Utility of Cell-Free DNA Detection in Transplant Oncology
Simple Summary
Abstract
1. Introduction to Transplant Oncology
2. Defining Transplant Oncology in Hepatocellular Carcinoma
3. The Concept of Circulating Free DNA
4. Characteristics of cfDNA Biology
5. History of cfDNA
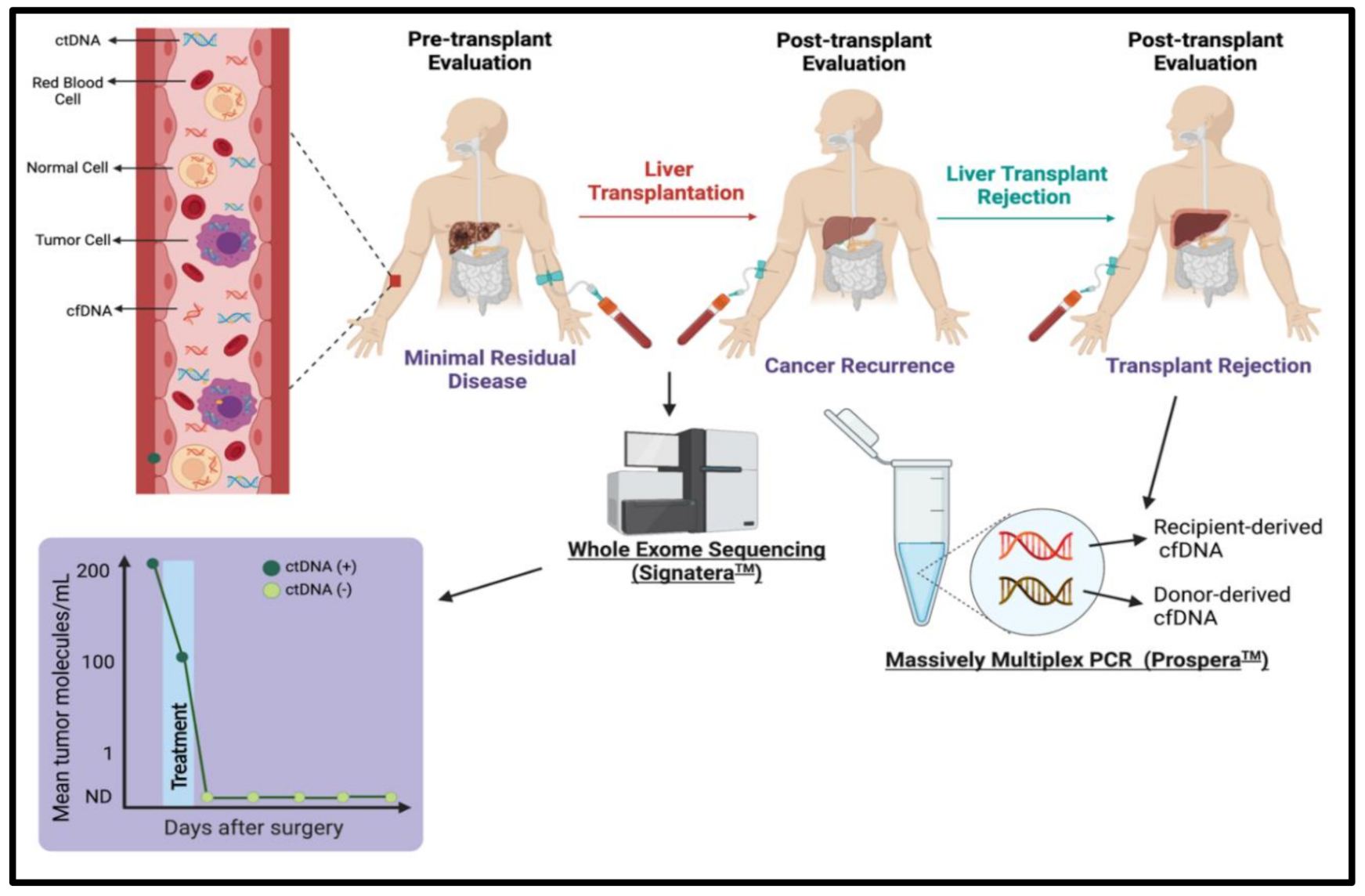
6. Utilizing cfDNA in Transplant Oncology
7. Emerging Genomic Tests in Transplant Oncology
7.1. Guardant360® CDx
7.2. FoundationOne
7.3. Signatera™
7.4. Prospera™
8. Utilizing cfDNA to Evaluate Cancer Occurrence Pre-Transplant
cfDNA to Evaluate Hepatocellular Carcinoma Recurrence Post-Transplant/Hepatectomy
9. cfDNA to Evaluate Liver Transplant Rejection
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Abdelrahim, M.; Esmail, A.; Abudayyeh, A.; Murakami, N.; Saharia, A.; McMillan, R.; Victor, D.; Kodali, S.; Shetty, A.; Fong, J.V.N.; et al. Transplant Oncology: An Evolving Field in Cancer Care. Cancers 2021, 13, 4911. [Google Scholar] [CrossRef] [PubMed]
- Sapisochin, G.; Hibi, T.; Ghobrial, M.; Man, K. The ILTS Consensus Conference on Transplant Oncology: Setting the Stage. Transplantation 2020, 104, 1119–1120. [Google Scholar] [CrossRef] [PubMed]
- Hibi, T.; Sapisochin, G. What is transplant oncology? Surgery 2019, 165, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Siddique, O.; Yoo, E.R.; Perumpail, R.B.; Perumpail, B.J.; Liu, A.; Cholankeril, G.; Ahmed, A. The importance of a multidisciplinary approach to hepatocellular carcinoma. J. Multidiscip. Healthc. 2017, 10, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Mehta, N.; Bhangui, P.; Yao, F.Y.; Mazzaferro, V.; Toso, C.; Akamatsu, N.; Durand, F.; Ijzermans, J.; Polak, W.; Zheng, S.; et al. Liver Transplantation for Hepatocellular Carcinoma. Working Group Report from the ILTS Transplant Oncology Consensus Conference. Transplantation 2020, 104, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- Balogh, J.; Victor, D., 3rd; Asham, E.H.; Burroughs, S.G.; Boktour, M.; Saharia, A.; Li, X.; Ghobrial, R.M.; Monsour, H.P., Jr. Hepatocellular carcinoma: A review. J. Hepatocell. Carcinoma 2016, 3, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Mazzaferro, V.; Regalia, E.; Doci, R.; Andreola, S.; Pulvirenti, A.; Bozzetti, F.; Montalto, F.; Ammatuna, M.; Morabito, A.; Gennari, L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N. Engl. J. Med. 1996, 334, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Chaiteerakij, R.; Zhang, X.; Addissie, B.D.; Mohamed, E.A.; Harmsen, W.S.; Theobald, P.J.; Peters, B.E.; Balsanek, J.G.; Ward, M.M.; Giama, N.H.; et al. Combinations of biomarkers and Milan criteria for predicting hepatocellular carcinoma recurrence after liver transplantation. Liver Transpl. 2015, 21, 599–606. [Google Scholar] [CrossRef]
- Halazun, K.J.; Najjar, M.; Abdelmessih, R.M.; Samstein, B.; Griesemer, A.D.; Guarrera, J.V.; Kato, T.; Verna, E.C.; Emond, J.C.; Brown, R.S., Jr. Recurrence After Liver Transplantation for Hepatocellular Carcinoma: A New MORAL to the Story. Ann. Surg. 2017, 265, 557–564. [Google Scholar] [CrossRef]
- Kaido, T.; Ogawa, K.; Mori, A.; Fujimoto, Y.; Ito, T.; Tomiyama, K.; Takada, Y.; Uemoto, S. Usefulness of the Kyoto criteria as expanded selection criteria for liver transplantation for hepatocellular carcinoma. Surgery 2013, 154, 1053–1060. [Google Scholar] [CrossRef]
- Pavel, M.C.; Fuster, J. Expansion of the hepatocellular carcinoma Milan criteria in liver transplantation: Future directions. World J. Gastroenterol. WJG 2018, 24, 3626–3636. [Google Scholar] [CrossRef] [PubMed]
- Lai, Q.; Iesari, S.; Melandro, F.; Mennini, G.; Rossi, M.; Lerut, J. The growing impact of alpha-fetoprotein in the field of liver transplantation for hepatocellular cancer: Time for a revolution. Transl. Gastroenterol. Hepatol. 2017, 2, 72. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.Y.; Ferrell, L.; Bass, N.M.; Watson, J.J.; Bacchetti, P.; Venook, A.; Ascher, N.L.; Roberts, J.P. Liver transplantation for hepatocellular carcinoma: Expansion of the tumor size limits does not adversely impact survival. Hepatology 2001, 33, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Mazzaferro, V.; Sposito, C.; Zhou, J.; Pinna, A.D.; De Carlis, L.; Fan, J.; Cescon, M.; Di Sandro, S.; Yi-Feng, H.; Lauterio, A.; et al. Metroticket 2.0 Model for Analysis of Competing Risks of Death After Liver Transplantation for Hepatocellular Carcinoma. Gastroenterology 2018, 154, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Sapisochin, G.; Goldaracena, N.; Laurence, J.M.; Dib, M.; Barbas, A.; Ghanekar, A.; Cleary, S.P.; Lilly, L.; Cattral, M.S.; Marquez, M.; et al. The extended Toronto criteria for liver transplantation in patients with hepatocellular carcinoma: A prospective validation study. Hepatology 2016, 64, 2077–2088. [Google Scholar] [CrossRef]
- Silva, M.; Moya, A.; Berenguer, M.; Sanjuan, F.; Lopez-Andujar, R.; Pareja, E.; Torres-Quevedo, R.; Aguilera, V.; Montalva, E.; De Juan, M.; et al. Expanded criteria for liver transplantation in patients with cirrhosis and hepatocellular carcinoma. Liver Transpl. 2008, 14, 1449–1460. [Google Scholar] [CrossRef]
- Qu, Z.; Ling, Q.; Gwiasda, J.; Xu, X.; Schrem, H.; Beneke, J.; Kaltenborn, A.; Krauth, C.; Mix, H.; Klempnauer, J.; et al. Hangzhou criteria are more accurate than Milan criteria in predicting long-term survival after liver transplantation for HCC in Germany. Langenbecks Arch. Surg. 2018, 403, 643–654. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2012, 56, 908–943. [Google Scholar] [CrossRef]
- Colombo, F.; Baldan, F.; Mazzucchelli, S.; Martin-Padura, I.; Marighetti, P.; Cattaneo, A.; Foglieni, B.; Spreafico, M.; Guerneri, S.; Baccarin, M.; et al. Evidence of distinct tumour-propagating cell populations with different properties in primary human hepatocellular carcinoma. PLoS ONE 2011, 6, e21369. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, J.; Luo, F. Serum tumor markers for detection of hepatocellular carcinoma. World J. Gastroenterol. WJG 2006, 12, 1175–1181. [Google Scholar] [CrossRef]
- Ye, Q.; Ling, S.; Zheng, S.; Xu, X. Liquid biopsy in hepatocellular carcinoma: Circulating tumor cells and circulating tumor DNA. Mol. Cancer 2019, 18, 114. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef]
- Smith, R.A.; Lam, A.K. Liquid Biopsy for Investigation of Cancer DNA in Esophageal Squamous Cell Carcinoma. Methods Mol. Biol. 2020, 2129, 203–215. [Google Scholar] [CrossRef]
- Schwarzenbach, H. Clinical Relevance of Circulating, Cell-Free and Exosomal microRNAs in Plasma and Serum of Breast Cancer Patients. Oncol. Res. Treat. 2017, 40, 423–429. [Google Scholar] [CrossRef]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10, 1758835918794630. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Craig, A.J.; von Felden, J.; Garcia-Lezana, T.; Sarcognato, S.; Villanueva, A. Tumour evolution in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 139–152. [Google Scholar] [CrossRef]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef]
- Underhill, H.R.; Kitzman, J.O.; Hellwig, S.; Welker, N.C.; Daza, R.; Baker, D.N.; Gligorich, K.M.; Rostomily, R.C.; Bronner, M.P.; Shendure, J. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016, 12, e1006162. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.; Zhang, J.; Leung, T.N.; Lau, T.K.; Chang, A.M.; Hjelm, N.M. Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 1999, 64, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Giacona, M.B.; Ruben, G.C.; Iczkowski, K.A.; Roos, T.B.; Porter, D.M.; Sorenson, G.D. Cell-free DNA in human blood plasma: Length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1998, 17, 89–97. [Google Scholar] [CrossRef]
- Thierry, A.R.; Mouliere, F.; Gongora, C.; Ollier, J.; Robert, B.; Ychou, M.; Del Rio, M.; Molina, F. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010, 38, 6159–6175. [Google Scholar] [CrossRef]
- Lo, Y.M.; Chan, K.C.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef]
- Volik, S.; Alcaide, M.; Morin, R.D.; Collins, C. Cell-free DNA (cfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Mol. Cancer Res. 2016, 14, 898–908. [Google Scholar] [CrossRef]
- Tsui, N.B.; Jiang, P.; Chow, K.C.; Su, X.; Leung, T.Y.; Sun, H.; Chan, K.C.; Chiu, R.W.; Lo, Y.M. High resolution size analysis of fetal DNA in the urine of pregnant women by paired-end massively parallel sequencing. PLoS ONE 2012, 7, e48319. [Google Scholar] [CrossRef]
- Nadano, D.; Yasuda, T.; Kishi, K. Measurement of deoxyribonuclease I activity in human tissues and body fluids by a single radial enzyme-diffusion method. Clin. Chem. 1993, 39, 448–452. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear Acids In Human Blood Plasma. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Tan, E.M.; Schur, P.H.; Carr, R.I.; Kunkel, H.G. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J. Clin. Invest. 1966, 45, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Bendich, A.; Wilczok, T.; Borenfreund, E. Circulating Dna as a Possible Factor in Oncogenesis. Science 1965, 148, 374–376. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Vasioukhin, V.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Stroun, M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br. J. Haematol. 1994, 86, 774–779. [Google Scholar] [CrossRef]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Prev. Biomark. 1994, 3, 67–71. [Google Scholar]
- Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef]
- Lo, Y.M.; Tein, M.S.; Lau, T.K.; Haines, C.J.; Leung, T.N.; Poon, P.M.; Wainscoat, J.S.; Johnson, P.J.; Chang, A.M.; Hjelm, N.M. Quantitative analysis of fetal DNA in maternal plasma and serum: Implications for noninvasive prenatal diagnosis. Am. J. Hum. Genet. 1998, 62, 768–775. [Google Scholar] [CrossRef]
- Lo, Y.M. Fetal DNA in maternal plasma: Biology and diagnostic applications. Clin. Chem. 2000, 46, 1903–1906. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A., Jr.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef]
- Gonzalez-Cao, M.; Mayo-de-Las-Casas, C.; Molina-Vila, M.A.; De Mattos-Arruda, L.; Muñoz-Couselo, E.; Manzano, J.L.; Cortes, J.; Berros, J.P.; Drozdowskyj, A.; Sanmamed, M.; et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res. 2015, 25, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Bae, B.N.; Sohn, B.S.; Pyo, J.S.; Kang, G.H.; Kim, K.M. Detection of KIT and PDGFRA mutations in the plasma of patients with gastrointestinal stromal tumor. Target. Oncol. 2015, 10, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Lubitz, C.C.; Parangi, S.; Holm, T.M.; Bernasconi, M.J.; Schalck, A.P.; Suh, H.; Economopoulos, K.P.; Gunda, V.; Donovan, S.E.; Sadow, P.M.; et al. Detection of Circulating BRAF(V600E) in Patients with Papillary Thyroid Carcinoma. J. Mol. Diagn. 2016, 18, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, Z.; Niederst, M.J.; Karlovich, C.A.; Wakelee, H.A.; Neal, J.W.; Mino-Kenudson, M.; Fulton, L.; Hata, A.N.; Lockerman, E.L.; Kalsy, A.; et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov. 2015, 5, 713–722. [Google Scholar] [CrossRef]
- Roschewski, M.; Dunleavy, K.; Pittaluga, S.; Moorhead, M.; Pepin, F.; Kong, K.; Shovlin, M.; Jaffe, E.S.; Staudt, L.M.; Lai, C.; et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: A correlative biomarker study. Lancet Oncol. 2015, 16, 541–549. [Google Scholar] [CrossRef]
- Schiavon, G.; Hrebien, S.; Garcia-Murillas, I.; Cutts, R.J.; Pearson, A.; Tarazona, N.; Fenwick, K.; Kozarewa, I.; Lopez-Knowles, E.; Ribas, R.; et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl. Med. 2015, 7, 313ra182. [Google Scholar] [CrossRef]
- Sefrioui, D.; Perdrix, A.; Sarafan-Vasseur, N.; Dolfus, C.; Dujon, A.; Picquenot, J.M.; Delacour, J.; Cornic, M.; Bohers, E.; Leheurteur, M.; et al. Short report: Monitoring ESR1 mutations by circulating tumor DNA in aromatase inhibitor resistant metastatic breast cancer. Int. J. Cancer 2015, 137, 2513–2519. [Google Scholar] [CrossRef]
- Spindler, K.L.; Pallisgaard, N.; Andersen, R.F.; Brandslund, I.; Jakobsen, A. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS ONE 2015, 10, e0108247. [Google Scholar] [CrossRef]
- Xu, S.; Lou, F.; Wu, Y.; Sun, D.Q.; Zhang, J.B.; Chen, W.; Ye, H.; Liu, J.H.; Wei, S.; Zhao, M.Y.; et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett. 2016, 370, 324–331. [Google Scholar] [CrossRef]
- Yoo, C.; Ryu, M.H.; Na, Y.S.; Ryoo, B.Y.; Park, S.R.; Kang, Y.K. Analysis of serum protein biomarkers, circulating tumor DNA, and dovitinib activity in patients with tyrosine kinase inhibitor-refractory gastrointestinal stromal tumors. Ann. Oncol. 2014, 25, 2272–2277. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Polivka, J., Jr.; Pesta, M.; Janku, F. Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: Are we there yet? Expert Rev. Mol. Diagn. 2015, 15, 1631–1644. [Google Scholar] [CrossRef] [PubMed]
- Arnedos, M.; Vicier, C.; Loi, S.; Lefebvre, C.; Michiels, S.; Bonnefoi, H.; Andre, F. Precision medicine for metastatic breast cancer--limitations and solutions. Nat. Rev. Clin. Oncol. 2015, 12, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; Wu, X.; Wang, C.; Wang, G.; Li, C.; Zhao, J.; Zhang, Y.; Xiang, J.; Xu, Y.; Nian, W.; et al. Discovery and validation of methylation signatures in blood-based circulating tumor cell-free DNA in early detection of colorectal carcinoma: A case-control study. Clin. Epigenet. 2021, 13, 26. [Google Scholar] [CrossRef]
- Luo, H.; Zhao, Q.; Wei, W.; Zheng, L.; Yi, S.; Li, G.; Wang, W.; Sheng, H.; Pu, H.; Mo, H.; et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci. Transl. Med. 2020, 12, 524. [Google Scholar] [CrossRef]
- Nassar, F.J.; Msheik, Z.S.; Nasr, R.R.; Temraz, S.N. Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction. Clin. Epigenet. 2021, 13, 111. [Google Scholar] [CrossRef]
- Laird, P.W. The power and the promise of DNA methylation markers. Nat. Rev. Cancer 2003, 3, 253–266. [Google Scholar] [CrossRef]
- Heyn, H.; Esteller, M. DNA methylation profiling in the clinic: Applications and challenges. Nat. Rev. Genet. 2012, 13, 679–692. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Marczynski, G.T.; Laus, A.C.; Dos Reis, M.B.; Reis, R.M.; Vazquez, V.L. Circulating tumor DNA (ctDNA) detection is associated with shorter progression-free survival in advanced melanoma patients. Sci. Rep. 2020, 10, 18682. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.C.; Weigelt, B.; Reis-Filho, J.S. Going with the flow: From circulating tumor cells to DNA. Sci. Transl. Med. 2013, 5, 207ps14. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.J.; Oxnard, G.R.; Paweletz, C.P.; Camidge, D.R.; Heymach, J.V.; Solit, D.B.; Johnson, B.E. Realizing the potential of plasma genotyping in an age of genotype-directed therapies. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef]
- Taavitsainen, S.; Annala, M.; Ledet, E.; Beja, K.; Miller, P.J.; Moses, M.; Nykter, M.; Chi, K.N.; Sartor, O.; Wyatt, A.W. Evaluation of Commercial Circulating Tumor DNA Test in Metastatic Prostate Cancer. JCO Precis. Oncol. 2019, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Natera. Natera Launches Initiative to Transform the Management of Cancer Patients in Organ Transplantation. 14 September 2020. Available online: https://www.natera.com/company/news/natera-launches-initiative-to-transform-the-management-of-cancer-patients-in-organ-transplantation/ (accessed on 20 August 2021).
- Health, G. Guardant Health Launches Guardant Reveal™ Liquid Biopsy Test for Residual Disease and Recurrence Monitoring in Patients with Early-Stage Colorectal Cancer. Available online: https://investors.guardanthealth.com/press-releases/press-releases/2021/Guardant-Health-Launches-Guardant-Reveal-Liquid-Biopsy-Test-for-Residual-Disease-and-Recurrence-Monitoring-in-Patients-with-Early-Stage-Colorectal-Cancer/default.aspx (accessed on 29 October 2021).
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Vincent, J.J.; Mortimer, S.; Vowles, J.V.; Ulrich, B.C.; Banks, K.C.; Fairclough, S.R.; Zill, O.A.; Sikora, M.; Mokhtari, R.; et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 3539–3549. [Google Scholar] [CrossRef]
- Jarrell, K. FDA Approves First Liquid Biopsy Next-Generation Sequencing Companion Diagnostic Test. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-liquid-biopsy-next-generation-sequencing-companion-diagnostic-test (accessed on 25 October 2021).
- Kaseb, A.O.; Sánchez, N.S.; Sen, S.; Kelley, R.K.; Tan, B.; Bocobo, A.G.; Lim, K.H.; Abdel-Wahab, R.; Uemura, M.; Pestana, R.C.; et al. Molecular Profiling of Hepatocellular Carcinoma Using Circulating Cell-Free DNA. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 6107–6118. [Google Scholar] [CrossRef]
- Fujii, Y.; Ono, A.; Hayes, C.N.; Aikata, H.; Yamauchi, M.; Uchikawa, S.; Kodama, K.; Teraoka, Y.; Fujino, H.; Nakahara, T.; et al. Identification and monitoring of mutations in circulating cell-free tumor DNA in hepatocellular carcinoma treated with lenvatinib. J. Exp. Clin. Cancer Res. 2021, 40, 215. [Google Scholar] [CrossRef]
- Ikeda, S.; Tsigelny, I.F.; Skjevik, Å., A.; Kono, Y.; Mendler, M.; Kuo, A.; Sicklick, J.K.; Heestand, G.; Banks, K.C.; Talasaz, A.; et al. Next-Generation Sequencing of Circulating Tumor DNA Reveals Frequent Alterations in Advanced Hepatocellular Carcinoma. Oncologist 2018, 23, 586–593. [Google Scholar] [CrossRef]
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol. 2019, 5, 1124–1131. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra392. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; McKendrick, J.; et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019, 5, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Li, Y.; Mo, S.; Ma, X.; Hu, X.; Zhang, L.; Huang, D.; Cai, S. Prognostic value of circulating tumor DNA (ctDNA) detection during adjuvant chemotherapy in patients with stage III colorectal cancer: The interim report of a prospective, observational study. J. Clin. Oncol. 2020, 38, 29. [Google Scholar] [CrossRef]
- Tarazona, N.; Gimeno-Valiente, F.; Gambardella, V.; Zuñiga, S.; Rentero-Garrido, P.; Huerta, M.; Roselló, S.; Martinez-Ciarpaglini, C.; Carbonell-Asins, J.A.; Carrasco, F.; et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann. Oncol. 2019, 30, 1804–1812. [Google Scholar] [CrossRef]
- FDA. FoundationOne Liquid CDx—P190032. Available online: https://www.fda.gov/medical-devices/recently-approved-devices/foundationone-liquid-cdx-p190032 (accessed on 25 October 2021).
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef]
- Natera. Signatera™. Available online: https://www.natera.com/oncology/signatera-advanced-cancer-detection/ (accessed on 20 August 2021).
- Domínguez-Vigil, I.G.; Moreno-Martínez, A.K.; Wang, J.Y.; Roehrl, M.H.A.; Barrera-Saldaña, H.A. The dawn of the liquid biopsy in the fight against cancer. Oncotarget 2018, 9, 2912–2922. [Google Scholar] [CrossRef]
- Lanman, R.B.; Mortimer, S.A.; Zill, O.A.; Sebisanovic, D.; Lopez, R.; Blau, S.; Collisson, E.A.; Divers, S.G.; Hoon, D.S.; Kopetz, E.S.; et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS ONE 2015, 10, e0140712. [Google Scholar] [CrossRef]
- Plagnol, V.; Woodhouse, S.; Howarth, K.; Lensing, S.; Smith, M.; Epstein, M.; Madi, M.; Smalley, S.; Leroy, C.; Hinton, J.; et al. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS ONE 2018, 13, e0193802. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Coombes, R.C.; Page, K.; Salari, R.; Hastings, R.K.; Armstrong, A.; Ahmed, S.; Ali, S.; Cleator, S.; Kenny, L.; Stebbing, J.; et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 4255–4263. [Google Scholar] [CrossRef]
- Christensen, E.; Birkenkamp-Demtröder, K.; Sethi, H.; Shchegrova, S.; Salari, R.; Nordentoft, I.; Wu, H.T.; Knudsen, M.; Lamy, P.; Lindskrog, S.V.; et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, X.; Wang, J.; Zhou, S.; Wang, C.L.; Ye, M.Z.; Wang, X.Y.; Song, Y.; Wang, Y.Q.; Zhang, L.T.; et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann. Oncol. 2019, 30, 464–470. [Google Scholar] [CrossRef]
- Yang, J.D.; Larson, J.J.; Watt, K.D.; Allen, A.M.; Wiesner, R.H.; Gores, G.J.; Roberts, L.R.; Heimbach, J.A.; Leise, M.D. Hepatocellular Carcinoma Is the Most Common Indication for Liver Transplantation and Placement on the Waitlist in the United States. Clin. Gastroenterol. Hepatol. 2017, 15, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Altuğ, Y.; Liang, N.; Ram, R.; Ravi, H.; Ahmed, E.; Brevnov, M.; Swenerton, R.K.; Zimmermann, B.; Malhotra, M.; Demko, Z.P.; et al. Analytical Validation of a Single-nucleotide Polymorphism-based Donor-derived Cell-free DNA Assay for Detecting Rejection in Kidney Transplant Patients. Transplantation 2019, 103, 2657–2665. [Google Scholar] [CrossRef]
- Grskovic, M.; Hiller, D.J.; Eubank, L.A.; Sninsky, J.J.; Christopherson, C.; Collins, J.P.; Thompson, K.; Song, M.; Wang, Y.S.; Ross, D.; et al. Validation of a Clinical-Grade Assay to Measure Donor-Derived Cell-Free DNA in Solid Organ Transplant Recipients. J. Mol. Diagn. 2016, 18, 890–902. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Archila, F.A.; Constantin, T.; Prins, S.A.; Liberto, J.; Damm, I.; Towfighi, P.; Navarro, S.; Kirkizlar, E.; Demko, Z.P.; et al. Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR. J. Clin. Med. 2018, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Bloom, R.D.; Bromberg, J.S.; Poggio, E.D.; Bunnapradist, S.; Langone, A.J.; Sood, P.; Matas, A.J.; Mehta, S.; Mannon, R.B.; Sharfuddin, A.; et al. Cell-Free DNA and Active Rejection in Kidney Allografts. J. Am. Soc. Nephrol. 2017, 28, 2221–2232. [Google Scholar] [CrossRef]
- Huang, E.; Sethi, S.; Peng, A.; Najjar, R.; Mirocha, J.; Haas, M.; Vo, A.; Jordan, S.C. Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients. Am. J. Transplant. 2019, 19, 1663–1670. [Google Scholar] [CrossRef]
- Liao, W.; Yang, H.; Xu, H.; Wang, Y.; Ge, P.; Ren, J.; Xu, W.; Lu, X.; Sang, X.; Zhong, S.; et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget 2016, 7, 40481–40490. [Google Scholar] [CrossRef]
- Cai, Z.; Chen, G.; Zeng, Y.; Dong, X.; Li, Z.; Huang, Y.; Xin, F.; Qiu, L.; Xu, H.; Zhang, W.; et al. Comprehensive Liquid Profiling of Circulating Tumor DNA and Protein Biomarkers in Long-Term Follow-Up Patients with Hepatocellular Carcinoma. Clin. Cancer Res. 2019, 25, 5284–5294. [Google Scholar] [CrossRef]
- Oh, C.R.; Kong, S.Y.; Im, H.S.; Kim, H.J.; Kim, M.K.; Yoon, K.A.; Cho, E.H.; Jang, J.H.; Lee, J.; Kang, J.; et al. Genome-wide copy number alteration and VEGFA amplification of circulating cell-free DNA as a biomarker in advanced hepatocellular carcinoma patients treated with Sorafenib. BMC Cancer 2019, 19, 292. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Watt, G.P.; Stevenson, H.L.; Calderone, T.L.; Fisher-Hoch, S.P.; Ye, Y.; Wu, X.; Vierling, J.M.; Beretta, L. Telomerase reverse transcriptase mutations in plasma DNA in patients with hepatocellular carcinoma or cirrhosis: Prevalence and risk factors. Hepatol. Commun. 2018, 2, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Oversoe, S.K.; Clement, M.S.; Pedersen, M.H.; Weber, B.; Aagaard, N.K.; Villadsen, G.E.; Grønbæk, H.; Hamilton-Dutoit, S.J.; Sorensen, B.S.; Kelsen, J. TERT promoter mutated circulating tumor DNA as a biomarker for prognosis in hepatocellular carcinoma. Scand. J. Gastroenterol. 2020, 55, 1433–1440. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Eun, J.W.; Choi, J.H.; Woo, H.G.; Cho, H.J.; Ahn, H.R.; Suh, C.W.; Baek, G.O.; Cho, S.W.; Cheong, J.Y. MLH1 single-nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma. Sci. Rep. 2020, 10, 17862. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.; Kinugasa, H.; Nouso, K.; Yamamoto, S.; Terasawa, H.; Onishi, Y.; Oyama, A.; Adachi, T.; Wada, N.; Sakata, M.; et al. Prediction of the prognosis of advanced hepatocellular carcinoma by TERT promoter mutations in circulating tumor DNA. J. Gastroenterol. Hepatol. 2021, 36, 1118–1125. [Google Scholar] [CrossRef]
- Shen, T.; Li, S.F.; Wang, J.L.; Zhang, T.; Zhang, S.; Chen, H.T.; Xiao, Q.Y.; Ren, W.H.; Liu, C.; Peng, B.; et al. TP53 R249S mutation detected in circulating tumour DNA is associated with Prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int. 2020, 40, 2834–2847. [Google Scholar] [CrossRef]
- Chan, K.C.; Jiang, P.; Chan, C.W.; Sun, K.; Wong, J.; Hui, E.P.; Chan, S.L.; Chan, W.C.; Hui, D.S.; Ng, S.S.; et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 18761–18768. [Google Scholar] [CrossRef]
- Hann, H.W.; Jain, S.; Park, G.; Steffen, J.D.; Song, W.; Su, Y.H. Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma. Hepatoma Res. 2017, 3, 105–111. [Google Scholar] [CrossRef]
- Wang, J.; Huang, A.; Wang, Y.P.; Yin, Y.; Fu, P.Y.; Zhang, X.; Zhou, J. Circulating tumor DNA correlates with microvascular invasion and predicts tumor recurrence of hepatocellular carcinoma. Ann. Transl. Med. 2020, 8, 237. [Google Scholar] [CrossRef]
- Long, G.; Fang, T.; Su, W.; Mi, X.; Zhou, L. The prognostic value of postoperative circulating cell-free DNA in operable hepatocellular carcinoma. Scand. J. Gastroenterol. 2020, 55, 1441–1446. [Google Scholar] [CrossRef]
- Ono, A.; Fujimoto, A.; Yamamoto, Y.; Akamatsu, S.; Hiraga, N.; Imamura, M.; Kawaoka, T.; Tsuge, M.; Abe, H.; Hayes, C.N.; et al. Circulating Tumor DNA Analysis for Liver Cancers and Its Usefulness as a Liquid Biopsy. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 516–534. [Google Scholar] [CrossRef] [PubMed]
- Lehmann-Werman, R.; Magenheim, J.; Moss, J.; Neiman, D.; Abraham, O.; Piyanzin, S.; Zemmour, H.; Fox, I.; Dor, T.; Grompe, M.; et al. Monitoring liver damage using hepatocyte-specific methylation markers in cell-free circulating DNA. JCI Insight 2018, 3, e120687. [Google Scholar] [CrossRef]
- Ng, H.I.; Sun, L.Y.; Zhu, Z.J. Detecting Graft-Derived Cell-Free DNA Through Amplification Refractory Mutation System Polymerase Chain Reaction in Living-Donor Liver Transplantation: Report of 2 Cases. Transplant. Proc. 2019, 51, 820–822. [Google Scholar] [CrossRef]
- Macher, H.C.; Suárez-Artacho, G.; Jiménez-Arriscado, P.; Álvarez-Gómez, S.; García-Fernández, N.; Guerrero, J.M.; Molinero, P.; Trujillo-Arribas, E.; Gómez-Bravo, M.A.; Rubio, A. Evaluation of the State of Transplanted Liver Health by Monitoring of Organ-Specific Genomic Marker in Circulating DNA from Receptor. Adv. Exp. Med. Biol. 2016, 924, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.; Bierau, S.; Balzer, S.; Andag, R.; Kanzow, P.; Schmitz, J.; Gaedcke, J.; Moerer, O.; Slotta, J.E.; Walson, P.; et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin. Chem. 2013, 59, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Schütz, E.; Fischer, A.; Beck, J.; Harden, M.; Koch, M.; Wuensch, T.; Stockmann, M.; Nashan, B.; Kollmar, O.; Matthaei, J.; et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study. PLoS Med. 2017, 14, e1002286. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.I.; Zhu, X.; Xuan, L.; Long, Y.; Mao, Y.; Shi, Y.; Sun, L.; Liang, B.; Scaglia, F.; Choy, K.W.; et al. Analysis of fragment size distribution of cell-free DNA: A potential non-invasive marker to monitor graft damage in living-related liver transplantation for inborn errors of metabolism. Mol. Genet. Metab. 2019, 127, 45–50. [Google Scholar] [CrossRef]
- Ng, H.I.; Sun, L.Y.; Zhu, Z.J. Application of graft-derived cell-free DNA in ornithine transcarbamylase deficiency patient after living donor liver transplantation: Two case reports. Medicine 2018, 97, e13843. [Google Scholar] [CrossRef]
- Lo, Y.M.; Tein, M.S.; Pang, C.C.; Yeung, C.K.; Tong, K.L.; Hjelm, N.M. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet 1998, 351, 1329–1330. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Cohen, J.; Li, L.; Hong, W.; Christie, M.; Wong, H.L.; Kosmider, S.; Wong, R.; Thomson, B.; et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: A prospective cohort study. PLoS Med. 2021, 18, e1003620. [Google Scholar] [CrossRef]
- Goh, S.K.; Do, H.; Testro, A.; Pavlovic, J.; Vago, A.; Lokan, J.; Jones, R.M.; Christophi, C.; Dobrovic, A.; Muralidharan, V. The Measurement of Donor-Specific Cell-Free DNA Identifies Recipients With Biopsy-Proven Acute Rejection Requiring Treatment After Liver Transplantation. Transplant. Direct 2019, 5, e462. [Google Scholar] [CrossRef] [PubMed]
| Study | Number of HCC Patients | Technique | Biomarkers | Outcomes |
|---|---|---|---|---|
| Liao et al., 2016 [104] | 41 | Illumina miSeq NGS | Hotspot mutations of TP53, TERT, and CTNNB1 |
|
| Cai et al., 2019 [105] | 34 | Target sequencing and low-coverage whole-genome sequencing | ctDNA (harboring copy number variants [CNV] or single-nucleotide variants [SNV]) |
|
| Oh et al., 2019 [106] | 151 HCC, 14 healthy controls | Low-depth whole-genome sequencing | CNV, VEGF amplification |
|
| Jiao et al., 2018 [107] | 218 HCC, 81 cirrhotic | Droplet digital PCR (ddPCR) | TERT C228T and C250T promoter mutations |
|
| Oversoe et al., 2020 [108] | 95 HCC, 45 liver cirrhotic | ddPCR | TERT C228T mutation |
|
| Kim et al., 2020 [109] | 107 HCC | Target deep sequencing, ddPCR | Deep sequencing of SNVs in 69 genes |
|
| Hirai et al., 2021 [110] | 130 HCC | ddPCR | TERT promoter mutation |
|
| Shen et al., 2020 [111] | 895 HCC | ddPCR, genomic sequencing | TP53 mutations (recurrent missense mutations R100L, V157F, A159P, and R249S) |
|
| Chan et al., 2013 [112] | 26 | Massively parallel bisulfite sequencing | Methylation density (MD) per 1 million base pairs (Mb) [bin], copy number aberrations (CNA) |
|
| Tie et al., 2021 [125] | 54 patients with colorectal cancer liver metastasis (CRLM) | Safe-sequencing (Safe-SeqS) assay | Somatic mutations in 15 genes recurrently mutated in CRC (SMAD4, TP53, AKT1, APC, BRAF, CTNNB1, ERBB3, FBXW7, HRAS, KRAS, NRAS, PIK3CA, PPP2R1A, RNF43, and POLE). |
|
| Hann et al., 2017 [113] | 10 HCC | Bisulfite treatment of DNA and quantitative PCR, magnetic resonance imaging (MRI) | TP53 249T mutations and aberrant methylation of RASSF1A and GSTP1 genes |
|
| Wang et al., 2020 [114] | 81 HCC | ddPCR | Four hotspot mutations: TP53-rs28934571 (c.747G > T), TRETrs1242535815 (c.1-124C > T), CTNNB1-rs121913412 (c.121A > G), and CTNNB1-rs121913407 (c.133T > C) |
|
| Ono et al., 2015 [116] | 46 HCC | Whole exome sequencing, PCR | ctDNA, a-fetoprotein (AFP), and des-g-carboxy prothrombin (DCP) |
|
| Long et al., 2020 [115] | 82 HCC patients | Fluorometric Qubit dsDNA BR assay kit | Cell-free dsDNA |
|
| Lo et al., 1998 [124] | 8 female transplant patients | PCR, gel electrophoresis | Y-chromosome specific genetic sequences |
|
| Lehmann-Werman et al., 2018 [117] | 18 transplant patients | Bisulfite conversion, PCR, and massively parallel sequencing | 3 genomic loci, adjacent to the ITIH4, IGF2R, and VTN genes, which were unmethylated in the liver compared with other tissues and cell types. |
|
| Ng et al., 2019 [118] | 2 transplant patients diagnosed with propionic acidemia | Amplification refractory mutation system PCR (ARMS-PCR) | Graft-derived cell-free DNA (Gcf-DNA), liver enzymes (alanine transaminase [ALT], aspartate transaminase [AST]) |
|
| Macher et al., 2016 [119] | 17 transplant patients | RT-PCR | Rh gene |
|
| Beck et al., 2013 [120] | Stable liver (n = 10), heart (n = 8), and kidney (n = 9) transplant recipients, and seven additional patients directly after transplantation | ddPCR | SNPs from graft-derived cell-free DNA (GcfDNA) |
|
| Schutz et al., 2017 [121] | 115 liver transplant patients | ddPCR | SNP loci with known high population minor allelic frequency |
|
| Ng et al., 2019 [122] | 11 liver transplant recipients | Y-chromosome capture methodology and sequencing read lengths | GcfDNA was defined by DNA fragment sizes (105–145 bp, 160–170 bp). The ratio of short fragments/long fragments (S/L) were calculated. |
|
| Goh et al., 2019 [126] | 20 liver transplant recipients | ddPCR | Deletion/insertion polymorphisms |
|
| Ng et al., 2018 [123] | Two liver transplant recipients | PCR of Y-chromosome specific genes | ALT, AST, and GcfDNA |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reddy, T.; Esmail, A.; Chang, J.C.; Ghobrial, R.M.; Abdelrahim, M. Utility of Cell-Free DNA Detection in Transplant Oncology. Cancers 2022, 14, 743. https://doi.org/10.3390/cancers14030743
Reddy T, Esmail A, Chang JC, Ghobrial RM, Abdelrahim M. Utility of Cell-Free DNA Detection in Transplant Oncology. Cancers. 2022; 14(3):743. https://doi.org/10.3390/cancers14030743
Chicago/Turabian StyleReddy, Tejaswini, Abdullah Esmail, Jenny C. Chang, Rafik Mark Ghobrial, and Maen Abdelrahim. 2022. "Utility of Cell-Free DNA Detection in Transplant Oncology" Cancers 14, no. 3: 743. https://doi.org/10.3390/cancers14030743
APA StyleReddy, T., Esmail, A., Chang, J. C., Ghobrial, R. M., & Abdelrahim, M. (2022). Utility of Cell-Free DNA Detection in Transplant Oncology. Cancers, 14(3), 743. https://doi.org/10.3390/cancers14030743








