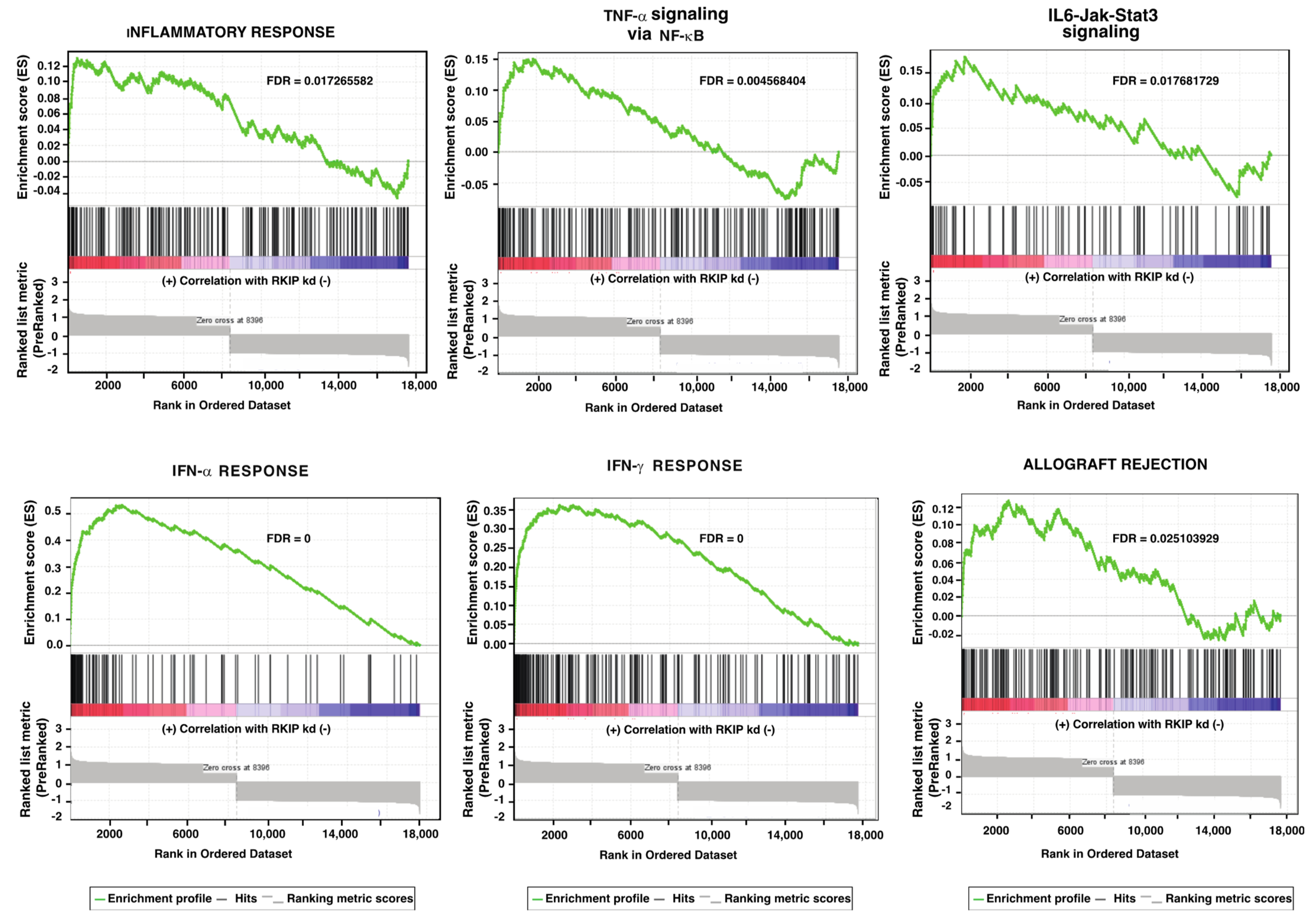
| RKIP kd vs. Control Up | NES | NOM p-Value | FDR q-Value | Negative Correlation RKIP | NES | NOM p-Value | FDR q-Value |
|---|---|---|---|---|---|---|---|
| INTERFERON_ALPHA_RESPONSE | 6.040572 | 0 | 0 | INFLAMMATORY_RESPONSE | −2.80127 | 0 | 0 |
| INTERFERON_GAMMA_RESPONSE | 5.630654 | 0 | 0 | EPITHELIAL_MESENCHYMAL_TRANSITION | −2.6795 | 0 | 0 |
| EPITHELIAL_MESENCHYMAL_TRANSITION | 3.491317 | 0 | 0 | INTERFERON_GAMMA_RESPONSE | −2.62019 | 0 | 0 |
| TNFA_SIGNALING_VIA_NFKB | 2.311845 | 0 | 0.004568 | ALLOGRAFT_REJECTION | −2.52816 | 0 | 0 |
| UV_RESPONSE_DN | 2.304864 | 0.001919 | 0.003655 | TNFA_SIGNALING_VIA_NFKB | −2.49759 | 0 | 0 |
| INFLAMMATORY_RESPONSE | 2.031128 | 0.005556 | 0.017266 | UV_RESPONSE_DN | −2.44412 | 0 | 0 |
| COMPLEMENT | 1.950489 | 0.006085 | 0.022258 | KRAS_SIGNALING_UP | −2.44371 | 0 | 0 |
| ALLOGRAFT_REJECTION | 1.911962 | 0.006061 | 0.025104 | IL6_JAK_STAT3_SIGNALING | −2.39554 | 0 | 0 |
| IL6_JAK_STAT3_SIGNALING | 1.890129 | 0.017682 | 0.025151 | COMPLEMENT | −2.29257 | 0 | 0 |
| MITOTIC_SPINDLE | −2.27098 | 0 | 0 | ||||
| INTERFERON_ALPHA_RESPONSE | −2.11048 | 0 | 0 |
| RKIP kd vs. Control Down | NES | NOM p-Value | FDR q-Value | Positive Correlation RKIP | NES | NOM p-Value | FDR q-Value |
|---|---|---|---|---|---|---|---|
| MYC_TARGETS_V1 | −3.33323 | 0 | 0 | OXIDATIVE_PHOSPHORYLATION | 3.495134 | 0 | 0 |
| E2F_TARGETS | −2.42757 | 0 | 0.002051 | MYC_TARGETS_V1 | 2.596274 | 0 | 0 |
| MYC_TARGETS_V2 | −2.24774 | 0 | 0.00527 | DNA_REPAIR | 2.395833 | 0 | 0 |
| REACTIVE_OXYGEN_SPECIES | −2.09321 | 0.004107 | 0.013857 | MYC_TARGETS_V2 | 2.364108 | 0 | 0 |
| DNA_REPAIR | −1.83964 | 0.008949 | 0.051236 | ADIPOGENESIS | 1.905392 | 0 | 1.45 × 10−4 |
| KRAS_SIGNALING_UP | −1.63499 | 0.036403 | 0.12295 | FATTY_ACID_METABOLISM | 1.816652 | 0 | 4.45 × 10−4 |
| OXIDATIVE_PHOSPHORYLATION | −1.51916 | 0.06993 | 0.18503 | REACTIVE_OXYGEN_SPECIES | 1.696399 | 0 | 0.001005 |
| ESTROGEN_RESPONSE_EARLY | −1.50451 | 0.052863 | 0.1742 | PEROXISOME | 1.475564 | 0 | 0.015246 |
| ESTROGEN_RESPONSE_LATE | 1.38756 | 0 | 0.033137 | ||||
| UV_RESPONSE_UP | 1.293321 | 0.026144 | 0.072677 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bach, V.N.; Ding, J.; Yeung, M.; Conrad, T.; Odeh, H.N.; Cubberly, P.; Figy, C.; Ding, H.-F.; Trumbly, R.; Yeung, K.C. A Negative Regulatory Role for RKIP in Breast Cancer Immune Response. Cancers 2022, 14, 3605. https://doi.org/10.3390/cancers14153605
Bach VN, Ding J, Yeung M, Conrad T, Odeh HN, Cubberly P, Figy C, Ding H-F, Trumbly R, Yeung KC. A Negative Regulatory Role for RKIP in Breast Cancer Immune Response. Cancers. 2022; 14(15):3605. https://doi.org/10.3390/cancers14153605
Chicago/Turabian StyleBach, Vu N., Jane Ding, Miranda Yeung, Taylor Conrad, Hussain N. Odeh, Paige Cubberly, Christopher Figy, Han-Fei Ding, Robert Trumbly, and Kam C. Yeung. 2022. "A Negative Regulatory Role for RKIP in Breast Cancer Immune Response" Cancers 14, no. 15: 3605. https://doi.org/10.3390/cancers14153605
APA StyleBach, V. N., Ding, J., Yeung, M., Conrad, T., Odeh, H. N., Cubberly, P., Figy, C., Ding, H.-F., Trumbly, R., & Yeung, K. C. (2022). A Negative Regulatory Role for RKIP in Breast Cancer Immune Response. Cancers, 14(15), 3605. https://doi.org/10.3390/cancers14153605








