Whole Blood Transcriptional Fingerprints of High-Grade Glioma and Longitudinal Tumor Evolution under Carbon Ion Radiotherapy
Abstract
Simple Summary
Abstract
1. Introduction
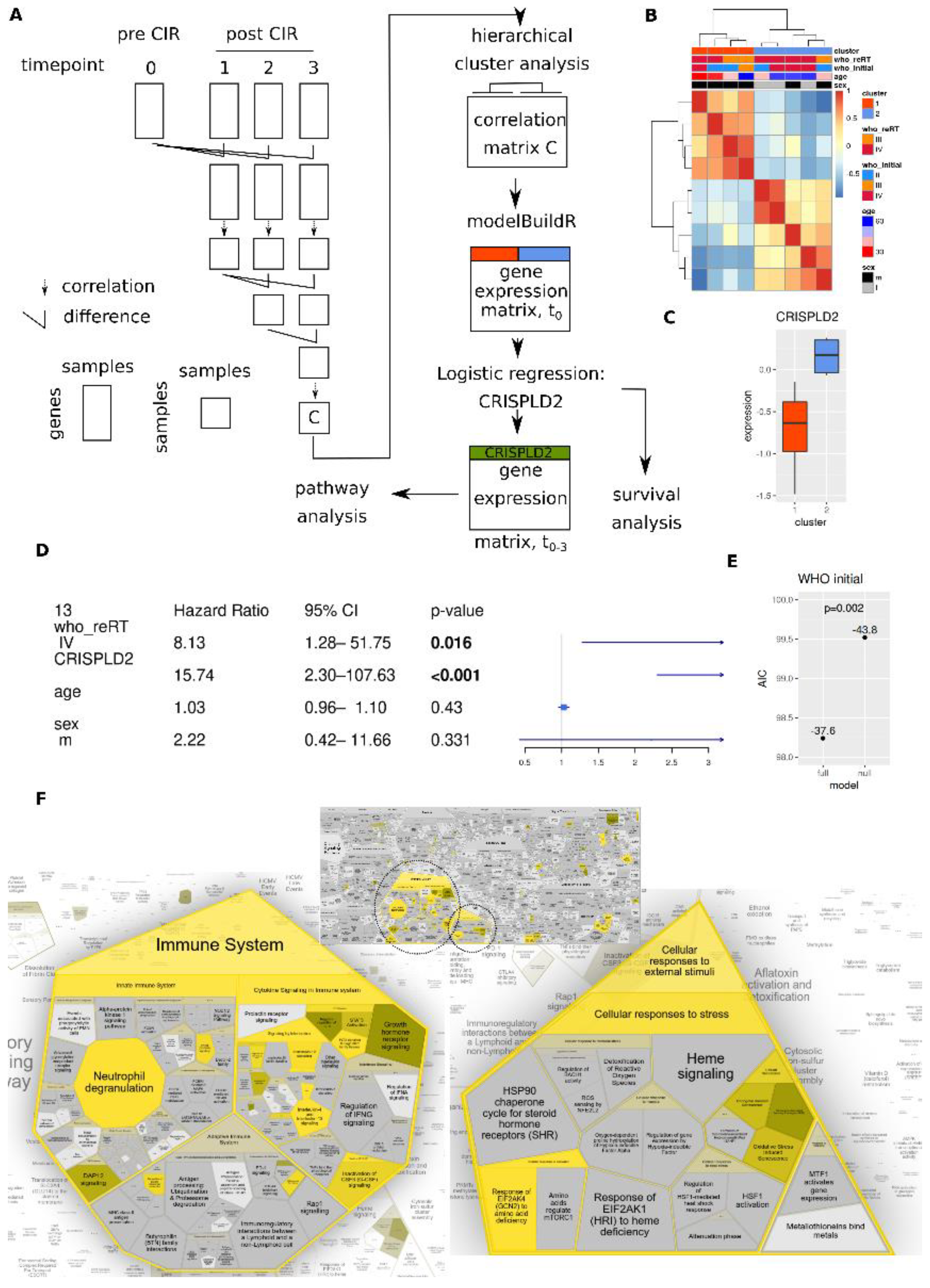
2. Materials and Methods
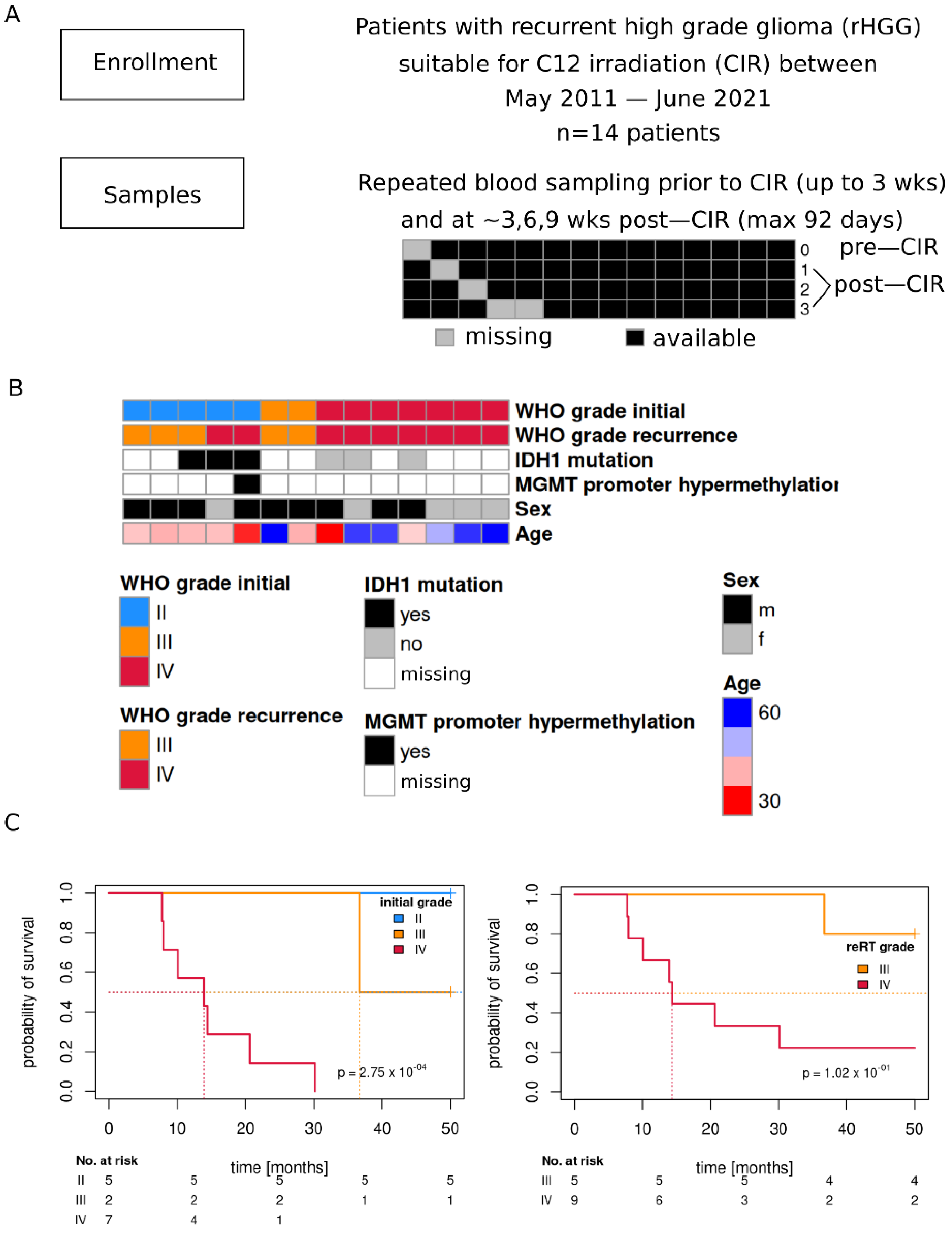
2.1. Study Cohort
2.2. Ethics Approval and Consent to Participate
2.3. Blood Processing
2.4. CIBERSORT
2.5. Statistical Analysis
3. Results
3.1. Study Cohort
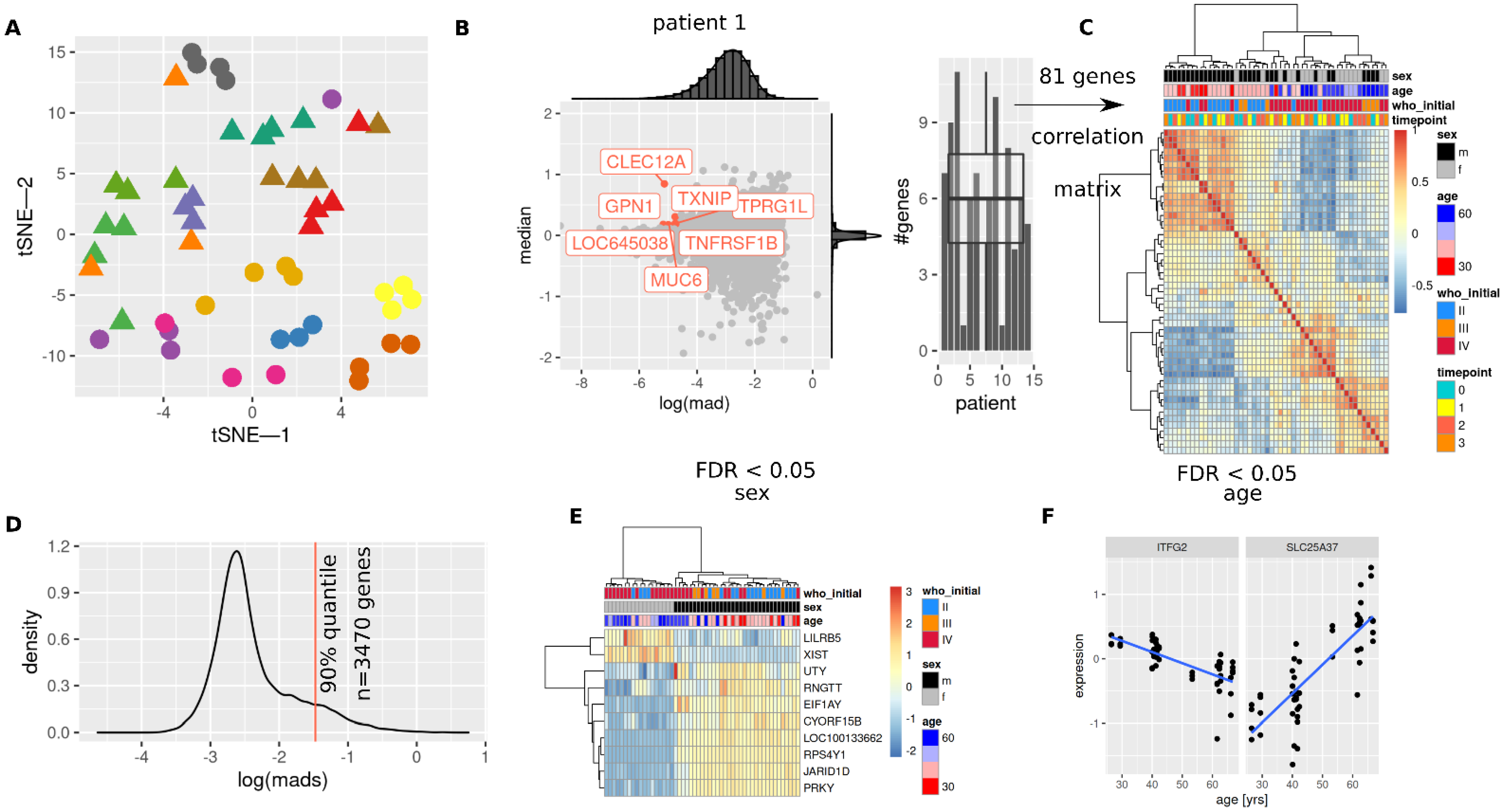
3.2. Patient Characteristics Sex and Age Are Mirrored in Transcriptome
3.3. Initial and Re-RT Glioma Grade Are Associated with Transcriptome Profiles
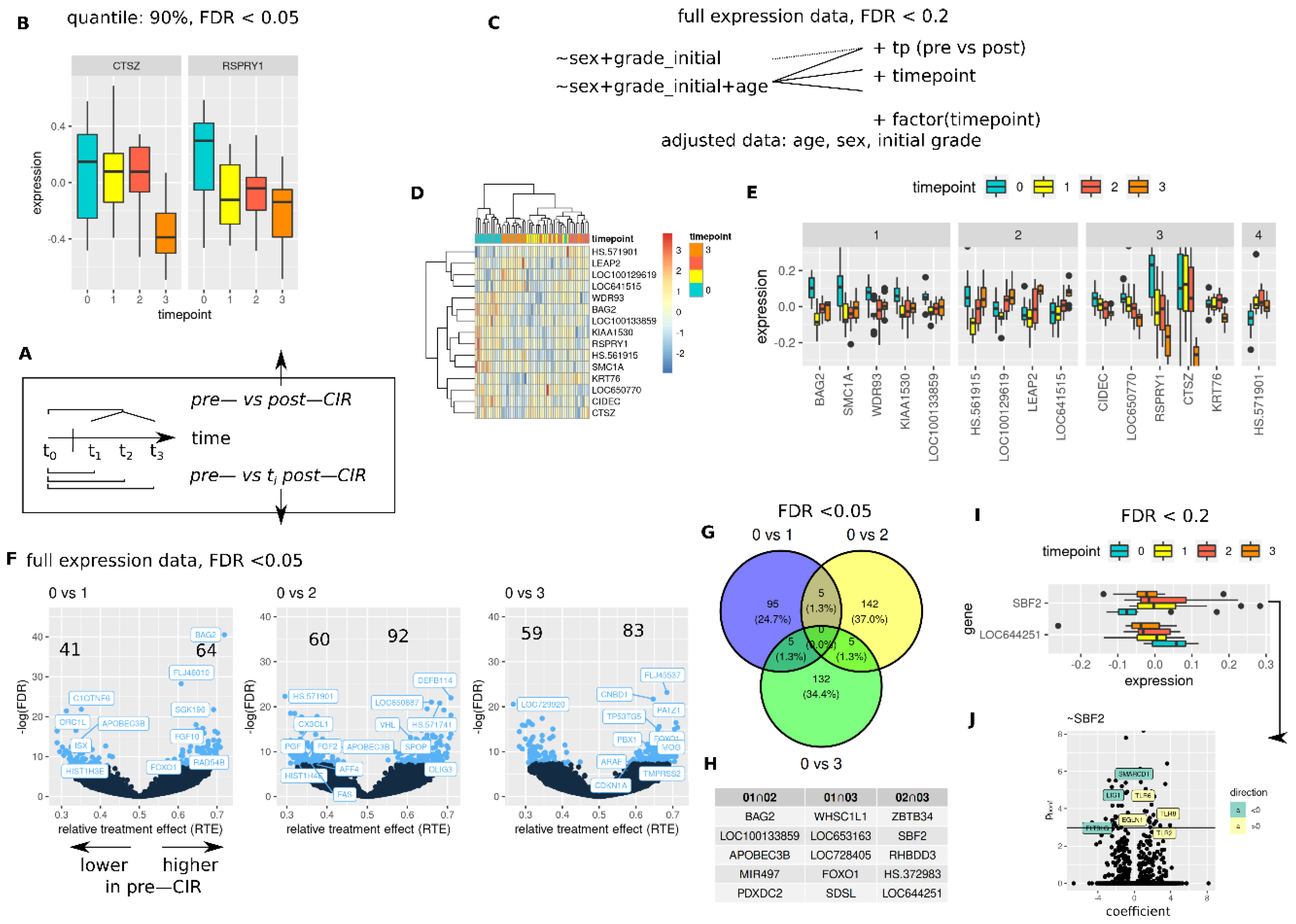
3.4. CIR Induces Sustained and Transient Changes in Transcriptome
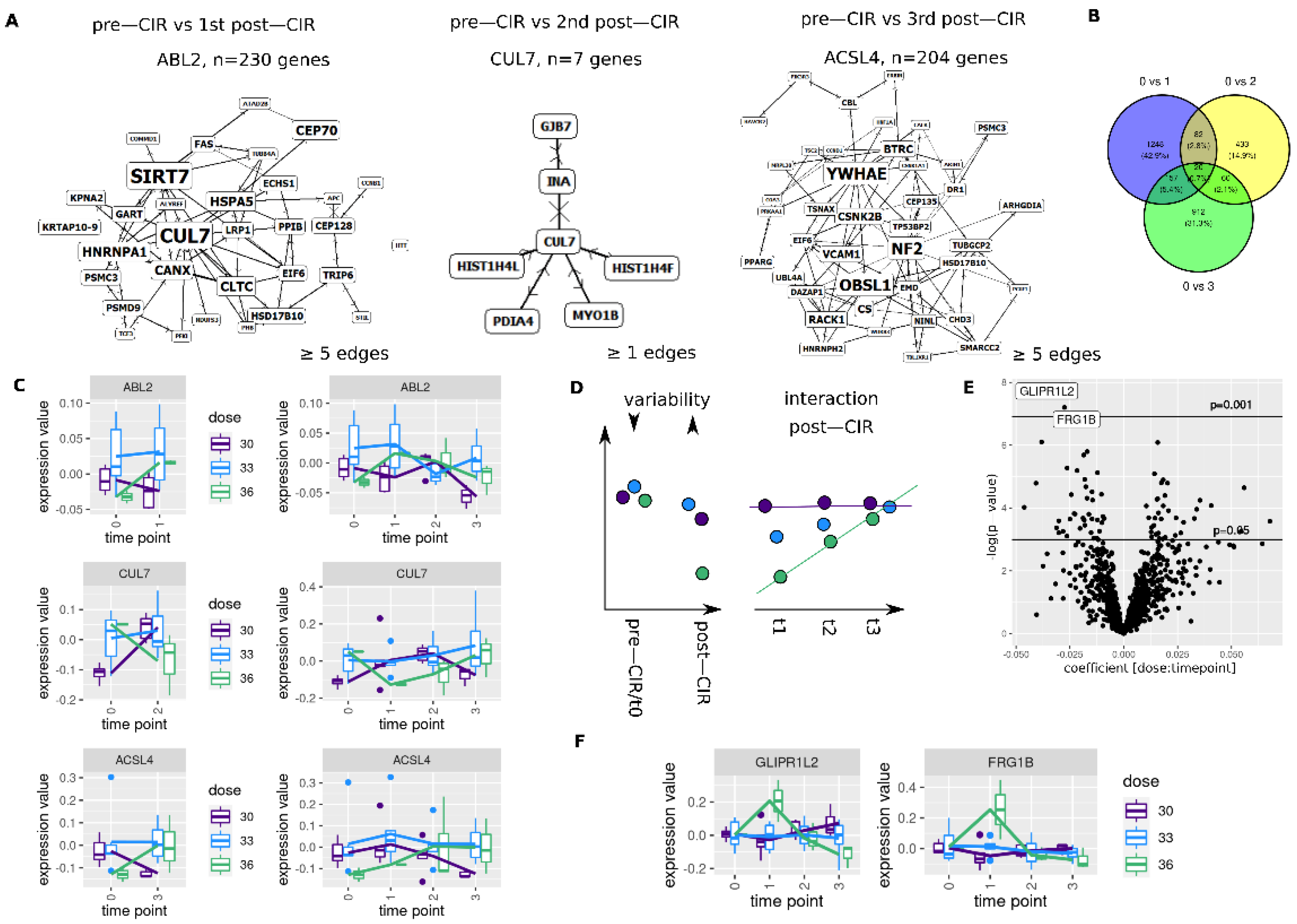
3.5. CIR Leads to Dose Dependent Transcriptome Alterations
3.6. Longitudinal Transcriptome Analysis Identifies Two Trajectories
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knoll, M.; Waltenberger, M.; Bougatf, N.; Bernhard, D.; Adeberg, S.; Budach, V.; Baumann, M.; Stuschke, M.; Fokas, E.; Grosu, A.; et al. Efficacy of re-irradiation with carbon ions (RiCi) in patients with recurrent high-grade glioma (rHGG) compared to the standard re-irradiation with photons (RiP): The reference multicenter cohort of the German Cancer Consortium Radiation Oncology Group (DKTK-ROG). J. Clin. Oncol. 2019, 37, 2057. [Google Scholar]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Knoll, M.; Debus, J.; Furkel, J.; Warta, R.; Bougatf, N.; Rapp, C.; Brors, B.; Wick, W.; Unterberg, A.; Herold-Mende, C.; et al. Glioblastoma evolution pattern under surgery and radio(chemo)therapy (RCHT) to identify novel methylome based glioma subtypes. J. Clin. Oncol. 2019, 37, 2012. [Google Scholar] [CrossRef]
- Mrugala, M.M. Advances and challenges in the treatment of glioblastoma: A clinician’s perspective. Discov. Med. 2013, 15, 221–230. [Google Scholar] [PubMed]
- Arvold, N.D.; Shi, D.D.; Aizer, A.A.; Norden, A.D.; Reardon, D.A.; Lee, E.Q.; Nayak, L.; Dunn, I.F.; Golby, A.J.; Johnson, M.D.; et al. Salvage re-irradiation for recurrent high-grade glioma and comparison to bevacizumab alone. J. NeuroOncol. 2017, 135, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Niyazi, M.; Adeberg, S.; Kaul, D.; Boulesteix, A.L.; Bougatf, N.; Fleischmann, D.F.; Grun, A.; Kramer, A.; Rodel, C.; Eckert, F.; et al. Independent validation of a new reirradiation risk score (RRRS) for glioma patients predicting post-recurrence survival: A multicenter DKTK/ROG analysis. Radiother. Oncol. 2018, 127, 121–127. [Google Scholar] [CrossRef]
- Dokic, I.; Mairani, A.; Niklas, M.; Zimmermann, F.; Chaudhri, N.; Krunic, D.; Tessonnier, T.; Ferrari, A.; Parodi, K.; Jakel, O.; et al. Next generation multi-scale biophysical characterization of high precision cancer particle radiotherapy using clinical proton, helium-, carbon- and oxygen ion beams. Oncotarget 2016, 7, 56676–56689. [Google Scholar] [CrossRef] [PubMed]
- Chiblak, S.; Tang, Z.; Campos, B.; Gal, Z.; Unterberg, A.; Debus, J.; Herold-Mende, C.; Abdollahi, A. Radiosensitivity of Patient-Derived Glioma Stem Cell 3-Dimensional Cultures to Photon, Proton, and Carbon Irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2016, 95, 112–119. [Google Scholar] [CrossRef]
- Chiblak, S.; Tang, Z.; Lemke, D.; Knoll, M.; Dokic, I.; Warta, R.; Moustafa, M.; Mier, W.; Brons, S.; Rapp, C.; et al. Carbon irradiation overcomes glioma radioresistance by eradicating stem cells and forming an antiangiogenic and immunopermissive niche. JCI Insight 2019, 4, 3837. [Google Scholar] [CrossRef]
- Winter, M.; Dokic, I.; Schlegel, J.; Warnken, U.; Debus, J.; Abdollahi, A.; Schnolzer, M. Deciphering the Acute Cellular Phosphoproteome Response to Irradiation with X-rays, Protons and Carbon Ions. Mol. Cell Proteom. 2017, 16, 855–872. [Google Scholar] [CrossRef]
- Dokic, I.; Niklas, M.; Zimmermann, F.; Mairani, A.; Seidel, P.; Krunic, D.; Jakel, O.; Debus, J.; Greilich, S.; Abdollahi, A. Correlation of Particle Traversals with Clonogenic Survival Using Cell-Fluorescent Ion Track Hybrid Detector. Front. Oncol. 2015, 5, 275. [Google Scholar] [CrossRef] [PubMed]
- Sharungbam, G.D.; Schwager, C.; Chiblak, S.; Brons, S.; Hlatky, L.; Haberer, T.; Debus, J.; Abdollahi, A. Identification of stable endogenous control genes for transcriptional profiling of photon, proton and carbon-ion irradiated cells. Radiat. Oncol. 2012, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Dokic, I.; Mairani, A.; Mein, S.; Brons, S.; Haring, P.; Haberer, T.; Jakel, O.; Zimmermann, A.; Zenke, F.; et al. Overcoming hypoxia-induced tumor radioresistance in non-small cell lung cancer by targeting DNA-dependent protein kinase in combination with carbon ion irradiation. Radiat. Oncol. 2017, 12, 208. [Google Scholar] [CrossRef] [PubMed]
- Debus, C.; Waltenberger, M.; Floca, R.; Afshar-Oromieh, A.; Bougatf, N.; Adeberg, S.; Heiland, S.; Bendszus, M.; Wick, W.; Rieken, S.; et al. Impact of (18)F-FET PET on Target Volume Definition and Tumor Progression of Recurrent High Grade Glioma Treated with Carbon-Ion Radiotherapy. Sci. Rep. 2018, 8, 7201. [Google Scholar] [CrossRef]
- Combs, S.E.; Burkholder, I.; Edler, L.; Rieken, S.; Habermehl, D.; Jakel, O.; Haberer, T.; Haselmann, R.; Unterberg, A.; Wick, W.; et al. Randomised phase I/II study to evaluate carbon ion radiotherapy versus fractionated stereotactic radiotherapy in patients with recurrent or progressive gliomas: The CINDERELLA trial. BMC Cancer 2010, 10, 533. [Google Scholar] [CrossRef] [PubMed]
- Zabel-du Bois, A.; Wagner-Ecker, M.; Milker-Zabel, S.; Schwager, C.; Wirkner, U.; Debus, J.; Abdollahi, A.; Huber, P.E. Gene expression signatures in the peripheral blood after radiosurgery of human cerebral arteriovenous malformations. Strahlenther. Onkol. 2010, 186, 91–98. [Google Scholar] [CrossRef]
- Liangos, O.; Domhan, S.; Schwager, C.; Zeier, M.; Huber, P.E.; Addabbo, F.; Goligorsky, M.S.; Hlatky, L.; Jaber, B.L.; Abdollahi, A. Whole blood transcriptomics in cardiac surgery identifies a gene regulatory network connecting ischemia reperfusion with systemic inflammation. PLoS ONE 2010, 5, e13658. [Google Scholar] [CrossRef]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Nassiri, F.; Chakravarthy, A.; Feng, S.; Shen, S.Y.; Nejad, R.; Zuccato, J.A.; Voisin, M.R.; Patil, V.; Horbinski, C.; Aldape, K.; et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat. Med. 2020, 26, 1044–1047. [Google Scholar] [CrossRef]
- Scholz, M.; Kellerer, A.M.; Kraft-Weyrather, W.; Kraft, G. Computation of cell survival in heavy ion beams for therapy. The model and its approximation. Radiat. Environ. BioPhys. 1997, 36, 59–66. [Google Scholar] [CrossRef]
- Schramm, K.; Marzi, C.; Schurmann, C.; Carstensen, M.; Reinmaa, E.; Biffar, R.; Eckstein, G.; Gieger, C.; Grabe, H.J.; Homuth, G.; et al. Mapping the genetic architecture of gene regulation in whole blood. PLoS ONE 2014, 9, e93844. [Google Scholar] [CrossRef] [PubMed]
- Homuth, G.; Wahl, S.; Müller, C.; Schurmann, C.; Mäder, U.; Blankenberg, S.; Carstensen, M.; Dörr, M.; Endlich, K.; Englbrecht, C.; et al. Extensive alterations of the whole-blood transcriptome are associated with body mass index: Results of an mRNA profiling study involving two large population-based cohorts. BMC Med. Genom. 2015, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 13 September 2021).
- Konopka, T. UMAP: Uniform Manifold Approximation and Projection. Available online: https://CRAN.R-project.org/package=umap (accessed on 13 September 2021).
- McInnes, L.; Healy, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Linderman, G.C.; Rachh, M.; Hoskins, J.G.; Steinerberger, S.; Kluger, Y. Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat. Methods 2019, 16, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Gel, Y.R.; Brunner, E.; Konietschke, F. nparLD: An R Software Package for the Nonparametric Analysis of Longitudinal Data in Factorial Experiments. J. Stat. Softw. 2012, 50, 1–23. [Google Scholar] [CrossRef]
- Konietschke, F.; Friedrich, S.; Brunner, E.; Pauly, M. rankFD: Rank-Based Tests for General Factorial Designs. Available online: https://CRAN.R-project.org/package=rankFD (accessed on 13 September 2021).
- Knoll, M. GeneSignatures: Computing Signature Scores for Omics Data. R Package Version 0.1. Available online: https://www.github.com/mknoll/geneSignatures (accessed on 13 September 2021).
- Therneau, T.M. A Package for Survival Analysis in S. Available online: http://CRAN.R-project.org/package=survival (accessed on 13 September 2021).
- Jawaid, W. EnrichR: Provides an R Interface to ‘Enrichr’. Available online: https://CRAN.R-project.org/package=enrichR (accessed on 13 September 2021).
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Schwager, C. SUMO-Statistical Utility for Micro array and Omics Data. Available online: http://angiogenesis.dkfz.de/oncoexpress/software/sumo/ (accessed on 13 September 2021).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Knoll, M.; Furkel, J.; Debus, J.; Abdollahi, A. modelBuildR: An R package for model building and feature selection with erroneous classifications. PeerJ 2021, 9, e10849. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Fertig, E.J.; Jaffe, A.E.; Zhang, Y.; Storey, J.D.; Torres, L.C. sva: Surrogate Variable Analysis. Available online: https://bioconductor.org/packages/release/bioc/html/sva.html (accessed on 11 October 2021).
- Chollet, F. Keras. Available online: https://github.com/fchollet/keras (accessed on 11 October 2021).
- Mayer, R.; Sminia, P. Reirradiation tolerance of the human brain. Int. J. Radiat. Oncol. Biol. Phys. 2008, 70, 1350–1360. [Google Scholar] [CrossRef]
- Mohan, R.; Liu, A.Y.; Brown, P.D.; Mahajan, A.; Dinh, J.; Chung, C.; McAvoy, S.; McAleer, M.F.; Lin, S.H.; Li, J.; et al. Proton therapy reduces the likelihood of high-grade radiation-induced lymphopenia in glioblastoma patients: Phase II randomized study of protons vs photons. Neuro-Oncology 2021, 23, 284–294. [Google Scholar] [CrossRef]
- Zhu, C.; Mohan, R.; Lin, S.H.; Jun, G.; Yaseen, A.; Jiang, X.; Wang, Q.; Cao, W.; Hobbs, B.P. Identifying Individualized Risk Profiles for Radiotherapy-Induced Lymphopenia Among Patients with Esophageal Cancer Using Machine Learning. JCO Clin. Cancer Inform. 2021, 5, 1044–1053. [Google Scholar] [CrossRef]
- Han, Z.; Feng, W.; Hu, R.; Ge, Q.; Ma, W.; Zhang, W.; Xu, S.; Zhan, B.; Zhang, L.; Sun, X.; et al. RNA-seq profiling reveals PBMC RNA as a potential biomarker for hepatocellular carcinoma. Sci. Rep. 2021, 11, 17797. [Google Scholar] [CrossRef]
- Hou, H.; Lyu, Y.; Jiang, J.; Wang, M.; Zhang, R.; Liew, C.C.; Wang, B.; Cheng, C. Peripheral blood transcriptome identifies high-risk benign and malignant breast lesions. PLoS ONE 2020, 15, e0233713. [Google Scholar] [CrossRef]
- van Wilpe, S.; Wosika, V.; Ciarloni, L.; Hosseinian Ehrensberger, S.; Jeitziner, R.; Angelino, P.; Duiveman-de Boer, T.; Koornstra, R.H.T.; de Vries, I.J.M.; Gerritsen, W.R.; et al. Whole Blood Transcriptome Profiling Identifies DNA Replication and Cell Cycle Regulation as Early Marker of Response to Anti-PD-1 in Patients with Urothelial Cancer. Cancers 2021, 13, 4660. [Google Scholar] [CrossRef]
- Yang, S.; Yao, Y.; Dong, Y.; Liu, J.; Li, Y.; Yi, L.; Huang, Y.; Gao, Y.; Yin, J.; Li, Q.; et al. Prediction of Radiation Pneumonitis Using Genome-Scale Flux Analysis of RNA-Seq Derived from Peripheral Blood. Front. Med. 2021, 8, 715961. [Google Scholar] [CrossRef]
- Yang, S.J.; Rafla, S.; Youssef, E.; Selim, H.; Salloum, N.; Chuang, J.Y. Changes in T-cell subsets after radiation therapy. Radiology 1988, 168, 537–540. [Google Scholar] [CrossRef]
- Rotstein, S.; Blomgren, H.; Petrini, B.; Wasserman, J.; Baral, E. Long term effects on the immune system following local radiation therapy for breast cancer. I. Cellular composition of the peripheral blood lymphocyte population. Int. J. Radiat. Oncol. Biol. Phys. 1985, 11, 921–925. [Google Scholar] [CrossRef]
- Petrini, B.; Wasserman, J.; Blomgren, H.; Glas, U. Changes of blood T cell subsets following radiation therapy for breast cancer. Cancer Lett. 1983, 19, 27–31. [Google Scholar] [CrossRef]
- De Ruysscher, D.; Waer, M.; Vandeputte, M.; Aerts, R.; Vantongelen, K.; van der Schueren, E. Changes of lymphocyte subsets after local irradiation for early stage breast cancer and seminoma testis: Long-term increase of activated (HLA-DR+) T cells and decrease of “naïve” (CD4-CD45R) T lymphocytes. Eur. J. Cancer 1992, 28A, 1729–1734. [Google Scholar] [CrossRef]
- Weakley, S.M.; Wang, H.; Yao, Q.; Chen, C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J. Surg. 2011, 35, 1751–1756. [Google Scholar] [CrossRef]
- Hu, S.; Chang, J.; Li, Y.; Wang, W.; Guo, M.; Zou, E.C.; Wang, Y.; Yang, Y. Long non-coding RNA XIST as a potential prognostic biomarker in human cancers: A meta-analysis. Oncotarget 2018, 9, 13911–13919. [Google Scholar] [CrossRef][Green Version]
- Kaasinen, E.; Kuismin, O.; Rajamaki, K.; Ristolainen, H.; Aavikko, M.; Kondelin, J.; Saarinen, S.; Berta, D.G.; Katainen, R.; Hirvonen, E.A.M.; et al. Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat. Commun. 2019, 10, 1252. [Google Scholar] [CrossRef]
- Antikainen, H.; Driscoll, M.; Haspel, G.; Dobrowolski, R. TOR-mediated regulation of metabolism in aging. Aging Cell 2017, 16, 1219–1233. [Google Scholar] [CrossRef]
- Kananen, L.; Marttila, S.; Nevalainen, T.; Jylhava, J.; Mononen, N.; Kahonen, M.; Raitakari, O.T.; Lehtimaki, T.; Hurme, M. Aging-associated DNA methylation changes in middle-aged individuals: The Young Finns study. BMC Genom. 2016, 17, 103. [Google Scholar] [CrossRef]
- Alvarez-Diaz, S.; Valle, N.; Ferrer-Mayorga, G.; Lombardia, L.; Herrera, M.; Dominguez, O.; Segura, M.F.; Bonilla, F.; Hernando, E.; Munoz, A. MicroRNA-22 is induced by vitamin D and contributes to its antiproliferative, antimigratory and gene regulatory effects in colon cancer cells. Hum. Mol. Genet. 2012, 21, 2157–2165. [Google Scholar] [CrossRef]
- Hope, N.R.; Murray, G.I. The expression profile of RNA-binding proteins in primary and metastatic colorectal cancer: Relationship of heterogeneous nuclear ribonucleoproteins with prognosis. Hum. Pathol. 2011, 42, 393–402. [Google Scholar] [CrossRef]
- Lefave, C.V.; Squatrito, M.; Vorlova, S.; Rocco, G.L.; Brennan, C.W.; Holland, E.C.; Pan, Y.X.; Cartegni, L. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 2011, 30, 4084–4097. [Google Scholar] [CrossRef]
- Ohkuri, T.; Ghosh, A.; Kosaka, A.; Zhu, J.; Ikeura, M.; David, M.; Watkins, S.C.; Sarkar, S.N.; Okada, H. STING contributes to antiglioma immunity via triggering type I IFN signals in the tumor microenvironment. Cancer Immunol. Res. 2014, 2, 1199–1208. [Google Scholar] [CrossRef]
- Kaur, B.; Khwaja, F.W.; Severson, E.A.; Matheny, S.L.; Brat, D.J.; Van Meir, E.G. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro-Oncology 2005, 7, 134–153. [Google Scholar] [CrossRef]
- Heddleston, J.M.; Li, Z.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 2009, 8, 3274–3284. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, J.; Fu, S.; Wang, Z. Knockdown of CDK2AP1 by RNA interference inhibits cell growth and tumorigenesis of human glioma. Neurol. Res. 2014, 36, 659–665. [Google Scholar] [CrossRef]
- Zeiner, P.S.; Preusse, C.; Blank, A.E.; Zachskorn, C.; Baumgarten, P.; Caspary, L.; Braczynski, A.K.; Weissenberger, J.; Bratzke, H.; Reiss, S.; et al. MIF Receptor CD74 is Restricted to Microglia/Macrophages, associated with a M1-Polarized Immune Milieu and Prolonged Patient Survival in Gliomas. Brain Pathol. 2015, 25, 491–504. [Google Scholar] [CrossRef]
- Kitange, G.J.; Carlson, B.L.; Schroeder, M.A.; Decker, P.A.; Morlan, B.W.; Wu, W.; Ballman, K.V.; Giannini, C.; Sarkaria, J.N. Expression of CD74 in high grade gliomas: A potential role in temozolomide resistance. J. NeuroOncol. 2010, 100, 177–186. [Google Scholar] [CrossRef][Green Version]
- Massara, M.; Persico, P.; Bonavita, O.; Mollica Poeta, V.; Locati, M.; Simonelli, M.; Bonecchi, R. Neutrophils in Gliomas. Front. Immunol. 2017, 8, 1349. [Google Scholar] [CrossRef]
- Pinton, L.; Masetto, E.; Vettore, M.; Solito, S.; Magri, S.; D’Andolfi, M.; Del Bianco, P.; Lollo, G.; Benoit, J.P.; Okada, H.; et al. The immune suppressive microenvironment of human gliomas depends on the accumulation of bone marrow-derived macrophages in the center of the lesion. J. Immunother. Cancer 2019, 7, 58. [Google Scholar] [CrossRef]
- Yue, X.; Zhao, Y.; Liu, J.; Zhang, C.; Yu, H.; Wang, J.; Zheng, T.; Liu, L.; Li, J.; Feng, Z.; et al. BAG2 promotes tumorigenesis through enhancing mutant p53 protein levels and function. eLife 2015, 4, 8401. [Google Scholar] [CrossRef]
- Driscoll, C.B.; Schuelke, M.R.; Kottke, T.; Thompson, J.M.; Wongthida, P.; Tonne, J.M.; Huff, A.L.; Miller, A.; Shim, K.G.; Molan, A.; et al. APOBEC3B-mediated corruption of the tumor cell immunopeptidome induces heteroclitic neoepitopes for cancer immunotherapy. Nat. Commun. 2020, 11, 790. [Google Scholar] [CrossRef]
- Olson, M.E.; Harris, R.S.; Harki, D.A. APOBEC Enzymes as Targets for Virus and Cancer Therapy. Cell Chem. Biol. 2018, 25, 36–49. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.; Xia, M.; Xu, S.; Wang, C.; Bao, Y.; Jiang, M.; Wu, Y.; Xu, T.; Cao, X. Rhomboid domain-containing protein 3 is a negative regulator of TLR3-triggered natural killer cell activation. Proc. Natl. Acad. Sci. USA 2013, 110, 7814–7819. [Google Scholar] [CrossRef] [PubMed]
- Calissi, G.; Lam, E.W.; Link, W. Therapeutic strategies targeting FOXO transcription factors. Nat. Rev. Drug Discov. 2021, 20, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; King, G.D.; Curtin, J.F.; Candolfi, M.; Xiong, W.; Liu, C.; Puntel, M.; Cheng, Q.; Prieto, J.; Ribas, A.; et al. Combined immunostimulation and conditional cytotoxic gene therapy provide long-term survival in a large glioma model. Cancer Res. 2005, 65, 7194–7204. [Google Scholar] [CrossRef] [PubMed]
- Greuber, E.K.; Smith-Pearson, P.; Wang, J.; Pendergast, A.M. Role of ABL family kinases in cancer: From leukaemia to solid tumours. Nat. Rev. Cancer 2013, 13, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Bhatia, R. The controversial role of Sirtuins in tumorigenesis-SIRT7 joins the debate. Cell Res. 2013, 23, 10–12. [Google Scholar] [CrossRef]
- Carafa, V.; Altucci, L.; Nebbioso, A. Dual Tumor Suppressor and Tumor Promoter Action of Sirtuins in Determining Malignant Phenotype. Front. Pharmacol. 2019, 10, 38. [Google Scholar] [CrossRef]
- Lee, J.; Zhou, P. Cullins and cancer. Genes Cancer 2010, 1, 690–699. [Google Scholar] [CrossRef]
- Wang, C.Y.; Shahi, P.; Huang, J.T.; Phan, N.N.; Sun, Z.; Lin, Y.C.; Lai, M.D.; Werb, Z. Systematic analysis of the achaete-scute complex-like gene signature in clinical cancer patients. Mol. Clin. Oncol. 2017, 6, 7–18. [Google Scholar] [CrossRef]
- Petrilli, A.M.; Fernandez-Valle, C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene 2016, 35, 537–548. [Google Scholar] [CrossRef]
- Kong, Y.; Wang, Z.; Huang, M.; Zhou, Z.; Li, Y.; Miao, H.; Wan, X.; Huang, J.; Mao, X.; Chen, C. CUL7 promotes cancer cell survival through promoting Caspase-8 ubiquitination. Int. J. Cancer 2019, 145, 1371–1381. [Google Scholar] [CrossRef]
- Wustenhagen, E.; Hampe, L.; Boukhallouk, F.; Schneider, M.A.; Spoden, G.A.; Negwer, I.; Koynov, K.; Kast, W.M.; Florin, L. The Cytoskeletal Adaptor Obscurin-Like 1 Interacts with the Human Papillomavirus 16 (HPV16) Capsid Protein L2 and Is Required for HPV16 Endocytosis. J. Virol. 2016, 90, 10629–10641. [Google Scholar] [CrossRef] [PubMed]
- Ou, W.B.; Lundberg, M.Z.; Zhu, S.; Bahri, N.; Kyriazoglou, A.; Xu, L.; Chen, T.; Marino-Enriquez, A.; Fletcher, J.A. YWHAE-NUTM2 oncoprotein regulates proliferation and cyClin. D1 via RAF/MAPK and Hippo pathways. Oncogenesis 2021, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Ren, C.H.; Li, L.; Goltsov, A.A.; Thompson, T.C. Identification and characterization of RTVP1/GLIPR1-like genes, a novel p53 target gene cluster. Genomics 2006, 88, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kho, A.T.; Wu, Q.; Halayko, A.J.; Limbert Rempel, K.; Chase, R.P.; Sweezey, N.B.; Weiss, S.T.; Kaplan, F. CRISPLD2 (LGL1) inhibits proinflammatory mediators in human fetal, adult, and COPD lung fibroblasts and epithelial cells. Physiol. Rep. 2016, 4, 2942. [Google Scholar] [CrossRef] [PubMed]
- Gour, N.; Wills-Karp, M. IL-4 and IL-13 signaling in allergic airway disease. Cytokine 2015, 75, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Geisenberger, C.; Mock, A.; Warta, R.; Rapp, C.; Schwager, C.; Korshunov, A.; Nied, A.K.; Capper, D.; Brors, B.; Jungk, C.; et al. Molecular profiling of long-term survivors identifies a subgroup of glioblastoma characterized by chromosome 19/20 co-gain. Acta Neuropathol. 2015, 130, 419–434. [Google Scholar] [CrossRef] [PubMed]

| Feature | N (%)/Time [Month] | |
|---|---|---|
| All | 14 (100) | |
| Sex | ||
| Male | 9 (64) | |
| Female | 5 (36) | |
| Age [yr] % | ||
| <40 | 2 (14) | |
| 40–49 | 6 (43) | |
| ≥50 | 6 (43) | |
| Grade ! | ||
| II | 5 (36) | |
| III | 2 (14) | |
| IV | 7 (50) | |
| Grade % | ||
| III | 5 (36) | |
| IV | 9 (64) | |
| Grade ! -> % | ||
| II -> III | 3 (21) | |
| II -> IV | 2 (14) | |
| Time to reRT | ||
| II | 5 (36)/80.9 | |
| III | 2 (14)/86.8 | |
| IV | 7 (50)/11.4 | |
| C12 irradiation | ||
| Total dose [GyRBE]/#fx | 30/10 | 5 (36) |
| 33/11 | 6 (43) | |
| 36/12 | 3 (21) | |
| Single dose [GyRBE] | 3 | 14 (100) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knoll, M.; Waltenberger, M.; Furkel, J.; Wirkner, U.; Gahlawat, A.W.; Dokic, I.; Schwager, C.; Adeberg, S.; Rieken, S.; Kessler, T.; et al. Whole Blood Transcriptional Fingerprints of High-Grade Glioma and Longitudinal Tumor Evolution under Carbon Ion Radiotherapy. Cancers 2022, 14, 684. https://doi.org/10.3390/cancers14030684
Knoll M, Waltenberger M, Furkel J, Wirkner U, Gahlawat AW, Dokic I, Schwager C, Adeberg S, Rieken S, Kessler T, et al. Whole Blood Transcriptional Fingerprints of High-Grade Glioma and Longitudinal Tumor Evolution under Carbon Ion Radiotherapy. Cancers. 2022; 14(3):684. https://doi.org/10.3390/cancers14030684
Chicago/Turabian StyleKnoll, Maximilian, Maria Waltenberger, Jennifer Furkel, Ute Wirkner, Aoife Ward Gahlawat, Ivana Dokic, Christian Schwager, Sebastian Adeberg, Stefan Rieken, Tobias Kessler, and et al. 2022. "Whole Blood Transcriptional Fingerprints of High-Grade Glioma and Longitudinal Tumor Evolution under Carbon Ion Radiotherapy" Cancers 14, no. 3: 684. https://doi.org/10.3390/cancers14030684
APA StyleKnoll, M., Waltenberger, M., Furkel, J., Wirkner, U., Gahlawat, A. W., Dokic, I., Schwager, C., Adeberg, S., Rieken, S., Kessler, T., Sahm, F., König, L., Herold-Mende, C., Combs, S. E., Debus, J., & Abdollahi, A. (2022). Whole Blood Transcriptional Fingerprints of High-Grade Glioma and Longitudinal Tumor Evolution under Carbon Ion Radiotherapy. Cancers, 14(3), 684. https://doi.org/10.3390/cancers14030684








