Repair-Assisted Damage Detection Reveals Biological Disparities in Prostate Cancer between African Americans and European Americans
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
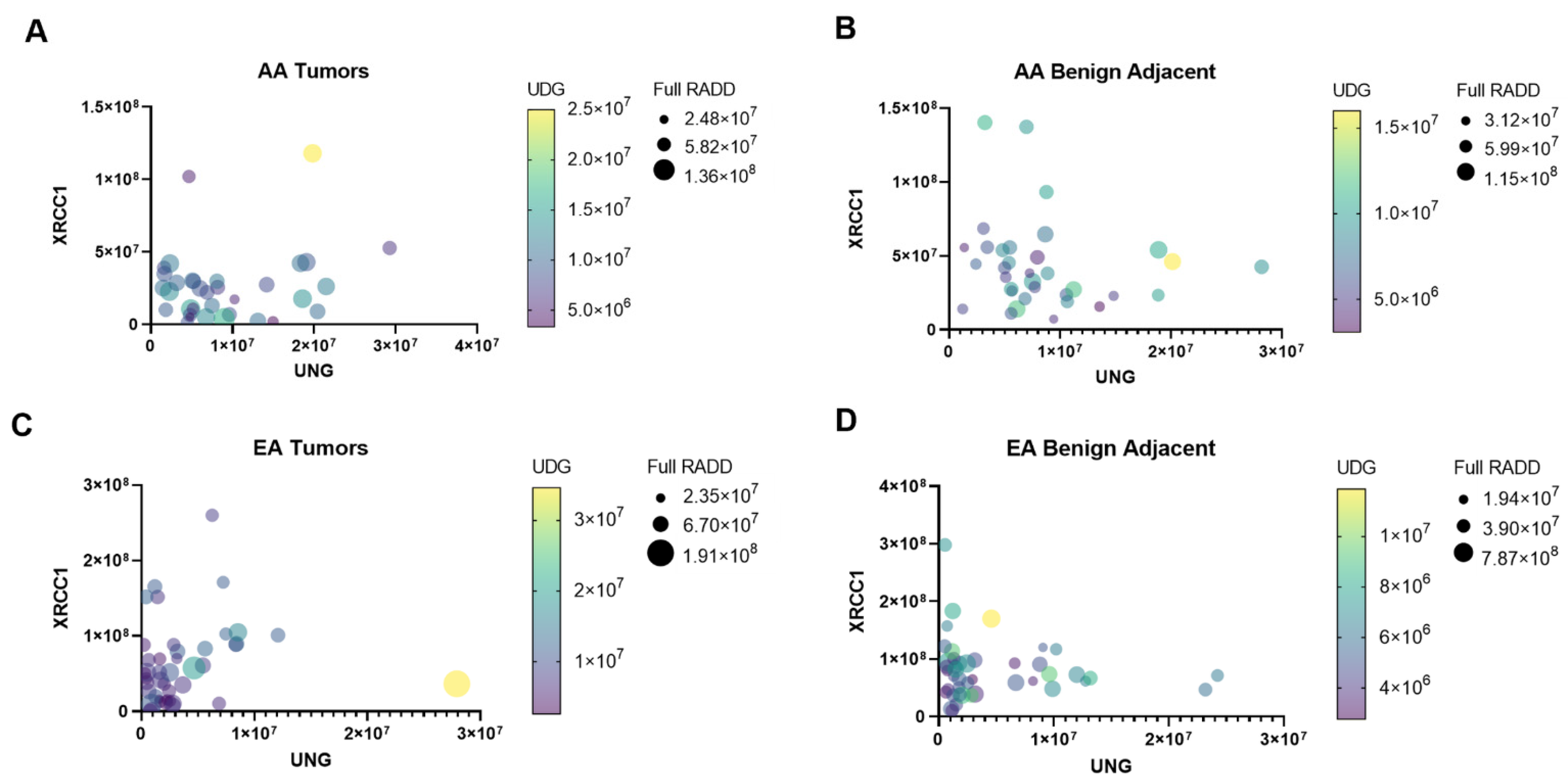
2.1. RADD Analysis
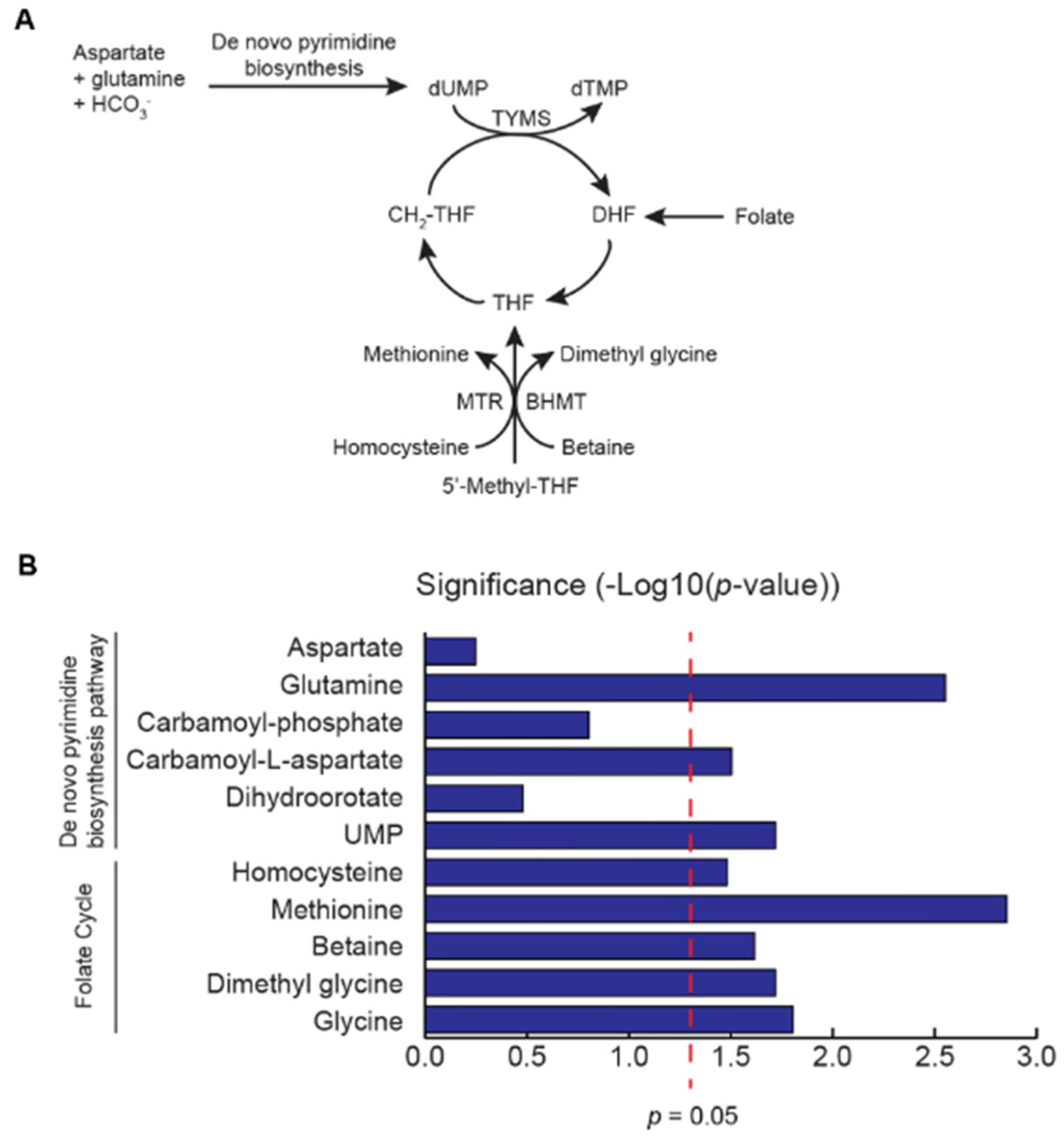
2.2. Tissue Metabolite Profiling
2.3. Statistics
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Mckay, R.R.; Sarkar, R.R.; Kumar, A.; Einck, J.P.; Garraway, I.P.; Lynch, J.A.; Mundt, A.J.; Murphy, J.D.; Stewart, T.F.; Yamoah, K.; et al. Outcomes of black men with prostate cancer treated with radiation therapy in the veterans health administration. Cancer 2021, 127, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Gohlke, J.H.; Lloyd, S.M.; Basu, S.; Putluri, V.; Vareed, S.K.; Rasaily, U.; Piyarathna, D.W.B.; Fuentes, H.; Rajendiran, T.M.; Dorsey, T.H.; et al. Methionine-homocysteine pathway in african-american prostate cancer. JNCI Cancer Spectr. 2019, 3, Pkz019. [Google Scholar] [CrossRef] [PubMed]
- Keith, S.W.; Kwabi-Addo, B.; Zeigler-Johnson, C. Interactions between obesity and one-carbon metabolism genes in predicting prostate cancer outcomes among white and black patients. J. Racial Ethn. Health Disparities 2021, 9, 305–314. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, S.L.; Na, R.; Wei, L.; Sun, J.; Gallagher, J.; Wei, J.; Resurreccion, W.K.; Ernst, S.; Sfanos, K.S.; et al. Distinct genomic alterations in prostate tumors derived from african american men. Mol. Cancer Res. 2020, 18, 1815–1824. [Google Scholar] [CrossRef]
- Mitchell, K.A.; Williams, H. Emerging genomic biomarkers for improving kidney, prostate, and bladder cancer health disparities outcomes. Urol. Oncol. 2019. [Google Scholar] [CrossRef]
- Rayford, W.; Beksac, A.T.; Alger, J.; Alshalalfa, M.; Ahmed, M.; Khan, I.; Falagario, U.G.; Liu, Y.; Davicioni, E.; Spratt, D.E.; et al. Comparative analysis of 1152 african-american and european-american men with prostate cancer identifies distinct genomic and immunological differences. Commun. Biol. 2021, 4, 670. [Google Scholar] [CrossRef]
- Tsai, C.W.; Chang, W.S.; Xu, J.; Xu, Y.; Huang, M.; Pettaway, C.; Bau, D.T.; Gu, J. Leukocyte telomere length is associated with aggressive prostate cancer in localized african american prostate cancer patients. Carcinogenesis 2020, 41, 1213–1218. [Google Scholar] [CrossRef]
- Yadav, S.; Anbalagan, M.; Baddoo, M.; Chellamuthu, V.K.; Mukhopadhyay, S.; Woods, C.; Jiang, W.; Moroz, K.; Flemington, E.K.; Makridakis, N. Somatic mutations in the DNA repairome in prostate cancers in african americans and caucasians. Oncogene 2020, 39, 4299–4311. [Google Scholar] [CrossRef]
- Yamoah, K.; Asamoah, F.A.; Abrahams, A.O.D.; Awasthi, S.; Mensah, J.E.; Dhillon, J.; Mahal, B.A.; Gueye, S.M.; Jalloh, M.; Farahani, S.J.; et al. Prostate tumors of native men from west africa show biologically distinct pathways-a comparative genomic study. Prostate 2021, 81, 1402–1410. [Google Scholar] [CrossRef]
- Yuan, J.; Hu, Z.; Mahal, B.A.; Zhao, S.D.; Kensler, K.H.; Pi, J.; Hu, X.; Zhang, Y.; Wang, Y.; Jiang, J.; et al. Integrated analysis of genetic ancestry and genomic alterations across cancers. Cancer Cell 2018, 34, 549–560.e9. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Mei, H.; Agee, J.; Brown, T.; Mao, J. Racial differences in distribution of fatty acids in prostate cancer and benign prostatic tissues. Lipids Health Dis. 2019, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Piyarathna, D.W.B.; Balasubramanian, A.; Arnold, J.M.; Lloyd, S.M.; Karanam, B.; Castro, P.; Ittmann, M.M.; Putluri, N.; Navone, N.; Jones, J.A.; et al. Err1 and pgc1alpha associated mitochondrial alterations correlate with pan-cancer disparity in african americans. J. Clin. Invest. 2019, 129, 2351–2356. [Google Scholar] [CrossRef] [PubMed]
- Holton, N.W.; Ebenstein, Y.; Gassman, N.R. broad spectrum detection of DNA damage by Repair Assisted Damage Detection (RADD). DNA Repair 2018, 66–67, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.J.; Mann, E.; Da Silva, L.M.; Scalici, J.; Gassman, N.R. DNA damage measurements within tissue samples with Repair Assisted Damage Detection (RADD). Curr. Res. Biotechnol. 2019, 1, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Mann, E.K.; Lee, K.J.; Chen, D.; Da Silva, L.M.; Dal Zotto, V.L.; Scalici, J.; Gassman, N.R. Associations between DNA damage and pd-l1 expression in ovarian cancer, a potential biomarker for clinical response. Biology 2021, 10, 385. [Google Scholar] [CrossRef]
- Torchinsky, D.; Michaeli, Y.; Gassman, N.R.; Ebenstein, Y. Simultaneous detection of multiple DNA damage types by multi-colour fluorescent labelling. Chem. Commun. 2019, 55, 11414–11417. [Google Scholar] [CrossRef]
- Gilat, N.; Fridman, D.; Sharim, H.; Margalit, S.; Gassman, N.R.; Michaeli, Y.; Ebenstein, Y. From Single-molecule to genome-wide mapping of DNA lesions: Repair-assisted damage detection sequencing. Biophys. Rep. 2021, 1, 100017. [Google Scholar] [CrossRef]
- Lee, K.J.; Piett, C.G.; Andrews, J.F.; Mann, E.; Nagel, Z.D.; Gassman, N.R. Defective Base Excision Repair In The Response To DNA damaging agents in triple negative breast cancer. PLoS ONE 2019, 14, e0223725. [Google Scholar] [CrossRef]
- Lee, K.J.; Mann, E.; Wright, G.; Piett, C.G.; Nagel, Z.D.; Gassman, N.R. Exploiting DNA repair defects in triple negative breast cancer to improve cell killing. Ther. Adv. Med. Oncol. 2020, 12, 1758835920958354. [Google Scholar] [CrossRef]
- Burdak-Rothkamm, S.; Mansour, W.Y.; Rothkamm, K. DNA damage repair deficiency in prostate cancer. Trends Cancer 2020, 6, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.H.; Pittman, D.L.; Wyatt, M.D. Uracil in DNA: Consequences for carcinogenesis and chemotherapy. Biochem. Pharmacol. 2008, 76, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Chon, J.; Field, M.S.; Stover, P.J. Deoxyuracil in DNA and disease: Genomic signal or managed situation? DNA Repair 2019, 77, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Field, M.S.; Szebenyi, D.M.; Stover, P.J. Regulation of de novo purine biosynthesis by methenyltetrahydrofolate synthetase in neuroblastoma. J. Biol. Chem. 2006, 281, 4215–4221. [Google Scholar] [CrossRef]
- Vanderwall, C.M.; Tangney, C.C.; Kwasny, M.J.; Gustashaw, K.A. Examination of circulating folate levels as a reflection of folate intakes among older adult supplement users and nonusers in the national health and nutrition examination survey 2003–2004. J. Acad. Nutr. Diet. 2012, 112, 285–290. [Google Scholar] [CrossRef]
- Krokan, H.E.; Drablos, F.; Slupphaug, G. Uracil in DNA—occurrence, consequences and repair. Oncogene 2002, 21, 8935–8948. [Google Scholar] [CrossRef]
- Sultana, R.; Abdel-Fatah, T.; Abbotts, R.; Hawkes, C.; Albarakati, N.; Seedhouse, C.; Ball, G.; Chan, S.; Rakha, E.A.; Ellis, I.O.; et al. Targeting XRCC1 deficiency in breast cancer for personalized therapy. Cancer Res. 2013, 73, 1621–1634. [Google Scholar] [CrossRef]
- Pulukuri, S.M.; Knost, J.A.; Estes, N.; Rao, J.S. Small Interfering rna-directed knockdown of uracil DNA glycosylase induces apoptosis and sensitizes human prostate cancer cells to genotoxic stress. Mol. Cancer Res. 2009, 7, 1285–1293. [Google Scholar] [CrossRef]
- Weeks, L.D.; Zentner, G.E.; Scacheri, P.C.; Gerson, S.L. Uracil DNA glycosylase (ung) loss enhances DNA double strand break formation in human cancer cells exposed to pemetrexed. Cell Death Dis. 2014, 5, e1045. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357. [Google Scholar] [CrossRef]
- Ali, R.; Alblihy, A.; Toss, M.S.; Algethami, M.; Al Sunni, R.; Green, A.R.; Rakha, E.A.; Madhusudan, S. XRCC1 deficient triple negative breast cancers are sensitive to atr, atm and wee1 inhibitor either alone or in combination with olaparib. Ther. Adv. Med. Oncol. 2020, 12, 1758835920974201. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Jiu, T.; Wang, D. Impact of polymorphisms of the DNA repair gene XRCC1 and their role in the risk of prostate cancer. Pak. J. Med. Sci. 2015, 31, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Zhou, Y.; Xu, Z.; Ruan, J.; Zhu, M.; Jin, K.; Zhou, D.; Hu, Q.; Wang, Q.; Wang, Z.; et al. XRCC1 Arg399Gln and Arg194Trp polymorphisms in prostate cancer risk: A meta-analysis. Prostate Cancer Prostatic Dis. 2011, 14, 225–231. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, R.; Price, D.K.; Dahut, W.L.; Reed, E.; Figg, W.D. genetic polymorphisms in XRCC1 associated with radiation therapy in prostate cancer. Cancer Biol. Ther. 2010, 10, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Henriquez-Hernandez, L.A.; Valenciano, A.; Foro-Arnalot, P.; Alvarez-Cubero, M.J.; Cozar, J.M.; Suarez-Novo, J.F.; Castells-Esteve, M.; Fernandez-Gonzalo, P.; De-Paula-Carranza, B.; Ferrer, M.; et al. Association between single-nucleotide polymorphisms in DNA double-strand break repair genes and prostate cancer aggressiveness in the spanish population. Prostate Cancer Prostatic Dis. 2016, 19, 28–34. [Google Scholar] [CrossRef]
- Henriquez-Hernandez, L.A.; Valenciano, A.; Foro-Arnalot, P.; Alvarez-Cubero, M.J.; Cozar, J.M.; Suarez-Novo, J.F.; Castells-Esteve, M.; Fernandez-Gonzalo, P.; De-Paula-Carranza, B.; Ferrer, M.; et al. Single nucleotide polymorphisms in DNA repair genes as risk factors associated to prostate cancer progression. BMC Med. Genet. 2014, 15, 143. [Google Scholar] [CrossRef]
- Ladiges, W.C. Mouse models of XRCC1 DNA repair polymorphisms and cancer. Oncogene 2006, 25, 1612–1619. [Google Scholar] [CrossRef]
- Takanami, T.; Nakamura, J.; Kubota, Y.; Horiuchi, S. The arg280his polymorphism in x-ray repair cross-complementing gene 1 impairs DNA repair ability. Mutat. Res. 2005, 582, 135–145. [Google Scholar] [CrossRef]
- Hanssen-Bauer, A.; Solvang-Garten, K.; Gilljam, K.M.; Torseth, K.; Wilson, D.M.; Akbari, M.; Otterlei, M. The region of XRCC1 which harbours the three most common nonsynonymous polymorphic variants, is essential for the scaffolding function of XRCC1. DNA Repair 2012, 11, 357–366. [Google Scholar] [CrossRef]
- Sizova, D.V.; Keh, A.; Taylor, B.F.; Sweasy, J.B. The r280h X-ray cross-complementing 1 germline variant induces genomic instability and cellular transformation. DNA Repair 2015, 31, 73–79. [Google Scholar] [CrossRef]


| Full RADD Lesion Processing Mix | Per 100 µL Reaction Volume | Gap-Filling Mix | Per 100 µL Reaction Volume |
|---|---|---|---|
| UDG (NEB M0280) | 2.5 U | Klenow exo- (Thermo Fisher, Waltham, MA, USA, EP0422) | 1 |
| FPG (NEB M0240) | 4 U | Digoxigenin dUTP (Sigma Aldrich, St. Louis, MO, USA, 11093088910) | 0.1 |
| T4 PDG (NEB M0308) | 5 U | Thermo Pol Buffer | 10 µL |
| (NEB B9004) | |||
| EndoIV (NEB M0304) | 5 U | ||
| AAG (NEB M0313) | 5 U | ||
| NAD+ (100x, NEB B9007) | 500 µM | ||
| BSA (Sigma Aldrich, St. Louis, MO, USA) | 200 µg/mL | ||
| Thermo Pol Buffer | 10 µL | ||
| (NEB B9004) | |||
| Cocktails | Lesion Processing Mix | Lesions |
|---|---|---|
| oxRADD | FPG + EndoIV + EndoVIII | Removes various types of oxidized purines, urea, 5, 6-dihydroxythymine, thymine glycol, 5-hydroxy-5-methylhydanton, 6-hydroxy-5,6-dihydrothymine and methyltartronylurea, abasic sites, and strand breaks. |
| T4PDG | T4 PDG + EndoIV | Removes cyclobutane pyrimidine dimers and 6-4 photoproducts along with abasic sites. |
| UDG | UDG + EndoIV | Removes uracil lesions and abasic sites. |
| Full RADD | AAG + FPG + T4 PDG + UDG + EndoIV | All of the above lesions plus the removal of various alkylated and oxidative DNA damaged sites, including 3-methyladenine, 7-methylguanine, 1, N6-ethenoadenine, and hypoxanthine. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krieger, K.L.; Gohlke, J.H.; Lee, K.J.; Piyarathna, D.W.B.; Castro, P.D.; Jones, J.A.; Ittmann, M.M.; Gassman, N.R.; Sreekumar, A. Repair-Assisted Damage Detection Reveals Biological Disparities in Prostate Cancer between African Americans and European Americans. Cancers 2022, 14, 1012. https://doi.org/10.3390/cancers14041012
Krieger KL, Gohlke JH, Lee KJ, Piyarathna DWB, Castro PD, Jones JA, Ittmann MM, Gassman NR, Sreekumar A. Repair-Assisted Damage Detection Reveals Biological Disparities in Prostate Cancer between African Americans and European Americans. Cancers. 2022; 14(4):1012. https://doi.org/10.3390/cancers14041012
Chicago/Turabian StyleKrieger, Kimiko L., Jie H. Gohlke, Kevin J. Lee, Danthasinghe Waduge Badrajee Piyarathna, Patricia D. Castro, Jeffrey A. Jones, Michael M. Ittmann, Natalie R. Gassman, and Arun Sreekumar. 2022. "Repair-Assisted Damage Detection Reveals Biological Disparities in Prostate Cancer between African Americans and European Americans" Cancers 14, no. 4: 1012. https://doi.org/10.3390/cancers14041012
APA StyleKrieger, K. L., Gohlke, J. H., Lee, K. J., Piyarathna, D. W. B., Castro, P. D., Jones, J. A., Ittmann, M. M., Gassman, N. R., & Sreekumar, A. (2022). Repair-Assisted Damage Detection Reveals Biological Disparities in Prostate Cancer between African Americans and European Americans. Cancers, 14(4), 1012. https://doi.org/10.3390/cancers14041012







