Exosomal Proteins and Lipids as Potential Biomarkers for Lung Cancer Diagnosis, Prognosis, and Treatment
Abstract
:Simple Summary
Abstract
1. Background
2. Tumor Microenvironment
3. Exosome Biogenesis, Secretion and Uptake
4. Purification and Characterization of Exosomes
5. Protein Composition of Lung Cancer Exosomes
6. Protein Function of Lung Cancer Exosomes
7. Lipid Composition and Function of Lung Cancer Exosomes
8. The Importance of CSCs and CSC-Derived Exosomes
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Maia, J.; Caja, S.; Moraes, M.C.S.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ren, L.; Li, S.; Li, W.; Zheng, X.; Yang, Y.; Fu, W.; Yi, J.; Wang, J.; Du, G. The biology, function, and applications of exosomes in cancer. Acta Pharm. Sin. B 2021, 11, 2783–2797. [Google Scholar] [CrossRef] [PubMed]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobb, R.; Lima, L.G.; Möller, A. Exosomes: Key mediators of metastasis and pre-metastatic niche formation. Semin. Cell Dev. Biol. 2017, 67, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, X.; Shi, H.; Wu, L.; Qian, H.; Xu, W. Exosomes in cancer: Small particle, big player. J. Hematol. Oncol. 2015, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Wu, D.; Wu, P.; Chen, Z.; Huang, J. The cancer stem cell niche: Cross talk between cancer stem cells and their microenvironment. Tumor Biol. 2014, 35, 3945–3951. [Google Scholar] [CrossRef]
- Albini, A.; Bruno, A.; Gallo, C.; Pajardi, G.E.; Noonan, U.M.; Dallaglio, K. Cancer stem cells and the tumor microenvironment: Interplay in tumor heterogeneity. Connect. Tissue Res. 2015, 56, 414–425. [Google Scholar] [CrossRef] [Green Version]
- Mosquera-Heredia, M.I.; Morales, L.C.; Vidal, O.M.; Barceló, E.; Silvera-Redondo, C.; Vélez, J.I.; Garavito-Galofre, P. Exosomes: Potential Disease Biomarkers and New Therapeutic Targets. Biomedicines 2021, 9, 1061. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Vader, P.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Emerging targets for cancer therapy. Trends Mol. Med. 2014, 20, 385–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marleau, A.M.; Chen, C.-S.; Joyce, J.A.; Tullis, R.H. Exosome removal as a therapeutic adjuvant in cancer. J. Transl. Med. 2012, 10, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnsen, K.B.; Gudbergsson, J.M.; Skov, M.N.; Pilgaard, L.; Moos, T.; Duroux, M. A comprehensive overview of exosomes as drug delivery vehicles—Endogenous nanocarriers for targeted cancer therapy. Biochim. Biophys. Acta 2014, 1846, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Popowski, K.; Lutz, H.; Hu, S.; George, A.; Dinh, P.; Cheng, K. Exosome therapeutics for lung regenerative medicine. J. Extracell. Vesicles 2020, 9, 1785161. [Google Scholar] [CrossRef]
- Bray, E.R.; Oropallo, A.R.; Grande, D.A.; Kirsner, R.S.; Badiavas, E.V. Extracellular Vesicles as Therapeutic Tools for the Treatment of Chronic Wounds. Pharmaceutics 2021, 13, 1543. [Google Scholar] [CrossRef]
- Santos, P.; Almeida, F. Exosome-Based Vaccines: History, Current State, and Clinical Trials. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Sandúa, A.; Alegre, E.; González, Á. Exosomes in Lung Cancer: Actors and Heralds of Tumor Development. Cancers 2021, 13, 4330. [Google Scholar] [CrossRef]
- Wang, H.; Lu, Z.; Zhao, X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J. Hematol. Oncol. 2019, 12, 133. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Wang, Q.; Kornmann, M.; Tian, X.; Yang, Y. The Role of exosomes in pancreatic cancer from bench to clinical application: An updated review. Front. Oncol. 2021, 11, 153. [Google Scholar] [CrossRef] [PubMed]
- Mannavola, F.; Salerno, T.; Passarelli, A.; Tucci, M.; Internò, V.; Silvestris, F. Revisiting the Role of Exosomes in Colorectal Cancer: Where Are We Now? Front. Oncol. 2019, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Gu, J.; Jiang, P.; Qian, H.; Xu, W.; Zhang, X. Exosomes in gastric cancer: Roles, mechanisms, and applications. Mol. Cancer 2019, 18, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, W.; Wang, K.; Wu, Z.; Xu, B.; Chen, M. Current status of research on exosomes in general, and for the diagnosis and treatment of kidney cancer in particular. J. Exp. Clin. Cancer Res. 2021, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Georgantzoglou, N.; Pergaris, A.; Masaoutis, C.; Theocharis, S. Extracellular Vesicles as Biomarkers Carriers in Bladder Cancer: Diagnosis, Surveillance, and Treatment. Int. J. Mol. Sci. 2021, 22, 2744. [Google Scholar] [CrossRef] [PubMed]
- Lorenc, T.; Klimczyk, K.; Michalczewska, I.; Słomka, M.; Kubiak-Tomaszewska, G.; Olejarz, W. Exosomes in Prostate Cancer Diagnosis, Prognosis and Therapy. Int. J. Mol. Sci. 2020, 21, 2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Sun, C.; Huang, X.; Li, J.; Fu, Z.; Li, W.; Yin, Y. The Advancing Roles of Exosomes in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 731062. [Google Scholar] [CrossRef]
- Shimizu, A.; Sawada, K.; Kimura, T. Pathophysiological Role and Potential Therapeutic Exploitation of Exosomes in Ovarian Cancer. Cells 2020, 9, 814. [Google Scholar] [CrossRef] [Green Version]
- Acevedo-Sánchez, V.; Rodríguez-Hernández, R.; Aguilar-Ruíz, S.; Torres-Aguilar, H.; Romero-Tlalolini, M. Extracellular Vesicles in Cervical Cancer and HPV Infection. Membranes 2021, 11, 453. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, M.; Xie, F.; Lou, J.; Zhou, X.; Zhang, L.; Fang, M.; Zhou, F. Exosomes in head and neck cancer: Roles, mechanisms and applications. Cancer Lett. 2020, 494, 7–16. [Google Scholar] [CrossRef]
- Feng, K.; Ma, R.; Zhang, L.; Li, H.; Tang, Y.; Du, G.; Niu, D.; Yin, D. The Role of Exosomes in Thyroid Cancer and Their Potential Clinical Application. Front. Oncol. 2020, 10, 596132. [Google Scholar] [CrossRef]
- Wu, X.; Wang, X.; Wang, J.; Hao, Y.; Liu, F.; Wang, X.; Yang, L.; Lu, Z. The Roles of Exosomes as Future Therapeutic Agents and Diagnostic Tools for Glioma. Front. Oncol. 2021, 11, 733529. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-C.; Chang, Y.-A.; Chen, Y.-J.; Sung, H.-M.; Bogeski, I.; Su, H.-L.; Hsu, Y.-L.; Wang, H.-M.D. The Roles of Extracellular Vesicles in Malignant Melanoma. Cells 2021, 10, 2740. [Google Scholar] [CrossRef] [PubMed]
- Khalife, J.; Sanchez, J.F.; Pichiorri, F. Extracellular Vesicles in Hematological Malignancies: From Biomarkers to Therapeutic Tools. Diagnostics 2020, 10, 1065. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.S.; Trindade, D.; Vaz, M.; Campelo, I.; Almeida, M.; Trigo, G.; da Cruz e Silva, O.A.B.; Henriques, A.G. Diagnostic and therapeutic potential of exosomes in Alzheimer’s disease. J. Neurochem. 2021, 156, 162–181. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Sun, T.; An, J.; Wen, L.; Liu, F.; Bu, Z.; Cui, Y.; Feng, J. Potential Roles of Exosomes in Parkinson’s Disease: From Pathogenesis, Diagnosis, and Treatment to Prognosis. Front. Cell Dev. Biol. 2020, 8, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ananbeh, H.; Vodicka, P.; Skalnikova, H.K. Emerging Roles of Exosomes in Huntington’s Disease. Int. J. Mol. Sci. 2021, 22, 4085. [Google Scholar] [CrossRef]
- Ferrara, D.; Pasetto, L.; Bonetto, V.; Basso, M. Role of Extracellular Vesicles in Amyotrophic Lateral Sclerosis. Front. Neurosci. 2018, 12, 574. [Google Scholar] [CrossRef]
- Saeedi, S.; Israel, S.; Nagy, C.; Turecki, G. The emerging role of exosomes in mental disorders. Transl. Psychiatry 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, X.; Guo, J.; Cao, C.; Zhang, Z.; Wang, B.; Zhang, L.; Shen, D.; Lim, K.; Woodfield, T.; et al. Small but significant: Insights and new perspectives of exosomes in cardiovascular disease. J. Cell. Mol. Med. 2020, 24, 8291–8303. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.; Luo, S.; Xiao, Y.; Xia, Y.; Li, X.; Huang, G.; Xie, Z.; Zhou, Z. Emerging Roles of Exosomes in T1DM. Front. Immunol. 2020, 11, 593348. [Google Scholar] [CrossRef]
- Liu, J.; Sun, X.; Zhang, F.-L.; Jin, H.; Yan, X.-L.; Huang, S.; Guo, Z.-N.; Yang, Y. Clinical Potential of Extracellular Vesicles in Type 2 Diabetes. Front. Endocrinol. 2021, 11, 596811. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.; Joo, H.; Hong, E.; Lee, H.; Lee, J. Therapeutic Application of Exosomes in Inflammatory Diseases. Int. J. Mol. Sci. 2021, 22, 1144. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 27 December 2021).
- World Health Organization. Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 27 December 2021).
- Liu, S.; Zhan, Y.; Luo, J.; Feng, J.; Lu, J.; Zheng, H.; Wen, Q.; Fan, S. Roles of exosomes in the carcinogenesis and clinical therapy of non-small cell lung cancer. Biomed. Pharmacother. 2019, 111, 338–346. [Google Scholar] [CrossRef]
- Kim, H.; Kim, D.W.; Cho, J.-Y. Exploring the key communicator role of exosomes in cancer microenvironment through proteomics. Proteome Sci. 2019, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Xu, X.; Qian, Z.; Zhang, C.; Niu, Y.; Wang, Z.; Sun, J.; Zhang, X.; Yu, Y. The biological functions and clinical applications of exosomes in lung cancer. Cell. Mol. Life Sci. 2019, 76, 4613–4633. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020, 5, 1–14. [Google Scholar] [CrossRef]
- Li, M.-Y.; Liu, L.-Z.; Dong, M. Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol. Cancer 2021, 20, 1–22. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, C.; Du, T.; Gabriel, A.N.A.; Wang, X.; Li, X.; Sun, L.; Wang, N.; Jiang, X.; Zhang, Y. Progress of exosomes in the diagnosis and treatment of lung cancer. Biomed. Pharmacother. 2021, 134, 111111. [Google Scholar] [CrossRef]
- Qi, R.; Zhao, Y.; Guo, Q.; Mi, X.; Cheng, M.; Hou, W.; Zheng, H.; Hua, B. Exosomes in the lung cancer microenvironment: Biological functions and potential use as clinical biomarkers. Cancer Cell Int. 2021, 21, 333. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Liu, X.; Shao, X.; Feng, T.; Xu, J.; Wang, Q.; Hua, S. The role of exosomes in lung cancer metastasis and clinical applications: An updated review. J. Transl. Med. 2021, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yin, Z.; Fan, J.; Zhang, S.; Yang, W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct. Target. Ther. 2019, 4, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, C.; Meiners, S.; Lukas, C.; Stathopoulos, G.T.; Chen, J. Role of exosomal microRNAs in lung cancer biology and clinical applications. Cell Prolif. 2020, 53, e12828. [Google Scholar] [CrossRef]
- Wu, J.; Shen, Z. Exosomal miRNAs as biomarkers for diagnostic and prognostic in lung cancer. Cancer Med. 2020, 9, 6909–6922. [Google Scholar] [CrossRef]
- He, X.; Park, S.; Chen, Y.; Lee, H. Extracellular Vesicle-Associated miRNAs as a Biomarker for Lung Cancer in Liquid Biopsy. Front. Mol. Biosci. 2021, 8, 630718. [Google Scholar] [CrossRef]
- Smolarz, M.; Widlak, P. Serum Exosomes and Their miRNA Load—A Potential Biomarker of Lung Cancer. Cancers 2021, 13, 1373. [Google Scholar] [CrossRef]
- Fan, T.; Sun, N.; He, J. Exosome-Derived LncRNAs in Lung Cancer. Front. Oncol. 2020, 10, 1728. [Google Scholar] [CrossRef]
- Reclusa, P.; Taverna, S.; Pucci, M.; Durendez, E.; Calabuig, S.; Manca, P.; Serrano, M.J.; Sober, L.; Pauwels, P.; Russo, A.; et al. Exosomes as diagnostic and predictive biomarkers in lung cancer. J. Thorac. Dis. 2017, 9 (Suppl. S13), S1373–S1382. [Google Scholar] [CrossRef] [Green Version]
- Masaoutis, C.; Mihailidou, C.; Tsourouflis, G.; Theocharis, S. Exosomes in lung cancer diagnosis and treatment. From the translating research into future clinical practice. Biochimie 2018, 151, 27–36. [Google Scholar] [CrossRef]
- Trappe, A.; Donnelly, S.C.; McNally, P.; Coppinger, J.A. Role of extracellular vesicles in chronic lung disease. Thorax 2021, 76, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Qing, B.; Wang, W.; Gu, L.; Chen, H.; Yuan, Y. Formation, contents, functions of exosomes and their potential in lung cancer diagnostics and therapeutics. Thorac. Cancer 2021, 12, 3088–3100. [Google Scholar] [CrossRef] [PubMed]
- Frawley, T.; Piskareva, O. Extracellular Vesicle Dissemination of Epidermal Growth Factor Receptor and Ligands and Its Role in Cancer Progression. Cancers 2020, 12, 3200. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin. Chem. 2016, 74, 103–141. [Google Scholar] [CrossRef] [Green Version]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, Z.Z. Mechanisms that mediate stem cell self-renewal and differentiation. J. Cell. Biochem. 2008, 103, 709–718. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [Green Version]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef]
- Hwang, I. Cell-cell communication via extracellular membrane vesicles and its role in the immune response. Mol. Cells 2013, 36, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. General Principles of Cell Communication. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. Available online: https://www.ncbi.nlm.nih.gov/books/NBK26813/ (accessed on 27 December 2021).
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, M.-F.; Nelson, C.M. Intercellular Communication, the Tumor Microenvironment, and Tumor Progression. In Intercellular Communication in Cancer; Kandouz, M., Ed.; Springer: Dordrecht, The Netherlands, 2015; pp. 343–362. [Google Scholar]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Suchorska, W.; Lach, M. The role of exosomes in tumor progression and metastasis (Review). Oncol. Rep. 2016, 35, 1237–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuffers, S.; Wegner, C.S.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, H.T.; Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011, 12, 517–533. [Google Scholar] [CrossRef]
- Parton, R.G.; Simons, K. The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 2007, 8, 185–194. [Google Scholar] [CrossRef]
- Swanson, J.A. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 2008, 9, 639–649. [Google Scholar] [CrossRef] [Green Version]
- Martens, S.; McMahon, H.T. Mechanisms of membrane fusion: Disparate players and common principles. Nat. Rev. Mol. Cell Biol. 2008, 9, 543–556. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017, 18, 1153. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.-S.; Kim, D.-K.; Kim, Y.-K.; Gho, Y.S. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics 2013, 13, 1554–1571. [Google Scholar] [CrossRef]
- Ferguson, S.; Nguyen, J. Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. J. Control. Release 2016, 228, 179–190. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [Green Version]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 1–18. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Green, T.M.; Alpaugh, M.L.; Barsky, S.H.; Rappa, G.; Lorico, A. Breast Cancer-Derived Extracellular Vesicles: Characterization and Contribution to the Metastatic Phenotype. BioMed Res. Int. 2015, 2015, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, H.; Greening, D.; Barnes, T.W.; Lim, J.W.; Tauro, B.J.; Rai, A.; Xu, R.; Adda, C.; Mathivanan, S.; Zhao, W.; et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 2013, 13, 1672–1686. [Google Scholar] [CrossRef] [PubMed]
- Welton, J.L.; Khanna, S.; Giles, P.J.; Brennan, P.; Brewis, I.A.; Staffurth, J.; Mason, M.D.; Clayton, A. Proteomics Analysis of Bladder Cancer Exosomes. Mol. Cell. Proteom. 2010, 9, 1324–1338. [Google Scholar] [CrossRef] [Green Version]
- Duijvesz, D.; Burnum-Johnson, K.E.; Gritsenko, M.A.; Hoogland, A.M.; Vredenbregt-van den Berg, M.S.; Willemsen, R.; Luider, T.; Paša-Tolić, L.; Jenster, G. Proteomic Profiling of Exosomes Leads to the Identification of Novel Biomarkers for Prostate Cancer. PLoS ONE 2013, 8, e82589. [Google Scholar] [CrossRef] [Green Version]
- Liang, B.; Peng, P.; Chen, S.; Li, L.; Zhang, M.; Cao, D.; Yang, J.; Li, H.; Gui, T.; Li, X.; et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J. Proteom. 2013, 80, 171–182. [Google Scholar] [CrossRef]
- Anderson, J.D.; Johansson, H.J.; Graham, C.S.; Vesterlund, M.; Pham, M.T.; Bramlett, C.S.; Montgomery, E.N.; Mellema, M.S.; Bardini, R.L.; Contreras, Z.; et al. Comprehensive Proteomic Analysis of Mesenchymal Stem Cell Exosomes Reveals Modulation of Angiogenesis via Nuclear Factor-KappaB Signaling. Stem Cells 2016, 34, 601–613. [Google Scholar] [CrossRef] [Green Version]
- Grange, C.; Tapparo, M.; Collino, F.; Vitillo, L.; Damasco, C.; Deregibus, M.C.; Tetta, C.; Bussolati, B.; Camussi, G. Microvesicles Released from Human Renal Cancer Stem Cells Stimulate Angiogenesis and Formation of Lung Premetastatic Niche. Cancer Res. 2011, 71, 5346–5356. [Google Scholar] [CrossRef] [Green Version]
- Lindoso, R.S.; Collino, F.; Camussi, G. Extracellular vesicles derived from renal cancer stem cells induce a pro-tumorigenic phenotype in mesenchymal stromal cells. Oncotarget 2015, 6, 7959–7969. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Guo, J.; Yang, M.; Zhu, X.; Cao, X. Chemokine-Containing Exosomes Are Released from Heat-Stressed Tumor Cells via Lipid Raft-Dependent Pathway and Act as Efficient Tumor Vaccine. J. Immunol. 2011, 186, 2219–2228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.-P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Investig. 2010, 120, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, S.; Zhu, R.; Li, H.; Han, Q.; Zhao, R.C. Lung tumor exosomes induce a pro-inflammatory phenotype in mesenchymal stem cells via NFκB-TLR signaling pathway. J. Hematol. Oncol. 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanteloup, G.; Cordonnier, M.; Isambert, N.; Bertaut, A.; Hervieu, A.; Hennequin, A.; Luu, M.; Zanetta, S.; Coudert, B.; Bengrine, L.; et al. Monitoring HSP70 exosomes in cancer patients’ follow up: A clinical prospective pilot study. J. Extracell. Vesicles 2020, 9, 1766192. [Google Scholar] [CrossRef]
- Al-Nedawi, K.; Meehan, B.; Kerbel, R.S.; Allison, A.C.; Rak, J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc. Natl. Acad. Sci. USA 2009, 106, 3794–3799. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.-H.; Li, Y.; Zhang, J.; Rong, J.; Ye, S. Epidermal Growth Factor Receptor-Containing Exosomes Induce Tumor-Specific Regulatory T Cells. Cancer Investig. 2013, 31, 330–335. [Google Scholar] [CrossRef]
- Yamashita, T.; Kamada, H.; Kanasaki, S.; Maeda, Y.; Nagano, K.; Abe, Y.; Yoshioka, Y.; Tsutsumi, Y.; Katayama, S.; Inoue, M.; et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Die Pharm. 2013, 68, 969–973. [Google Scholar]
- Xie, X.; Nie, H.; Zhou, Y.; Lian, S.; Mei, H.; Lu, Y.; Dong, H.; Li, F.; Li, T.; Li, B.; et al. Eliminating blood oncogenic exosomes into the small intestine with aptamer-functionalized nanoparticles. Nat. Commun. 2019, 10, 5476. [Google Scholar] [CrossRef] [Green Version]
- Taverna, S.; Pucci, M.; Giallombardo, M.; Di Bella, M.A.; Santarpia, M.; Reclusa, P.; Gil-Bazo, I.; Rolfo, C.; Alessandro, R. Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ueda, K.; Ishikawa, N.; Tatsuguchi, A.; Saichi, N.; Fujii, R.; Nakagawa, H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014, 4, srep06232. [Google Scholar] [CrossRef]
- Jakobsen, K.R.; Paulsen, B.S.; Bæk, R.; Varming, K.; Sorensen, B.; Jørgensen, M.M. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J. Extracell. Vesicles 2015, 4, 26659. [Google Scholar] [CrossRef] [PubMed]
- Sandfeld-Paulsen, B.; Jakobsen, K.R.; Bæk, R.; Folkersen, B.H.; Rasmussen, T.R.; Meldgaard, P.; Varming, K.; Jørgensen, M.M.; Sorensen, B. Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandfeld-Paulsen, B.; Aggerholm-Pedersen, N.; Baek, R.; Jakobsen, K.R.; Meldgaard, P.; Folkersen, B.H.; Rasmussen, T.R.; Varming, K.; Jørgensen, M.M.; Sorensen, B.S. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 2016, 10, 1595–1602. [Google Scholar] [CrossRef] [Green Version]
- Park, J.O.; Choi, D.-Y.; Choi, D.-S.; Kim, H.J.; Kang, J.W.; Jung, J.H.; Lee, J.H.; Kim, J.; Freeman, M.R.; Lee, K.Y.; et al. Identification and characterization of proteins isolated from microvesicles derived from human lung cancer pleural effusions. Proteomics 2013, 13, 2125–2134. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.J.; Fondrie, W.; Yang, A.; Mao, L. Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes. J. Proteom. 2016, 133, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xia, Z.; Shang, Z.; Sun, K.; Niu, X.; Qian, L.; Fan, L.-Y.; Cao, C.-X.; Xiao, H. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci. Rep. 2016, 6, 24669. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Liu, S.; Qiao, Z.; Shang, Z.; Xia, Z.; Niu, X.; Qian, L.; Zhang, Y.; Fan, L.; Cao, C.-X.; et al. Systematic comparison of exosomal proteomes from human saliva and serum for the detection of lung cancer. Anal. Chim. Acta 2017, 982, 84–95. [Google Scholar] [CrossRef]
- Sun, Y.; Huo, C.; Qiao, Z.; Shang, Z.; Uzzaman, A.; Liu, S.; Jiang, X.; Fan, L.Y.; Ji, L.; Guan, X.; et al. Comparative Proteomic Analysis of Exosomes and Microvesicles in Human Saliva for Lung Cancer. J. Proteome Res. 2018, 17, 1101–1107. [Google Scholar] [CrossRef]
- Qiao, Z.; Zhang, Y.; Ge, M.; Liu, S.; Jiang, X.; Shang, Z.; Liu, H.; Cao, C.; Xiao, H. Cancer Cell Derived Small Extracellular Vesicles Contribute to Recipient Cell Metastasis Through Promoting HGF/c-Met Pathway. Mol. Cell. Proteom. 2019, 18, 1619–1629. [Google Scholar] [CrossRef]
- Mo, F.; Xu, Y.; Zhang, J.; Zhu, L.; Wang, C.; Chu, X.; Pan, Y.; Bai, Y.; Shao, C.; Zhang, J. Effects of Hypoxia and Radiation-Induced Exosomes on Migration of Lung Cancer Cells and Angiogenesis of Umbilical Vein Endothelial Cells. Radiat. Res. 2020, 194, 71–80. [Google Scholar] [CrossRef]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yi, J.; Chen, X.; Zhang, Y.; Xu, M.; Yang, Z. The regulation of cancer cell migration by lung cancer cell-derived exosomes through TGF-β and IL-10. Oncol. Lett. 2016, 11, 1527–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.A.; Barger, J.F.; Lovat, F.; Gao, M.; Otterson, G.A.; Nana-Sinkam, P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget 2016, 7, 54852–54866. [Google Scholar] [CrossRef]
- Vykoukal, J.; Sun, N.; Aguilar-Bonavides, C.; Katayama, H.; Tanaka, I.; Fahrmann, J.F.; Capello, M.; Fujimoto, J.; Aguilar, M.; Wistuba, I.I.; et al. Plasma-derived extracellular vesicle proteins as a source of biomarkers for lung adenocarcinoma. Oncotarget 2017, 8, 95466–95480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Qiu, X.; Li, X.; Fan, H.; Zhang, F.; Lv, T.; Song, Y. Expression profiles and clinical value of plasma exosomal Tim-3 and Galectin-9 in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018, 498, 409–415. [Google Scholar] [CrossRef]
- Wang, N.; Song, X.; Liu, L.; Niu, L.; Wang, X.; Song, X.; Xie, L. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 2018, 109, 1701–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, L.; Song, X.; Wang, N.; Xue, L.; Song, X.; Xie, L. Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci. 2019, 110, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Pan, D.; Chen, J.; Feng, C.; Wu, W.; Wang, Y.; Tong, J.; Zhou, D. Preferential Localization of MUC1 Glycoprotein in Exosomes Secreted by Non-Small Cell Lung Carcinoma Cells. Int. J. Mol. Sci. 2019, 20, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, W.; Ma, W.; Wei, S.; Li, Q.; Liu, R.; Carney, R.P.; Yang, K.; Lee, J.; Nyugen, A.; Yoneda, K.Y.; et al. High-affinity peptide ligand LXY30 for targeting α3β1 integrin in non-small cell lung cancer. J. Hematol. Oncol. 2019, 12, 56. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, Y.; Qiu, F.; Qiu, Z. Proteomic identification of exosomal LRG1: A potential urinary biomarker for detecting NSCLC. Electrophoresis 2011, 32, 1976–1983. [Google Scholar] [CrossRef]
- Li, Z.; Zeng, C.; Nong, Q.; Long, F.; Liu, J.; Mu, Z.; Chen, B.; Wu, D.; Wu, H. Exosomal Leucine-Rich-Alpha2-Glycoprotein 1 Derived from Non-Small-Cell Lung Cancer Cells Promotes Angiogenesis via TGF-β Signal Pathway. Mol. Ther. Oncolytics 2019, 14, 313–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Yin, Z.; Yang, L.; Fan, J.; Xu, J.; Jin, Y.; Yu, J.; Zhang, D.; Yang, G. Smoking Induced Extracellular Vesicles Release and Their Distinct Properties in Non-Small Cell Lung Cancer. J. Cancer 2019, 10, 3435–3443. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Lee, J.-E.; Sung, K.J.; Sung, Y.H.; Pack, C.-G.; Jung, M.-K.; Han, B.; et al. Exosomal PD-L1 promotes tumor growth through immune escape in non-small cell lung cancer. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Chen, M.; Gu, J.; Niu, K.; Zhao, X.; Zheng, L.; Xu, Z.; Yu, Y.; Li, F.; Meng, L.; et al. Novel Biomarkers of Dynamic Blood PD-L1 Expression for Immune Checkpoint Inhibitors in Advanced Non-Small-Cell Lung Cancer Patients. Front. Immunol. 2021, 12, 665133. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, C.; Zhi, C.; Liang, W.; Wang, X.; Chen, X.; Lv, T.; Shen, Q.; Song, Y.; Lin, D.; et al. Clinical significance of PD-L1 expression in serum-derived exosomes in NSCLC patients. J. Transl. Med. 2019, 17, 3551. [Google Scholar] [CrossRef] [Green Version]
- Shimada, Y.; Matsubayashi, J.; Kudo, Y.; Maehara, S.; Takeuchi, S.; Hagiwara, M.; Kakihana, M.; Ohira, T.; Nagao, T.; Ikeda, N. Serum-derived exosomal PD-L1 expression to predict anti-PD-1 response and in patients with non-small cell lung cancer. Sci. Rep. 2021, 11, 7831. [Google Scholar] [CrossRef]
- Wu, F.; Gu, Y.; Kang, B.; Heskia, F.; Pachot, A.; Bonneville, M.; Wei, P.; Liang, J. PD-L1 detection on circulating tumor-derived extracellular vesicles (T-EVs) from patients with lung cancer. Transl. Lung Cancer Res. 2021, 10, 2441–2451. [Google Scholar] [CrossRef]
- Yoh, K.E.; Lowe, C.J.; Mahajan, S.; Suttmann, R.; Nguy, T.; Reichelt, M.; Yang, J.; Melendez, R.; Li, Y.; Molinero, L.; et al. Enrichment of circulating tumor-derived extracellular vesicles from human plasma. J. Immunol. Methods 2021, 490, 112936. [Google Scholar] [CrossRef]
- Choi, E.-S.; Faruque, H.; Kim, J.-H.; Kim, K.; Choi, J.; Kim, B.; Kim, B.; Kim, Y.; Woo, M.; Park, J.; et al. CD5L as an Extracellular Vesicle-Derived Biomarker for Liquid Biopsy of Lung Cancer. Diagnostics 2021, 11, 620. [Google Scholar] [CrossRef]
- Jeong, H.; Choi, B.H.; Park, J.; Jung, J.-H.; Shin, H.; Kang, K.-W.; Quan, Y.H.; Yu, J.; Park, J.-H.; Park, Y.; et al. GCC2 as a New Early Diagnostic Biomarker for Non-Small Cell Lung Cancer. Cancers 2021, 13, 5482. [Google Scholar] [CrossRef]
- Dong, Q.; Dong, L.; Liu, S.; Kong, Y.; Zhang, M.; Wang, X. Tumor-Derived Exosomal eIF4E as a Biomarker for Survival Prediction in Patients with Non-Small Cell Lung Cancer. Med. Sci. Monit. 2020, 26, e923210. [Google Scholar] [CrossRef]
- Jørgensen, M.; Bæk, R.; Pedersen, S.; Søndergaard, E.K.; Kristensen, S.R.; Varming, K. Extracellular Vesicle (EV) Array: Microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J. Extracell. Vesicles 2013, 2, 20920. [Google Scholar] [CrossRef] [PubMed]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Gocher, A.M.; Workman, C.J.; Vignali, D.A.A. Interferon-γ: Teammate or opponent in the tumour microenvironment? Nat. Rev. Immunol. 2021, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Yoshioka, Y.; Katsuda, T.; Ono, M.; Ochiya, T. Exosome in disease biology, diagnosis, and therapy. Inflamm. Regen. 2014, 34, 233–239. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [Green Version]
- Chairoungdua, A.; Smith, D.L.; Pochard, P.; Hull, M.; Caplan, M.J. Exosome release of β-catenin: A novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 2010, 190, 1079–1091. [Google Scholar] [CrossRef] [Green Version]
- Böker, K.O.; Diaz, N.L.; Ferreira, R.R.; Schiller, L.; Schneider, S.; Gruber, J. The Impact of the CD9 Tetraspanin on Lentivirus Infectivity and Exosome Secretion. Mol. Ther. 2018, 26, 634–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurwitz, S.N.; Conlon, M.M.; Rider, M.A.; Brownstein, N.; Meckes, D.G. Nanoparticle analysis sheds budding insights into genetic drivers of extracellular vesicle biogenesis. J. Extracell. Vesicles 2016, 5, 31295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappa, G.; Green, T.M.; Karbanová, J.; Corbeil, D.; Lorico, A. Tetraspanin CD9 determines invasiveness and tumorigenicity of human breast cancer cells. Oncotarget 2015, 6, 7970–7991. [Google Scholar] [CrossRef]
- Kischel, P.; Bellahcene, A.; Deux, B.; Lamour, V.; Dobson, R.; De Pauw, E.; Clézardin, P.; Castronovo, V. Overexpression of CD9 in human breast cancer cells promotes the development of bone metastases. Anticancer Res. 2012, 32, 5211–5220. [Google Scholar]
- Vences-Catalán, F.; Rajapaksa, R.; Srivastava, M.K.; Marabelle, A.; Kuo, C.-C.; Levy, R.; Levy, S. Tetraspanin CD81, a modulator of immune suppression in cancer and metastasis. OncoImmunology 2016, 5, e1120399. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Campanella, C.; Bavisotto, C.C.; Gammazza, A.M.; Nikolic, D.; Rappa, F.; David, S.; Cappello, F.; Bucchieri, F.; Fais, S. Exosomal Heat Shock Proteins as New Players in Tumour Cell-to-Cell Communication. J. Circ. Biomark. 2014, 3, 4. [Google Scholar] [CrossRef] [Green Version]
- McCready, J.; Sims, J.D.; Chan, D.; Jay, D.G. Secretion of extracellular hsp90α via exosomes increases cancer cell motility: A role for plasminogen activation. BMC Cancer 2010, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in Membrane Traffic and Cell Physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef] [Green Version]
- Tzeng, H.-T.; Wang, Y.-C. Rab-mediated vesicle trafficking in cancer. J. Biomed. Sci. 2016, 23, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lizarbe, M.A.; Barrasa, J.I.; Olmo, N.; Gavilanes, F.; Turnay, J. Annexin-Phospholipid Interactions. Functional Implications. Int. J. Mol. Sci. 2013, 14, 2652–2683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maji, S.; Chaudhary, P.; Akopova, I.; Nguyen, P.M.; Hare, R.J.; Gryczynski, I.; Vishwanatha, J. Exosomal Annexin II Promotes Angiogenesis and Breast Cancer Metastasis. Mol. Cancer Res. 2017, 15, 93–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madureira, P.; Hill, R.; Miller, V.A.; Giacomantonio, C.; Lee, P.W.K.; Waisman, D.M. Annexin A2 is a novel Cellular Redox Regulatory Protein involved in Tumorigenesis. Oncotarget 2011, 2, 1075–1093. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattila, P.; Batista, F.D.; Treanor, B. Dynamics of the actin cytoskeleton mediates receptor cross talk: An emerging concept in tuning receptor signaling. J. Cell Biol. 2016, 212, 267–280. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Hamidi, H.; Pietilä, M.; Ivaska, J. The complexity of integrins in cancer and new scopes for therapeutic targeting. Br. J. Cancer 2016, 115, 1017–1023. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Fedele, C.; Singh, A.; Zerlanko, B.J.; Iozzo, R.V.; Languino, L.R. The αvβ6 Integrin Is Transferred Intercellularly via Exosomes. J. Biol. Chem. 2015, 290, 4545–4551. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Fedele, C.; Lu, H.; Nevalainen, M.T.; Keen, J.H.; Languino, L.R. Exosome-mediated Transfer of αvβ3 Integrin from Tumorigenic to Nontumorigenic Cells Promotes a Migratory Phenotype. Mol. Cancer Res. 2016, 14, 1136–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Cheng, D.; Tu, Q.; Yang, H.; Sun, B.; Yan, L.; Dai, H.; Luo, J.; Mao, B.; Cao, Y.; et al. HUWE1 controls the development of non-small cell lung cancer through down-regulation of p53. Theranostics 2018, 8, 3517–3529. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, T.; Gao, H.; Yue, X.; Bian, W.; Mei, J.; Zhang, Y. The interplay between IQGAP1 and small GTPases in cancer metastasis. Biomed. Pharmacother. 2021, 135, 111243. [Google Scholar] [CrossRef] [PubMed]
- Higginbotham, J.N.; Beckler, M.D.; Gephart, J.D.; Franklin, J.L.; Bogatcheva, G.; Kremers, G.-J.; Piston, D.W.; Ayers, G.D.; McConnell, R.E.; Tyska, M.J.; et al. Amphiregulin Exosomes Increase Cancer Cell Invasion. Curr. Biol. 2011, 21, 779–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimondo, S.; Saieva, L.; Vicario, E.; Pucci, M.; Toscani, D.; Manno, M.; Raccosta, S.; Giuliani, N.; Alessandro, R.; Raccosta, S. Multiple myeloma-derived exosomes are enriched of amphiregulin (AREG) and activate the epidermal growth factor pathway in the bone microenvironment leading to osteoclastogenesis. J. Hematol. Oncol. 2019, 12, 2. [Google Scholar] [CrossRef]
- Shelke, G.V.; Yin, Y.; Jang, S.C.; Lässer, C.; Wennmalm, S.; Hoffmann, H.J.; Li, L.; Gho, Y.S.; Nilsson, J.A.; Lötvall, J. Endosomal signalling via exosome surface TGFβ-1. J. Extracell. Vesicles 2019, 8, 1650458. [Google Scholar] [CrossRef] [Green Version]
- Saito, A.; Horie, M.; Nagase, T. TGF-β Signaling in Lung Health and Disease. Int. J. Mol. Sci. 2018, 19, 2460. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Shen, C.; Wang, X.; Lai, Y.; Zhou, K.; Li, P.; Liu, L.; Che, G. Prognostic value of TGF-β in lung cancer: Systematic review and meta-analysis. BMC Cancer 2019, 19, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Akinleye, A.; Rasool, Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J. Hematol. Oncol. 2019, 12, 92. [Google Scholar] [CrossRef] [Green Version]
- Das, M.; Zhu, C.; Kuchroo, V.K. Tim-3 and its role in regulating anti-tumor immunity. Immunol. Rev. 2017, 276, 97–111. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 pathway: Current researches in cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar] [PubMed]
- Kandel, S.; Adhikary, P.; Li, G.; Cheng, K. The TIM3/Gal9 signaling pathway: An emerging target for cancer immunotherapy. Cancer Lett. 2021, 510, 67–78. [Google Scholar] [CrossRef]
- Tari, A. GRB2: A pivotal protein in signal transduction. Semin. Oncol. 2001, 28, 142–147. [Google Scholar] [CrossRef]
- Roberts, P.J.; Der, C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007, 26, 3291–3310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.T.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 2007, 1773, 1263–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220. [Google Scholar] [CrossRef]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orlowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2013, 1831, 1302–1309. [Google Scholar] [CrossRef]
- Fan, T.W.; Zhang, X.; Wang, C.; Yang, Y.; Kang, W.-Y.; Arnold, S.; Higashi, R.M.; Liu, J.; Lane, A.N. Exosomal lipids for classifying early and late stage non-small cell lung cancer. Anal. Chim. Acta 2018, 1037, 256–264. [Google Scholar] [CrossRef]
- Chakraborty, M.; Jiang, X.-C. Sphingomyelin and Its Role in Cellular Signaling. Adv. Exp. Med. Biol. 2013, 991, 1–14. [Google Scholar] [CrossRef] [PubMed]
- De Gassart, A.; Geminard, C.; Fevrier, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.W.; Lee, H.M.; Lee, T.H.; Kang, C.; Kleinman, H.K.; Gho, Y.S. Extracellular membrane vesicles from tumor cells promote angiogenesis via sphingomyelin. Cancer Res. 2002, 62, 6312–6317. [Google Scholar] [PubMed]
- Eramo, A.; Haas, T.L.; De Maria, R. Lung cancer stem cells: Tools and targets to fight lung cancer. Oncogene 2010, 29, 4625–4635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciardiello, C.; Leone, A.; Budillon, A. The Crosstalk between Cancer Stem Cells and Microenvironment Is Critical for Solid Tumor Progression: The Significant Contribution of Extracellular Vesicles. Stem Cells Int. 2018, 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Fatima, F.; Vallabhaneni, K.C.; Penfornis, P.; Valadi, H.; Ekström, K.; Kholia, S.; Whitt, J.D.; Fernandes, J.D.; Pochampally, R.; et al. Extracellular Vesicles: Evolving Factors in Stem Cell Biology. Stem Cells Int. 2016, 2016, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Akbarzadeh, M.; Maroufi, N.F.; Tazehkand, A.P.; Akbarzadeh, M.; Bastani, S.; Safdari, R.; Farzane, A.; Fattahi, A.; Nejabati, H.R.; Nouri, M.; et al. Current approaches in identification and isolation of cancer stem cells. J. Cell. Physiol. 2019, 234, 14759–14772. [Google Scholar] [CrossRef]
- Su, C.; Zhang, J.; Yarden, Y.; Fu, L. The key roles of cancer stem cell-derived extracellular vesicles. Signal Transduct. Target. Ther. 2021, 6, 109. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.-G.; Lee, S.-H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raniszewska, A.; Kwiecień, I.; Rutkowska, E.; Rzepecki, P.; Domagała-Kulawik, J. Lung Cancer Stem Cells—Origin, Diagnostic Techniques and Perspective for Therapies. Cancers 2021, 13, 2996. [Google Scholar] [CrossRef] [PubMed]
- Abu Halim, N.H.; Zakaria, N.; Satar, N.A.; Yahaya, B.H. Isolation and Characterization of Cancer Stem Cells of the Non-Small-Cell Lung Cancer (A549) Cell Line. Methods Mol. Biol. 2016, 1516, 371–388. [Google Scholar] [CrossRef]
- Wang, L.; He, J.; Hu, H.; Tu, L.; Sun, Z.; Liu, Y.; Luo, F. Lung CSC-derived exosomal miR-210-3p contributes to a pro-metastatic phenotype in lung cancer by targeting FGFRL1. J. Cell. Mol. Med. 2020, 24, 6324–6339. [Google Scholar] [CrossRef]
- Lee, N.-K.; Kothandan, V.; Kothandan, S.; Byun, Y.; Hwang, S.-R. Exosomes and Cancer Stem Cells in Cancer Immunity: Current Reports and Future Directions. Vaccines 2021, 9, 441. [Google Scholar] [CrossRef]
- Aramini, B.; Masciale, V.; Haider, K.H. Defining lung cancer stem cells exosomal payload of miRNAs in clinical perspective. World J. Stem Cells 2020, 12, 406–421. [Google Scholar] [CrossRef]
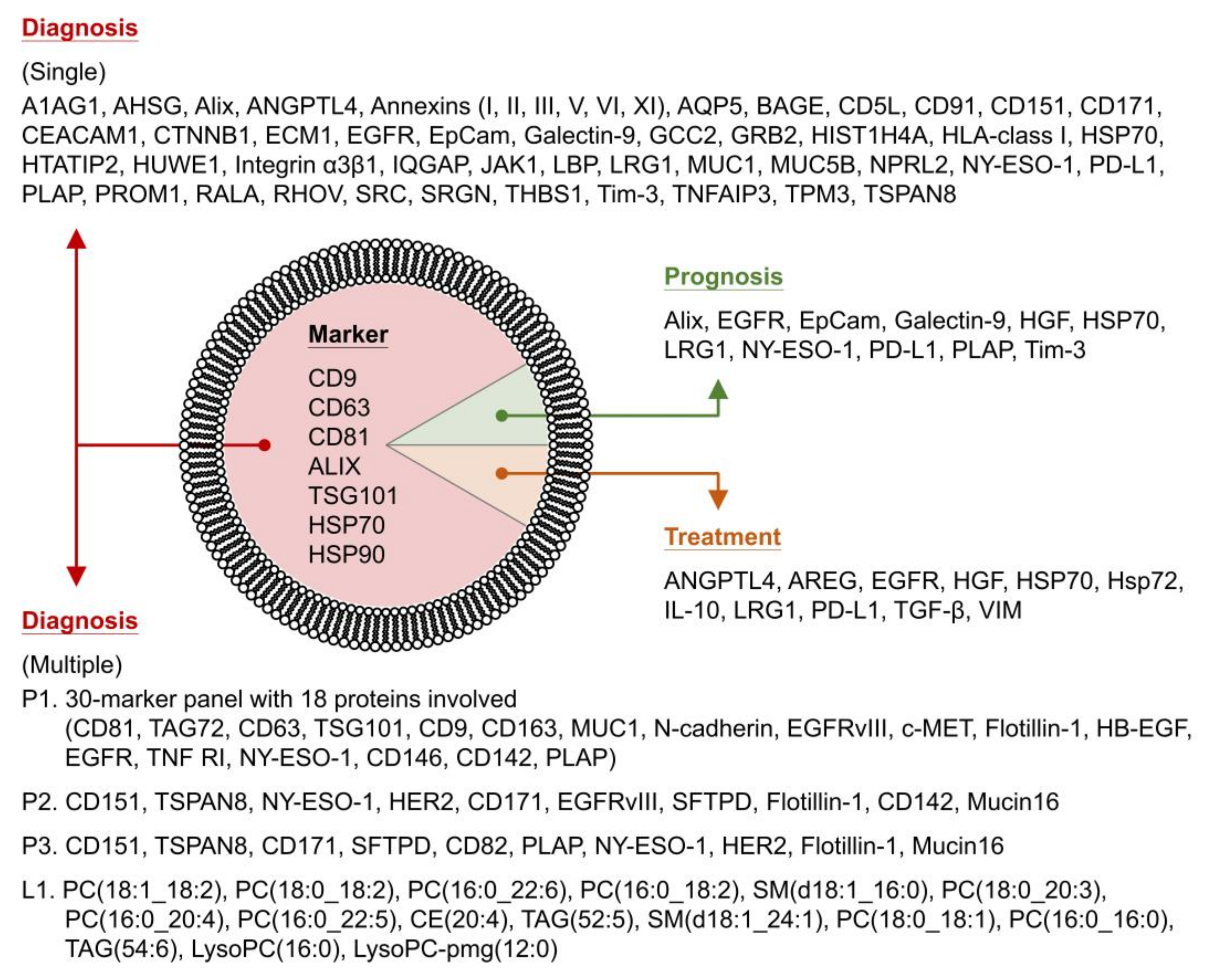
| Protein | Source | Application b | Year [Ref] | ||
|---|---|---|---|---|---|
| Marker | Lung Cancer a | Cell Line | Biopsy | ||
| CD63 | CD54, CD86, MHC-I, MHC-II, HSP60, HSC70, HSP70, HSP90 (ANXA2, CD18, CD71, CD80, CD107a) | 3LL (mouse) | - | Treatment | 2011 [105] |
| CCL2, CCL3, CCL4, CCL5, CCL20 c | |||||
| tsg101 d | Hsp72 (Hsc73) | H23 (human) | - | Treatment | 2010 [106] |
| CD63 | HSP70 | A549 (human) | - | Treatment | 2016 [107] |
| CD9 CD63 TSG101 | HSP70 | - | Plasma (AC) Plasma (SCC) | Diagnosis Prognosis | 2020 [108] |
| FLOT1 | EGFR | A549 (human) | - | Treatment | 2009 [109] |
| EEA1 | EGFR | - | Tissue (NSCLC) | Treatment | 2013 [110] |
| CD81 | EGFR | HARA (human) HARA-B (human) A549 (human) RERF-LC-MS (human) LU65 (human) | Plasma (xenograft mouse) Plasma (human) | Diagnosis | 2013 [111] |
| CD9 CD63 | EGFR | A549 (human) | Blood (NSCLC) | Diagnosis Treatment | 2019 [112] |
| CD63 ALIX TSG101 | AREG | CRL-2868 (human) A549 (human) | Plasma (NSCLC) | Treatment | 2017 [113] |
| CD9 CD81 | CD91 (CD317, BAIP2, CPNE1, FIBA, CO5, MASP1, APOB, CO8A, PLMN, PGBM, PERM, FCGBP, ITA2B, KV313, IC1, A2MG, HABP2) | - | Serum (AC) Serum (SCC) | Diagnosis | 2014 [114] |
| CD9 CD63 CD81 TSG101 Hsp90 | 30-marker panel: 11 markers, 3 markers (normalized value), 16 marker relations | - | Plasma (AC IIIa-IV) | Diagnosis | 2015 [115] |
| 18 proteins involved: CD81, TAG72, CD63, TSG101, CD9, CD163, MUC1, N-cadherin, EGFRvIII, c-MET, Flotillin-1, HB-EGF, EGFR, TNF RI, NY-ESO-1, CD146, CD142, PLAP | |||||
| CD9 CD63 CD81 | Single marker: (Cancer vs. noncancer patients)
| - | Plasma (AC) Plasma (SCC) Plasma (SCLC) | Diagnosis | 2016 [116] |
| Multiple markers: (Cancer vs. noncancer patients)
| |||||
| CD9 CD63 CD81 CD82 CD37 Alix TSG101 Hsp90 | Presence or not: CD171 (Flotillin-1, HER3, GRP78) | - | Plasma (NSCLC) | Diagnosis Prognosis | 2016 [117] |
| Expression level: NY-ESO-1, EGFR, PLAP (EpCam, CAIX, CD13, CD276) | |||||
| Measurable level: NY-ESO-1, EGFR, PLAP, EpCam, Alix (HER3, CAIX, CD13, CD276) | |||||
| Ezrin β-Actin CD63 | 912 proteins (e.g., EGFR, KRAS, CEACAM6, BSG, CLDN1, CLDN3, RAB family proteins) | - | Pleural effusion (NSCLC) | Diagnosis Treatment | 2013 [118] |
| CD63 ALIX | A549 & HCC827: EGFR, SRC, GRB2, JAK1 (DSG2, CD151, CNTN1, EDIL3, ITGB6, ITGB1, BSG, SLC3A2, LAMP2, CEACAM6, PAPPA, CTSA, GGT3P) | A549 (human) HCC827 (human) | - | Diagnosis | 2016 [119] |
| A549: (MFGE8, HSPG2, GPRC5A) | |||||
| HCC827: RALA, CTNNB1 (SDC1, EPCAM, MPZL1, MPZL2, TACSTD2, GNB1, TOM1L1) | |||||
| - | ANXA1, ANXA2, ANXA3, ANXA5, ANXA6, ANXA11, NPRL2, CEACAM1, MUC1, PROM1, HIST1H4A, TNFAIP3 | - | Saliva (cancer) | Diagnosis | 2016 [120] |
| CD63 TSG101 | A1AG1, AQP5, MUC5B | - | Saliva (cancer) Serum (cancer) | Diagnosis | 2017 [121] |
| CD63 CD81 TSG101 | MUC5B, IQGAP (ENO1, SPARCL1) | - | Saliva (cancer) | Diagnosis | 2018 [122] |
| CD9 CD63 CD81 TSG101 | HGF | 95C (LCC) 95D (LCC) | Plasma (xenograft mouse) Plasma (cancer) | Prognosis Treatment | 2019 [123] |
| CD9 CD81 HSP70 | ANGPTL4 (ACLY, LRP1, PSMD14, TKT, TTN, VCAN) | A549 (NSCLC) | - | Diagnosis Treatment | 2020 [124] |
| CD9 CD81 TSG101 | HTATIP2 RHOV | A549 and others (human) | Tissue (AC) Plasma (AC) | Diagnosis | 2020 [125] |
| - | TGF-β, IL-10 | NCI-H1688 (SCLC) NCI-H2228 (NSCLC) | - | Treatment | 2016 [126] |
| CD9 CD63 HSP70 | VIM | PC14 (nonmetastatic) PC14HM (highly metastatic) | Serum (early stage) Serum (late stage) | Treatment | 2016 [127] |
| CD9 TSG101 | SRGN, TPM3, THBS1, HUWE1 (CCDC18, ALDH1L1, HIST1H4A, NCCRP1, MED14, BHMT, GLUD1, PPIA, EXOC8) | H23 (human) H647 (human) H1573 (human) HCC4019 (human) | Plasma (AC) | Diagnosis | 2017 [128] |
| CD63 | Tim-3, Galectin-9 | - | Plasma (NSCLC) | Diagnosis Prognosis | 2018 [129] |
| HSP70 | LBP | - | Serum (nonmetastatic NSCLC) Serum (metastatic NSCLC) | Diagnosis | 2018 [130] |
| CD9 CD63 CD81 HSP70 | AHSG, ECM1 | - | Serum (NSCLC) | Diagnosis | 2019 [131] |
| CD63 | MUC1 (THBS1, ANXA6, HIST1H4A, COL18A1, MDK, CD151, SRGN, ENO1, TUBA4A, SLC3A2, GPI, MIF, TALDO1, SLC7A5, ICAM1, HSP90AA1, G6PD, LRP1) | NCI-H838 (NSCLC) | Plasma (NSCLC) | Diagnosis | 2019 [132] |
| CD63 | ITGA3, ITGB1 | A549 (AC) H1975 (AC) H3255 (AC) H1650 (AC) | Pericardial effusion (NSCLC) | Diagnosis | 2019 [133] |
| - | LRG1 | - | Urine (NSCLC) | Diagnosis | 2011 [134] |
| CD63 TSG101 | LRG1 | SPCA1 (NSCLC) A549 (NSCLC) PC9 (NSCLC) H1299 (NSCLC) H358 (NSCLC) | Plasma (NSCLC) | Diagnosis Prognosis Treatment | 2019 [135] |
| CD63 ANXA5 | HLA-class I, BAGE, PD-L1, ANXA2 | - | BAL (NSCLC) | Diagnosis | 2019 [136] |
| CD63 HRS Alix TSG101 | PD-L1 | H1299 (human) H358 (human) H1264 (human) | - | Prognosis Treatment | 2018 [137] |
| CD9 HSP70 HSP90 | PD-L1 | A549 (NSCLC) H460 (NSCLC) H1975 (NSCLC) | Plasma (NSCLC) | Diagnosis | 2019 [138] |
| CD9 CD63 | PD-L1 | - | Plasma (advanced NSCLC) | Prognosis | 2021 [139] |
| CD63 TSG101 | PD-L1 | - | Serum (NSCLC) | Diagnosis Prognosis | 2019 [140] |
| - | PD-L1 | - | Serum (NSCLC) | Prognosis | 2021 [141] |
| Epcam | PD-L1 | A549 (human) SK-MES1 (human) | Plasma (cancer) | Diagnosis Prognosis | 2021 [142] |
| EpCAM | PD-L1 | A549 (human) H1975 (human) | Plasma (NSCLC) | Diagnosis | 2021 [143] |
| CD9 CD63 CD81 Alix TSG101 | CD5L (CLEC3B, SERFINF1, ITIH4, SAA4, SERFINC1, C20ORF3) | - | Serum (AC) Serum (SCC) Serum (SCLC) | Diagnosis | 2021 [144] |
| CD9 CD63 CD81 | GCC2 (TUBA1C, GAPDH, KRT25, POTEKP) | A549 (NSCLC) H1299 (NSCLC) PC9 (NSCLC) H1650 (NSCLC) H522 (NSCLC) | Plasma (AC) | Diagnosis | 2021 [145] |
| CD9 CD63 TSG101 | eIF4E e | - | Serum (NSCLC) | Prognosis | 2020 [146] |
| Protein Class a | Protein Subclass a | Protein | Top100 c |
|---|---|---|---|
| Calcium-binding protein | - | ANXA1 | Yes |
| ANXA2 | Yes | ||
| ANXA3 | No | ||
| ANXA5 | Yes | ||
| ANXA6 | Yes | ||
| ANXA11 | Yes | ||
| Cell adhesion molecule | integrin | ITGA3 | No |
| ITGB1 | Yes | ||
| - | MUC1 | No | |
| Chaperone | Hsp90 family chaperone | HSP90/HSP90AA1 | Yes |
| Chromatin/chromatin-binding, or -regulatory protein | histone | HIST1H4A/H4C1 | Yes |
| Cytoskeletal protein | microtubule or microtubule-binding cytoskeletal protein | GCC2 | No |
| actin binding motor protein (actin or actin-binding cytoskeletal protein) | TPM3 | No | |
| Defense/immunity protein | immunoglobulin receptor superfamily | CD86 | No |
| PD-L1/CD274 | No | ||
| Tim-3/TIM3 | No | ||
| major histocompatibility complex protein | MHC-I | No | |
| MHC-II | No | ||
| Extracellular matrix protein | - | Galectin-9 | No |
| MUC5B | No | ||
| Intercellular signal molecule | - | ANGPTL4 | No |
| growth factor | AREG | No | |
| TGF-β/TGFB1 | No | ||
| Membrane traffic protein | - | ALIX/PDCD6IP | Yes |
| Metabolite interconversion enzyme | oxidoreductase | HTATIP2 | No |
| phosphatase (hydrolase) | PLAP/ALPP | No | |
| Protein modifying enzyme | serine protease (protease) | HGF | No |
| cysteine protease (protease) | TNFAIP3 | No | |
| ubiquitin-protein ligase | HUWE1 | No | |
| TSG101 | Yes | ||
| Protein-binding activity modulator | protease inhibitor | AHSG | No |
| GTPase-activating protein (G-protein modulator) | IQGAP/IQGAP1 | No d | |
| NPRL2 | No | ||
| small GTPase (G-protein) | RALA | No d | |
| RHOV | No | ||
| Transfer/carrier protein | apolipoprotein | CD91/LRP1 | No |
| Transmembrane signal receptor | - | EGFR | No |
| Transporter | - | AQP5 | No |
| NA b | NA b | CD9, CD63, CD81, FLOT1, HSC70/HSPA8, HSP70, Hsp72, THBS1 | Yes |
| A1AG1/ORM1, BAGE, CD5L, CD54/ICAM1, CD151, CD171/L1CAM, CEACAM1, CTNNB1, ECM1, EpCam, GRB2, HRS/HGS, HSP60, IL-10/IL10, JAK1, LBP, LRG1, NY-ESO-1/ESO1/CTAG1B, PROM1, SRC, SRGN, TSPAN8, VIM | No |
| Lipid | Source | Application b | Year [Ref] | ||
|---|---|---|---|---|---|
| Marker | Lung Cancer | Cell Line | Biopsy | ||
| - | 16 lipid features b: PC(18:1_18:2), PC(18:0_18:2), PC(16:0_22:6), PC(16:0_18:2), SM(d18:1_16:0), PC(18:0_20:3), PC(16:0_20:4), PC(16:0_22:5), CE(20:4), TAG(52:5), SM(d18:1_24:1), PC(18:0_18:1), PC(16:0_16:0), TAG(54:6), LysoPC(16:0), LysoPC–pmg(12:0) | - | Plasma (NSCLC I, II) Plasma (NSCLC III, IV) | Diagnosis | 2018 [195] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, M.-T.; Wang, Y.-K.; Tseng, Y.J. Exosomal Proteins and Lipids as Potential Biomarkers for Lung Cancer Diagnosis, Prognosis, and Treatment. Cancers 2022, 14, 732. https://doi.org/10.3390/cancers14030732
Hsu M-T, Wang Y-K, Tseng YJ. Exosomal Proteins and Lipids as Potential Biomarkers for Lung Cancer Diagnosis, Prognosis, and Treatment. Cancers. 2022; 14(3):732. https://doi.org/10.3390/cancers14030732
Chicago/Turabian StyleHsu, Ming-Tsung, Yu-Ke Wang, and Yufeng Jane Tseng. 2022. "Exosomal Proteins and Lipids as Potential Biomarkers for Lung Cancer Diagnosis, Prognosis, and Treatment" Cancers 14, no. 3: 732. https://doi.org/10.3390/cancers14030732
APA StyleHsu, M.-T., Wang, Y.-K., & Tseng, Y. J. (2022). Exosomal Proteins and Lipids as Potential Biomarkers for Lung Cancer Diagnosis, Prognosis, and Treatment. Cancers, 14(3), 732. https://doi.org/10.3390/cancers14030732





