Simple Summary
Hepatocellular carcinoma (HCC) is a major cause of cancer-related death worldwide. As the complex genetic landscape of HCC is considered to compromise the antitumor efficacy of targeted therapy, genetic landscape of hepatocarcinogenesis is necessary to be understood in detail. Here, we summarize the landscape of the liver cancer genome and its intratumor heterogeneity. Recent insight on early genetic alterations in hepatocarcinogenesis, especially those in early HCC and noncancerous liver tissues are also introduced. Importantly recent studies demonstrated that noncancerous liver tissues already possess a variety of somatic mutations. Finally, we summarize recent findings on the multistep accumulation of genetic aberrations throughout liver cancer progression. These genetic aberrations have been used for subtyping of liver cancers, which can be applied for clinical practice in the future.
Abstract
Hepatocellular carcinoma (HCC) is a major cause of cancer-related death worldwide. Although several targeted therapy agents are available for advanced HCC, their antitumor efficacy remains limited. As the complex genetic landscape of HCC would compromise the antitumor efficacy of targeted therapy, a deeper understanding of the genetic landscape of hepatocarcinogenesis is necessary. Recent comprehensive genetic analyses have revealed the driver genes of HCC, which accumulate during the multistage process of hepatocarcinogenesis, facilitating HCC genetic heterogeneity. In addition, as early genetic changes may represent key therapeutic targets, the genetic landscapes of early HCC and precancerous liver tissues have been characterized in recent years, in parallel with the advancement of next-generation sequencing analysis. In this review article, we first summarize the landscape of the liver cancer genome and its intratumor heterogeneity. We then introduce recent insight on early genetic alterations in hepatocarcinogenesis, especially those in early HCC and noncancerous liver tissues. Finally, we summarize the multistep accumulation of genetic aberrations throughout cancer progression and discuss the future perspective towards the clinical application of this genetic information.
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most lethal malignancies worldwide [1,2,3]. HCC therapy, including surgical resection, liver transplantation, transcatheter arterial chemoembolization (TACE), percutaneous radiofrequency ablation, percutaneous ethanol injection, and molecular targeted therapy (MTT), is selected based on tumor status and liver function [1,4,5,6]. Whereas the curative treatment strategies such as hepatectomy and liver transplantation have resulted in favorable outcomes in terms of overall or recurrence-free survival times, the survival rate of unresectable HCCs is quite poor, as shown in Table 1 [7,8,9,10,11,12,13,14,15,16,17,18]. Approximately 10 years after the introduction of sorafenib, the first approved targeted therapy agent for HCC [17], the non-inferiority of lenvatinib with regard to overall survival (OS) was demonstrated in a phase III trial, resulting in the latter’s inclusion in clinical practice as another choice of first-line systemic therapy [18,19,20]. Multi-kinase inhibitors regorafenib and cabozantinib, as well as the VEGFR2 inhibitor ramucirumab, have been approved as second-line MTT following sorafenib treatment [8,21].

Table 1.
Summary of treatment outcomes of recent treatment strategies.
Immune checkpoint inhibition has also been introduced, with PD-1 inhibitor nivolumab being the first checkpoint inhibitor approved by the FDA for the treatment of HCC in patients who have been previously treated with sorafenib. Of note, another PD-1 inhibitor, pembrolizumab, was approved for any type of cancer with high microsatellite instability (MSI-high), HCC included [22]. In addition to regimens utilizing immune checkpoint inhibitors (ICIs), recently developed regimens utilize a combination of ICI and other molecular targeting agents. The IMbrave150 trial demonstrated that dual therapy using atezolizumab, a PD-L1 antibody, with bevacizumab, which is an antibody directed against VEGF, significantly improved OS, progression-free survival (PFS), and the objective response rate (ORR) compared to monotherapy using sorafenib as the first-line chemotherapeutic agent for patients with advanced HCC [9].
Although the treatment options for patients with advanced HCC have been increasing [23], the antitumor efficacy of these approved agents remains limited. In the IMbrave150 trial that showed among the most favorable results for HCC to date, the ORR assessed using RECIST1.1 after the administration of atezolizumab with bevacizumab was 27.3% [9]. Among alternate choices for patients with advanced HCC, the ORR after administration of sorafenib was less than 5% in the SHARP trial [17] and was limited in clinical practice [24,25,26,27,28], whereas the ORR for lenvatinib was 18.8% in the REFLECT trial [18]. Although recent advances in systemic therapies for HCC are remarkable [29], there have been no established treatment strategies effective for every patient with advanced HCC to date [30,31].
While considering the development of a novel therapeutic strategy for liver cancer, it is essential to understand the landscape of accumulating genetic aberrations in liver cancer cells [32]. Utilizing recently developed next-generation sequencing (NGS) approaches, international projects have carried out comprehensive genetic analyses of liver cancer, identifying several driver genes associated with liver carcinogenesis [33,34,35,36,37,38]. Recent molecular biology studies have identified molecular pathways associated with liver cancer [3,39,40]. Additionally, mechanisms underlying the accumulation of several cancer-related genetic aberrations in tumor cells during multistep liver carcinogenesis have been investigated. In this review article, we summarize the recent investigations on the genetic alterations that accumulate during the multistep process of hepatocarcinogenesis, ranging from precancerous liver tissues and early HCC to advanced HCC (Table 2).

Table 2.
Summary of the multistep accumulation of genomic and transcriptomic aberrations during hepatocarcinogenesis.
2. Genetic Landscape of HCC
2.1. Comprehensive Genetic Analysis of HCC
As evidenced by the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) project [48,49], vast amounts of information on genetic aberrations across various types of tumor tissues have been collected worldwide. In addition, whole-exome and genome analyses of liver cancer, including HCC, have been recently carried out in different countries, elucidating the greater picture of genetic aberrations accumulated in transformed liver tissues [50].
Large-scale whole-exome sequencing (WES) of the liver cancer genome has been conducted mostly in the United States (TCGA) [50] as well as in Japan, China, and France (ICGC) [33,36,37,38,51], while whole-genome sequencing (WGS) has been performed mainly in Japan. WGS analyses revealed over 9000 point mutations per human liver cancer sample, with somatic mutations detected in approximately 40–80 protein-coding genes per specimen [34,35,52]. Previous genetic analyses have detected a number of candidate HCC driver genes (Figure 1).
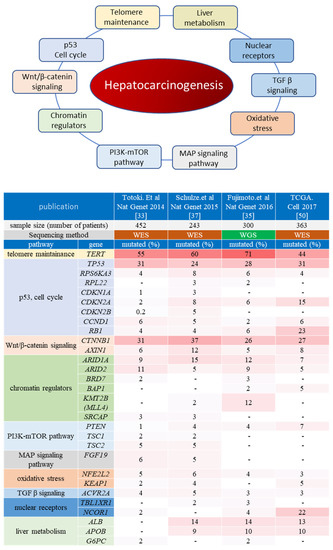
Figure 1.
Oncogenic pathways associated with hepatocarcinogenesis. The upper panel shows the major oncogenic pathways elucidated via comprehensive genome analysis projects. The lower heatmap indicates the genes which have been reported as putative liver cancer driver genes in at least two publications of ICGC/TCGA projects. The top row shows each publication, and the second row shows the number of liver cancer samples analyzed. The major pathways and representative cancer-related genes for each pathway are listed on the left. The mutation frequency of each gene (percentage of the cases with mutated genes among all cases analyzed in each cohort) is shown as a heatmap. WES: whole-exome sequencing; WGS: whole-genome sequencing.
In addition to coding mutations, noncoding mutations have also been detected, and some HCC candidate driver mutations have been identified among them, such as the promoter sequence of the TERT gene, as well as long intergenic noncoding RNAs [35,53]. Various aberrations can be observed in the TERT promoter region, including hotspot point mutations, structural alterations, as well as hepatitis B virus HBV genome integration. Importantly, TERT-associated genetic alterations are found in over 70% of HCC tissues, making them the most frequent aberrations associated with HCC [35].
WES/WGS analysis confirmed TP53 and CTNNB1 as the most frequently mutated coding genes in HCC, with chromatin modulators ARID1A and ARID2 also recurrently mutated [33,37]. Apart from frequently observed mutations in these driver genes, many HCC-related mutated genes have been identified in cancer patients, with a frequency of less than 5% among cases described by the so-called long-tail distribution of driver genes, which facilitates the genetic heterogeneity of HCC [33].
Mutational signature analyses following whole-exome/genome sequencing [54] have revealed that the most frequent patterns of single nucleotide changes in HCC are C>T/G>A as well as T>C/A>G. Some liver cancer-specific mutational signatures, such as signatures 12 and 16 in the Catalogue Of Somatic Mutations In Cancer (COSMIC) Mutational Signature, have been identified, although the etiologies of these signatures remain unknown [33,55]. Of note, the whole-exome analysis revealed differences in mutational signatures according to geographical origin among European, Asian American, and Japanese people. The correlation between specific driver gene mutations and genome-wide mutational signatures has also been demonstrated [33]. Thus, comprehensive genetic analyses in different patient populations have elucidated the landscape of genetic aberrations accumulated in liver cancer cells.
2.2. Molecular Pathways Associated with Hepatocarcinogenesis
Hepatocarcinogenesis-associated molecular pathways have been summarized in detail previously [56]. As described in the above section, TERT-associated genetic alterations are the most frequently detected in HCC. Nevertheless, various other molecular pathways have been demonstrated to contribute to hepatocarcinogenesis. For example, Schulze et al. summarized HCC-related genetic alterations into eleven categories, including telomere maintenance, Wnt/β-catenin signaling, p53/cell cycle, oxidative stress, epigenetic regulation, PI3K-Akt-mTOR, MAPK, and hepatic differentiation [37]. Among them, genes involved in Wnt/β-catenin and p53/cell cycle signaling are frequently aberrated in HCC. These findings suggested that HCC is not caused by one particular driver mutation but involves several oncogenic pathways, making tumors highly heterogeneous [56]. Herein, we summarize the main pathways and processes involved in hepatocarcinogenesis (Figure 1).
2.2.1. Telomere Maintenance
Telomerase reactivation is a major event in malignant transformation. As in various other malignancies, telomerase expression is known to be increased in most HCC tumors [35,53,57]. The causes of telomerase reactivation include hotspot mutations in the TERT promoter sequence (54–60%), gene amplification (5–6%), structural variation (4–5%), and HBV integration in the promoter or gene body (10–15%). In general, these aberrations are mutually exclusive [35].
Another mechanism of telomerase reactivation is through epigenetic changes. Hypermethylation of specific regions has been confirmed as associated with TERT mRNA expression in breast, colorectal, and other cancers [58,59,60]. We previously demonstrated the aberrant methylation status of the TERT promoter in HCC tissue, along with elevated TERT mRNA expression levels [46].
2.2.2. Wnt/β-Catenin Pathway
The Wnt/β-catenin pathway is one of the most frequently activated oncogenic pathways in HCC. It is usually upregulated by activating CTNNB1 mutations or inactivating mutations of the AXIN1, NCOR1, and APC genes [32,61,62]. These mutations were detected in a mutually exclusive manner. Interestingly, previous WES studies revealed that the CTNNB1 mutation tended to co-occur with TERT mutations [33]. Although AXIN1 has been generally described as a well-known negative regulator of the Wnt/β-catenin pathway, it is notable that Abitbol et al. recently reported that AXIN1-mutated HCCs occur independent of the Wnt/β-catenin pathway and involve Notch and YAP pathways [63].
CTNNB1 mutations are sometimes considered when molecular subclasses of HCC are classified. Importantly, CTNNB1 is the only gene with a mutation that defines a specific subclass of HCCs (the perivenous-type HCC subclass [64]), with a particular metabolic zonation phenotype or G5/G6 subtypes in Boyault’s classification [65]. Tumors carrying CTNNB1 exon three mutations are generally moderately differentiated and of low-to-intermediate aggressiveness [66].
2.2.3. p53/Cell Cycle
p53/cell cycle signaling is altered in at least half of HCC patients with TP53 mutations. In general, tumor suppressor genes are more frequently mutated in cancer. The retinoblastoma (RB) pathway, which controls progression from the G1 to the S phase of the cell cycle, is inactivated in HCC mainly as a result of homozygous CDKN2A deletions or RB1 mutations. Among the genes involved in this pathway, recurrent HBV insertions have been reported in CCNE1 (coding cyclin E1). In addition, the focal amplification of CCND1/FGF19, which is downstream of this oncogenic pathway, has been reported in over 10% of HCC tissues. A recent study demonstrated that some HCC subgroups exhibit cyclin activation through various mechanisms, including HBV and adeno-associated virus type 2 (AAV2) insertions and enhancer hijacking and recurrent CCNA2 fusions, defining a homogenous entity of aggressive HCC [67].
Importantly, TP53 mutations have been demonstrated to be related to the aggressiveness, some phenotypes, and etiologies of HCC. For example, TP53 mutations are associated with aggressive HCCs of the STEM (Désert’s classification) [64] S2 (Hoshida’s classification) [68] or G3 (Boyault classification) subclasses [65]. These tumors tend to be associated with HBV infection and genomic instability [69]. Moreover, there is an etiological link between TP53 mutations and aflatoxin DNA adducts in intertropical regions of the World [70].
2.2.4. Chromatin Remodeling Factors
Alterations in chromatin remodeling factors, such as ARID1A, ARID1B, ARID2, and BRD8, are also considerably associated with hepatocarcinogenesis [33,37]. In particular, mutations of ARID1A and ARID2, which encode key players in SWI/SNF chromatin remodeling complexes, are frequently detected in liver cancer [71]. These complexes modify the chromatin structure and nucleosome position, thus indirectly regulating the determination of cell fate. Recurrent somatic alterations in genes encoding histone methylation writer family proteins, mainly MLL1-4, are also observed in HCC. Notably, the MLL4 gene is known to be the second most frequent region of HBV integration in HCC [72,73].
2.2.5. PI3K/Akt/mTOR and RAS/RAF/MAPK Pathways
Activating mutations of PIK3CA and inactivating mutations of TSC1 as well as TSC2 lead to the activation of Akt/mTOR signaling in a subset of HCC tumors. In addition, homozygous deletion of PTEN, which encodes an inhibitor of the PI3K kinase, has been identified in 1–3% of HCC cases [33,35,37].
Activating mutations in genes belonging to the RAS family are rarely observed in HCC (<2%), and inactivating mutations of RP6SKA3, encoding RAS inhibitor RSK2, were identified in 2–9% of liver cancer cases. The inactivation of RSK2 releases associated negative feedback, inducing constitutive activation of the pathway. In approximately 5–10% of HCC cases, the PI3K/Akt/mTOR and RAS/RAF/MAPK pathways are upregulated through amplification of the FGF19/CCND1 locus [33,35,37].
2.2.6. Other Oncogenic Pathways in Hepatocarcinogenesis
Genes associated with oxidative stress are altered, such as activating mutations in NFE2L2 (encoding NRF2) or inactivating KEAP1 mutations in 5–15% of liver cancer cases [33,35,37]. Further, genes associated with the IL-6/JAK-STAT pathway, TGF-β signaling, and liver differentiation are also altered in some cases of liver cancer [33,35,37]. Interestingly, WGS revealed that mutational clusters accumulated in ALB (encoding albumin) or APOB (encoding apolipoprotein B), although these were not directly associated with cancer development. Most ALB mutations are short indels in the gene body, whose significance with regard to hepatocarcinogenesis has not been elucidated [35].
Some driver genes discovered by WGS projects were characterized based on their focal amplifications, including VEGF (coding vascular endothelial growth factor), MYC, and MET, leading to overexpression of their mRNA, which results in the activation of various oncogenic signaling pathways [74]. As one of the most typical steps during multistep hepatocarcinogenesis, the acquisition of a hypervascular feature comprising genes associated with angiogenesis, is considered important when examining its pathogenesis and molecular targets. Especially, VEGFA amplification has been discussed as a biomarker to predict the treatment effect of some molecular targeting therapies [24,75]. Notably, analyzing the WGS data has discovered some noncoding drivers in HCC. Fujimoto et al. reported two long intergenic noncoding RNAs (lincRNAs), NEAT1 and MALAT1, as significantly mutated drivers in HCC tissues [35], whereas a recent pan-cancer WGS analysis revealed that U1 small nuclear RNAs (snRNAs), the noncoding component of the spliceosome, are also mutated, suggesting that driver genes or locus can exist over a wider range of genomic regions [76].
3. Intratumor Genetic Heterogeneity
3.1. Intratumoral Heterogeneity Revealed by NGS
To date, phylogenetic analyses utilizing multiregional sequencing methodology have been employed for the analysis of various cancer types, and the modes of cancer evolution have been revealed to differ among various cancers [77]. The modes of evolution include some classical evolutional models, including a linear evolution model with sequentially acquired driver mutations (late diversification) [78] or branched evolution with many standing lineages (early diversification) [79,80,81]. For example, Gerlinger et al. explored the intratumor heterogeneity of metastatic renal cell carcinoma through multiregional sequencing [79]. They demonstrated the branched evolution of cancer cells, where some mutations acquired by the parental cell at the early stage of tumorigenesis are inherited by the majority of tumor cells and are called “ubiquitous” mutations, while mutations acquired by each subclone in the comparatively late phase of tumorigenesis are called “shared” or “private” mutations in a phylogenetic tree. Ubiquitous mutations are also described as trunk mutations, whereas private or shared mutations are also described as branch mutations [82]. These evolutionary processes lead to the formation of subclones in each tumor, generating intratumor clonal diversity, or so-called intratumor heterogeneity [79,81]. Similarly, intratumor and intertumor genetic heterogeneity have been identified in colorectal cancer, glioblastoma, and many other malignancies [80,83,84,85,86,87].
Intratumor heterogeneity might explain the difficulties encountered in the validation of oncology biomarkers owing to sampling bias. For example, in the multiregional sequencing analysis of RCC by Gerlinger et al., 63–69% of all somatic mutations found based on multiregion sequencing were heterogeneous and not detectable in every sequenced region [79]. Multiregional WES analysis of HCC by Lin et al. demonstrated a median of 38.9% (range: 5.5–91.7%) of heterogeneous variants, and the genetic heterogeneity could also be validated at the protein level by immunohistochemistry [83].
It should be noted that when branch mutations are targeted by certain drugs, the therapeutic effect may be limited to the subpopulations harboring the branch mutation. Importantly, if trunk mutations shared by most of the tumor cells are identified, the effect of therapy targeting these mutations is expected to be favorable. Gerlinger et al. suggested that reconstructing tumor clonal architectures and the identification of common mutations located in the trunk of the phylogenetic tree might contribute to more robust biomarkers and therapeutic approaches [79]. In this context, it is important to understand the hierarchy of driver mutations that facilitate tumor heterogeneity through cancer cell evolution.
3.2. Intratumoral Heterogeneity of HCC
Intratumor heterogeneity has been also explored in liver cancer including HCC [46,80,82,88,89,90,91,92]. Lin et al. performed multiregional WES on large tumor tissues from 10 HCC patients and constructed a phylogenetic tree of each tumor, showing that identical liver nodules harbored a variety of subclones [82]. Moreover, Zhai et al. reported a multiregional WGS or WES study on nine HCC patients, whereas Xue et al. conducted a phylogenetic analysis of 53 samples from 10 patients with HCCs. They described a variety of evolutionary trees with different degrees of intratumoral heterogeneity, most of which exhibited the branched evolution in HCC [80,88]. Every report demonstrates that the numbers of passenger mutations and some driver mutations are shared by all different regions of a single tumor, suggesting that intratumor heterogeneity in HCC can develop from a single origin cell.
Based on the branched evolution model of HCC, a schematic evolutionary tree is shown, along with radiological findings of multistep hepatocarcinogenesis, in Figure 2. In this evolutionary model, a normal parental cell acquires certain trunk mutations associated with the initiation of malignant transformation, followed by the emergence of a single parental cancer cell. Thereafter, this parental cell acquires additional genetic aberrations, including driver and passenger mutations, clonally expanding with the numbers of subclones [69,70,81]. During this multistep cancer cell evolution, many subclones emerge in a single nodule, contributing to intratumor heterogeneity [81].
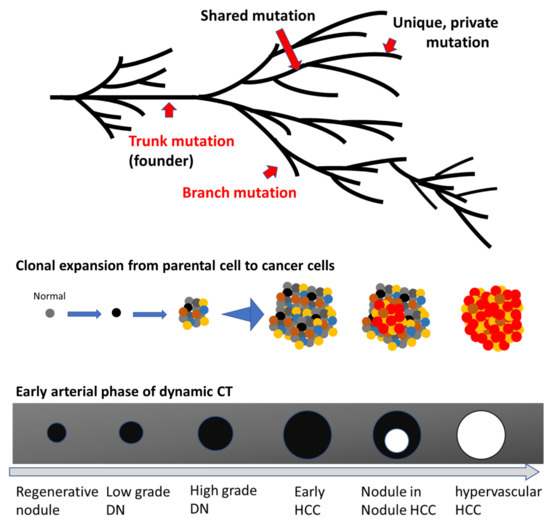
Figure 2.
Scheme of the multistep evolution of typical hepatocellular carcinoma (HCC) The upper scheme is a phylogenetic tree showing a scenario of the progression from a normal cell to an advanced HCC. Examples of trunk and branch mutations in addition to shared as well as unique mutations are shown by red arrows. The middle scheme depicts the evolutionary progression from parental cancer cell to dysplastic nodule (DN), early HCC, classical HCC, and, finally, metastatic HCC. Each colored dot indicates a tumor cell with a different mutational profile, and each nodule consists of various tumor cells with distinct genetic alterations, contributing to intratumoral heterogeneity. The red dots indicate progressive tumor cells. This scheme omits nonepithelial cells such as vessels and fibrotic tissues. The lower scheme describes the typical radiological change of liver nodules during multistep hepatocarcinogenesis via nodule-in-nodule HCC, shown in the early arterial phase of contrast-enhanced dynamic computed tomography (CT) imaging. The black areas indicate nodules described as hypovascular areas in the early arterial phase of dynamic CT, whereas white circles mark hypervascular nodules in the early arterial phase of dynamic CT.
Regarding driver genes associated with HCC, many mutations were detected heterogeneously in some reports of multiregional sequencing. However, by calculating the cancer cell fraction of all mutations, Lin et al. demonstrated that several driver mutations are fully clonal within some tumor regions. They reported that TP53 mutations are likely detected as trunk mutations, although less than half of HCC cases had TP53 mutations. TERT promoter mutations were almost always detected ubiquitously [80,82]. Other driver genes can be detected as trunk mutations, such as ARID1A and SETD2, but the mutated trunk mutations vary across cases, which can be considered intertumor genetic heterogeneity [80].
Multiregional analysis of HCC has also been conducted using RNA sequencing, as well as proteome analysis, revealing the complex nature of HCC tumor heterogeneity [80], which might complicate the treatment of advanced HCC via targeted therapy. Furthermore, recent progress in single-cell sequencing technologies has enabled an understanding of the heterogeneity of microenvironments of HCC tissues. These analyses have demonstrated the ecosystem of HCC, including not only cancer cells but also infiltrating immune cells. As the novel therapeutic strategies for HCC include immunocheckpoint inhibitors (ICIs), exploring the single-cell level heterogeneity, including immune cells, should have significance for translational research, such as the prediction of ICI treatment efficacy.
4. Genetic Alterations in Early Hepatocarcinogenesis
4.1. Genetic Landscape of Early HCC
The heterogeneity described in the previous section is based on the genetic landscape of classical surgically resected HCCs, arising as the result of multistep carcinogenesis. Considering the importance of early HCC detection, it is essential to understand the genetic landscape during the early stages of hepatocarcinogenesis.
Liver cancer usually develops from liver cirrhosis, and after a dysplastic premalignant nodule is initially formed, it becomes early HCC (eHCC), followed by progression into hypervascular classical HCC. Several studies have explored the relatively early stages of multistep hepatocarcinogenesis. In particular, the genetic characteristics of early HCCs were recently reported by Nault et al. [47]. Using Sanger sequencing, they analyzed TERT promoter hotspot mutations, observing that approximately 60% of eHCC cases harbored TERT mutations, while nearly 65% of classical HCC tumors had TERT promoter mutations. This clearly showed that many eHCC have already acquired point mutations in the TERT promoter region at this early stage. In contrast, mutations in other HCC-associated driver genes, such as TP53 and CTNNB1, were not detected in the eHCC samples. It should be noted that, due to the low sensitivity of Sanger sequencing, this analysis may have missed somatic mutations with low variant allele frequencies. Thus, it cannot be concluded whether certain cancer cells with driver mutations other than those of the TERT promoter mutations existed in the analyzed eHCC tissues.
4.2. Genetic Landscape of Precancerous Liver Tissues
To elucidate genetic aberrations developing in the earliest step of hepatocarcinogenesis, we previously conducted a genetic analysis of nontumor liver cirrhosis tissues, from which liver cancer cells originate [43]. To this end, we focused on the genetic alterations accumulated in the regenerative nodules (RNs) of cirrhotic livers. While the so-called bulk sampling from normal tissue is likely to fail in the detection of somatic mutations at the single-cell level, RNs have been previously demonstrated to possess clonal structures [93,94,95,96], which justified the genetic analysis of these structures. Based on WES, the somatic mutation rate of RNs was approximately one-tenth that of HCC, and the most enriched mutational signature was COSMIC signature 1 [97], mimicking that of HCC. The majority of the somatic mutations forming COSMIC signature 1 in the regenerative nodules did not include HCC-associated driver genes but passenger mutations, which is considered reasonable because COSMIC signature 1 is a clock-like or aging-related signature. Targeted deep sequencing of HCC-associated driver genes demonstrated that several damaging nonsynonymous mutations with low allele frequencies were acquired in RNs. One of the genes recurrently mutated in several nodules was major HCC-related chromatin remodeling factor ARID1A. None of the 205 RNs analyzed by targeted deep sequencing harbored TERT promoter mutations, and the TERT mRNA levels of RNs were as low as those in healthy liver tissue. These results suggested that RNs already accumulate several genetic alterations while their telomerase activity remains suppressed.
Similarly, two large-scaled genetic analyses have been conducted on cirrhotic liver tissues (Table 2). Zhu et al. carried out a genetic analysis of non-tumor cirrhotic liver tissues of METAVIR stage F1 to F4 [42]. Using WES as well as targeted sequencing, they identified PKD1, KMT2D, PPARGC1B, and ARID1A as significantly mutated genes in cirrhotic liver tissues. The loss of these genes was considered regeneration-promoting and hepatoprotective. This concept resembles the suggestion of a previous report stating that NOTCH1 mutations frequently detected in normal esophageal tissues exert a tumor-suppressive effect [98].
In addition, Brunner et al. conducted a WGS study on 482 microdissections from five normal and nine cirrhotic liver tissues [41].
In this study, multiregional sampling was performed using a single RN. The different regions of this RN shared common somatic mutations, which differed from those detected in neighboring RNs. These observations raised two important points, the first being that a hepatocyte acquiring a specific mutation proliferates in the RN and goes on to form a clonal cell population. Second, clonal expansion does not cross over the fibrotic tissues between RNs, creating the individual mutational landscape of each RN. Although regenerative nodules are not neoplastic tissues, multiregional WGS clearly revealed their clonal structures. Comparison of mutational profiles between regenerative nodules and normal livers revealed that the cirrhotic liver not only has a higher mutational burden but also harbors various structural variants, including ones that have undergone chromothripsis. These results indicated that inflamed nontumor liver tissues, such as the RNs, have already acquired various genetic aberrations prior to malignant transformation, although most of these somatic mutations are considered passenger mutations or mutations contributing to an inflammatory microenvironment adaptation. It should be noted that TERT promoter mutations or other TERT-associated genetic alterations were not observed in RNs, supporting the previously discussed multiregional genetic analysis of cirrhotic liver tissues. Considering these data, even if hepatocytes acquire somatic mutations under chronic inflammation, they may not evolve into malignant cells. As many researchers have indicated, genetic aberrations associated with the TERT gene should be the key event in malignant transformation, with the reactivation of telomerase being one of the first steps in hepatocarcinogenesis.
However, TERT promoter hotspot mutations can be detected in 6–19% of dysplastic nodules, whereas 10 other well-known driver gene mutations could not be detected by Sanger sequencing [47]. To date, only a little is known about the genetic landscape of dysplastic nodules, and there have been no whole-genome or whole-exome sequencing data for liver dysplastic nodules to date. Genetic analysis of low-grade and high-grade dysplastic nodules in comparison to regenerative nodules and chronic hepatitis tissues is therefore essential for a deeper understanding of the genetic landscape during early-stage hepatocarcinogenesis.
5. Multistep Acquisition of Genetic Aberrations during Hepatocarcinogenesis
5.1. The Significance of Comprehensive Analysis Using Clinical and Genetic Data
A multiregional tumor genetic analysis explored intratumoral heterogeneity in HCC [80,82], revealing that most tumor cells share trunk mutations, whereas subclones within the same tumor eventually attain various branch mutations. As these multiregional sequencing analyses were based on random sampling without consideration of the biological features of obtained tissue specimens, it is unclear whether the branch mutations detected in these multiregional sequencing analyses were driver mutations or simply “passenger” nucleotide alterations.
There are apparent differences in biological characteristics between early-stage HCCs, which are detected as hypovascular nodules in the early arterial phase of dynamic CT or EOB-MRI imaging, and classical HCCs, which are characterized by hypervascularity in the early arterial phase and hypovascularity in the portal phase of dynamic CT or EOB-MRI imaging. As the therapeutic strategies for hypovascular early HCC and hypervascular classical HCC differ in clinical practice, it is important to determine which branch mutations drive the progression from hypovascular early HCC to hypervascular classical HCC. To assess this, mutation data from each specimen should be accompanied by radiological, pathological, as well as clinical information.
5.2. Whole-Genome Mutational Analysis Using Nodule-in-Nodule HCCs
In order to elucidate the multistep accumulation of genetic aberrations in a single liver tumor, we focused on rare HCC phenotypes with nodule-in-nodule appearance (NIN-HCC), consisting of hypervascular (progressed) HCC surrounded by hypovascular (early) HCC arising from a common origin. Whereas there are various progression patterns for HCCs, such as the confluent multinodular type and infiltrative type for which the progression pattern could be different from the NIN evolutional pattern, NIN-HCC has been a good model to explore multistep carcinogenesis to date. Transcriptomic and genome-wide copy number analyses have been previously carried out in NIN-HCC samples, revealing differences in expression and copy number alterations between early and progressed HCC [44,99]. We conducted multiregional WGS of NIN-HCCs and compared genetic alterations between hypovascular and hypervascular HCC in identical nodules [46]. Based on the genetic landscape, together with pathological and radiological findings, we examined the stepwise evolution of cancer cells from slow-growing hypovascular HCC cells to classical HCC.
Phylogenetic analysis revealed thousands of point mutations, and even several structural variations were identified as trunk mutations at the early stage of hepatocarcinogenesis. Among them, TERT-associated aberrations were present as trunk mutations in all cases examined, including point mutations in the promoter region, chromosomal translocations, or HBV integration. Consequently, TERT mRNA levels were significantly elevated in almost all cases. Genetic aberrations and TERT reactivation are considered key events in the initiation of liver carcinogenesis. In some cases, chromothripsis, which is a large-scale genome rearrangement with a focal accumulation of structural variations such as translocations, duplications, inversions, and deletions, is detected at the trunk level. It is important that such a catastrophic genetic event can occur in the initiation step of early hepatocarcinogenesis.
In contrast, various genes involved in cell growth pathways are aberrantly expressed in hypervascular HCC. Further, these tend to differ between cases. One case harbored LOH of PTEN at the early stage of HCC, as well as a second hit of the PTEN gene in the form of a nonsense mutation, which was considered to have driven the progress toward hypervascular HCC. In another case, a CTNNB1 mutation was a hypervascular HCC-specific genetic alteration. Further, genes associated with the Wnt/β-catenin pathway, such as CTNNB1, AXIN1, or HNF4A, were mutated only in hypervascular aHCC regions with relatively high variant allele frequencies, suggesting that these were associated with tumor progression from hypovascular to hypervascular HCC in the cohort.
These results are of major relevance when considering therapeutic targets in different liver cancer patients. For example, telomerase could be a pivotal candidate drug target for the suppression of hypovascular early HCC development, while proteins associated with Wnt or PIK3CA-mTOR signaling can be targeted to inhibit the progression from hypovascular to hypervascular HCC. Taken together, our findings indicated that the pivotal targets associated with tumor progression differ between cases, highlighting the importance of personalized strategies for the identification of therapeutic targets and the respective MTT for each patient.
5.3. Genetic Profiles with Tumor Progression
The above-described multiregional sequencing study focused on the multistep accumulation of genetic aberrations in a single nodule during early disease, that is, within BCLC stages 0 or A. In contrast, Nault et al. analyzed the genetic profiles of hundreds of HCC tissues of BCLC stage 0, A, B, and C [100]. They compared genetic landscapes between advanced-stage HCC and BCLC 0/A tumors. Somatic alterations of SF3B1, RB1, and TP53 were enriched in advanced-stage tumors, whereas CTNNB1 mutations were more frequently detected in BCLC 0/A HCCs. Notably, the mutation frequencies of other classical genetic drivers, such as the TERT promoter, ARID1A, and ARID2, did not differ between advanced and early BCLC stage HCCs.
There have been several combined transcriptomic–genomic classifications of HCCs, some of which are tightly associated with tumor progression. Désert et al. recently analyzed a 1,133-HCC transcriptomic metadata set along with validation based on a 210-HCC RNA-sequencing set, classifying all HCCs into four categories, namely periportal (PP), perivenous (PV), extracellular matrix (ECM), and STEM (displaying stem cell features) subclasses [64]. From the molecular profiling, they demonstrated the existence of increasingly aggressive tumor phenotypes from PP through PV, ECM, and STEM subtypes. CTNNB1 mutations are frequently detected in the PV subtype, which is a well-differentiated HCC subclass carrying mutant CTNNB1 and expressing β-catenin target genes. CTNNB1 mutations are also enriched in the G5/G6 type in Boyault’s classification and Hoshida’s S3 subtype [68], either of which fit within the non-proliferative HCC class [32]. However, TP53 mutations are more frequent in poorly differentiated tumors, such as the STEM subtype in Désert’s classification [64], the S2 in Hoshida’s system [68], and G3 in Boyault’s classification [65]. Interestingly, BCLC classification, from stages A to C, also follows transcriptomics classification from the periportal-type to STEM-type of Désert’s classification [64]. In this manner, recent molecular subtype classifications are closely associated with clinical phenotypes associated with the multistep progression of HCC.
5.4. Multi-Omics Analyses of Multistep Hepatocarcinogenesis
Different levels of omics data on cancer development are intricately associated with each other. For example, the gene expression profile is influenced not only by mutations but also by epigenetic modifications as well as other mechanisms, and some reports have demonstrated that proteomic and transcriptomic HCC data often do not correspond to mutational data. In light of these observations, omics approaches other than DNA sequencing are essential in order to understand the molecular basis of multistep hepatocarcinogenesis. In this context, Midorikawa et al. comprehensively compared the whole-exome mutational landscape as well as transcriptomic and epigenetic profiles between early and advanced (overt) HCC. They classified 191 HCC cases using RNA-seq and BeadArray data observing that Wnt target genes, as well as target genes downstream of the p53/RB pathway (cell cycle-related genes), were rarely upregulated in early HCC, despite the frequent mutations of CTNNB1 and TP53 at that stage. These findings highlight that mutational data alone are not sufficient to explain changes in pathway activity. Downregulation of the CDH1 gene and the chromosomal deletion of the 4q and 16q arms were associated with the transcriptional activation of downstream targets of the Wnt and p53/RB pathways, respectively, during HCC progression. Multi-omics analysis using early and advanced tumor regions of NIN-HCC tissues was also conducted, and the multistep genetic and transcriptomic alterations during HCC progression were validated. In contrast, interestingly, the global methylation status was found to be maintained throughout HCC progression.
Based on the methylation status and mutational/transcriptional profiles, patients were categorized into four groups (G1–G4): the G1 (normal-like) grouping included patients with certain HCC-related gene mutations such as in TP53 and CTNNB1 during the progression phase, whereas the G2 (global hypomethylation) group showed mutations in TP53 at early stages and subsequent 4q or 16q loss during the progression phase. Patients in the G3 group (stem-like methylation) were characterized by mutations in TP53, EZH2 upregulation and showed poor survival. The G4 group (CpG island methylation) included patients with TP53 and CTNNB1 mutations during the progression phase. In each case, genetic and transcriptional characteristics in the progression phase included the upregulation of genes related to the cell cycle, Wnt target gene activation, and cellular growth. Further, the immortalization of cells by TERT upregulation was shown to be a common aberration at the initiation step in all subgroups evaluated [78].
Taken together, a two-step classification of genetic aberrations in multistep hepatocarcinogenesis can be performed; the first step includes TERT-aberration associated cell immortalization, and the second step involves the abnormal upregulation of genes associated with the cell cycle and cell growth pathways caused by genetic and transcriptional aberrations (Table 1).
6. Conclusions and Perspectives
In the current work, we summarized the genetic landscape throughout multistage hepatocarcinogenesis, including precancerous tissue, early HCC, and advanced HCC. In the early stages of tumorigenesis, hepatocytes acquire TERT-associated genetic aberrations and are immortalized. Subsequently, various genes associated with cell cycle regulation, chromatin remodeling, and growth-associated signaling pathways are dysregulated in the immortalized parental cell. The accumulation of these genetic and expression changes drives the progression of early to advanced HCC, complicating the tumor’s genetic profile. Intratumoral HCC heterogeneity is thus established, compromising targeted molecular therapy. In order to understand the molecular mechanisms underlying multistep hepatocarcinogenesis, the application of multiregional and multi-omics strategies is crucial, and thus further investigations utilizing novel sequencing technologies are warranted.
Importantly, genome-based precision medicine has already been introduced into the clinic, supporting the decision-making process of anti-cancer drug selection. Nevertheless, even though a number of agents that target the products of genetic aberrations have been developed, only a few patient subgroups benefit from those in the clinic. Recent whole-genome multi-omics analyses data have clearly answered for the reason why the current genome-based medicine has comparatively low efficacy. First, sequencing data obtained via targeted sequencing is only a small part of the whole-genome mutational profile, potentially overlooking various other genetic aberrations associated with tumor initiation and progression. Second, mutations detected by targeted sequencing include certain trunk mutations together with many branch mutations, some of which are not crucial for tumor progression. Further, bulk sampling makes it difficult to distinguish between trunk and branch mutations in clinical specimens. Third, expression data is just as important as mutation data regarding multistep carcinogenesis, necessitating the use of multi-omics approaches, including WES and WGS sequencing, in clinical genome-based treatment. Overcoming these limitations will allow for the realization of a much more qualified anti-HCC strategy based on the genome profiling of clinical specimens.
Author Contributions
Conceptualization, H.T. and A.T.; writing—original draft preparation, H.T.; writing—review and editing, A.T., Y.E. and K.T.; supervision, H.M. and H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We thank Akihiro Fujimoto and Yoshihide Ueda, for their helpful advice.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Forner, A.; Llovet, J.M.; Bruix, J. Hepatocellular carcinoma. Lancet 2012, 379, 1245–1255. [Google Scholar] [CrossRef]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, H.; Takai, A.; Inuzuka, T.; Marusawa, H. Genetic basis of hepatitis virus-associated hepatocellular carcinoma: Linkage between infection, inflammation, and tumorigenesis. J. Gastroenterol. 2017, 52, 26–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Serag, H.B. Hepatocellular carcinoma. N. Engl. J. Med. 2011, 365, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012, 142, 1264–1273.e1261. [Google Scholar] [CrossRef] [Green Version]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Meyer, T.; Cheng, A.L.; El-Khoueiry, A.B.; Rimassa, L.; Ryoo, B.Y.; Cicin, I.; Merle, P.; Chen, Y.; Park, J.W.; et al. Cabozantinib in Patients with Advanced and Progressing Hepatocellular Carcinoma. N. Engl. J. Med. 2018, 379, 54–63. [Google Scholar] [CrossRef]
- Bruix, J.; Qin, S.; Merle, P.; Granito, A.; Huang, Y.H.; Bodoky, G.; Pracht, M.; Yokosuka, O.; Rosmorduc, O.; Breder, V.; et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 389, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Han, G.; Finn, R.S.; Poon, R.T.; Blanc, J.F.; Yan, L.; Yang, J.; Lu, L.; Tak, W.Y.; Yu, X.; et al. Brivanib as adjuvant therapy to transarterial chemoembolization in patients with hepatocellular carcinoma: A randomized phase III trial. Hepatology 2014, 60, 1697–1707. [Google Scholar] [CrossRef]
- Lencioni, R.; Llovet, J.M.; Han, G.; Tak, W.Y.; Yang, J.; Guglielmi, A.; Paik, S.W.; Reig, M.; Kim, D.Y.; Chau, G.Y.; et al. Sorafenib or placebo plus TACE with doxorubicin-eluting beads for intermediate stage HCC: The SPACE trial. J. Hepatol. 2016, 64, 1090–1098. [Google Scholar] [CrossRef] [Green Version]
- Mazzaferro, V.; Regalia, E.; Doci, R.; Andreola, S.; Pulvirenti, A.; Bozzetti, F.; Montalto, F.; Ammatuna, M.; Morabito, A.; Gennari, L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N. Engl. J. Med. 1996, 334, 693–699. [Google Scholar] [CrossRef]
- Ng, K.K.C.; Chok, K.S.H.; Chan, A.C.Y.; Cheung, T.T.; Wong, T.C.L.; Fung, J.Y.Y.; Yuen, J.; Poon, R.T.P.; Fan, S.T.; Lo, C.M. Randomized clinical trial of hepatic resection versus radiofrequency ablation for early-stage hepatocellular carcinoma. Br. J. Surg. 2017, 104, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Shiina, S.; Teratani, T.; Obi, S.; Sato, S.; Tateishi, R.; Fujishima, T.; Ishikawa, T.; Koike, Y.; Yoshida, H.; Kawabe, T.; et al. A randomized controlled trial of radiofrequency ablation with ethanol injection for small hepatocellular carcinoma. Gastroenterology 2005, 129, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.D.; Tang, Z.Y.; Yang, B.H.; Lin, Z.Y.; Ma, Z.C.; Ye, S.L.; Wu, Z.Q.; Fan, J.; Qin, L.X.; Zheng, B.H. Experience of 1000 patients who underwent hepatectomy for small hepatocellular carcinoma. Cancer 2001, 91, 1479–1486. [Google Scholar] [CrossRef]
- Zhu, A.X.; Kang, Y.K.; Yen, C.J.; Finn, R.S.; Galle, P.R.; Llovet, J.M.; Assenat, E.; Brandi, G.; Pracht, M.; Lim, H.Y.; et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019, 20, 282–296. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.; Finn, R.S.; Qin, S.; Han, K.; Ikeda, K.; Piscaglia, F.; Baron, A.D.; Park, J.W.; Han, G.; Jassem, J.; et al. Phase III trial of lenvatinib (LEN) vs. sorafenib (SOR) in first-line treatment of patients (pts) with unresectable hepatocellular carcinoma (uHCC). J. Clin. Oncol. 2017, 35, 116. [Google Scholar] [CrossRef]
- Kudo, M. Lenvatinib in Advanced Hepatocellular Carcinoma. Liver Cancer 2017, 6, 253–263. [Google Scholar] [CrossRef]
- Kudo, M. A New Era of Systemic Therapy for Hepatocellular Carcinoma with Regorafenib and Lenvatinib. Liver Cancer 2017, 6, 177–184. [Google Scholar] [CrossRef]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef]
- Llovet, J.M.; Villanueva, A.; Lachenmayer, A.; Finn, R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 2015, 12, 408–424. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M. Focal gains of VEGFA: Candidate predictors of sorafenib response in hepatocellular carcinoma. Cochrane Database Syst. Rev. 2018, 12, CD013188. [Google Scholar] [CrossRef]
- Horwitz, E.; Stein, I.; Andreozzi, M.; Nemeth, J.; Shoham, A.; Pappo, O.; Schweitzer, N.; Tornillo, L.; Kanarek, N.; Quagliata, L.; et al. Human and mouse VEGFA-amplified hepatocellular carcinomas are highly sensitive to sorafenib treatment. Cancer Discov. 2014, 4, 730–743. [Google Scholar] [CrossRef] [Green Version]
- Arao, T.; Ueshima, K.; Matsumoto, K.; Nagai, T.; Kimura, H.; Hagiwara, S.; Sakurai, T.; Haji, S.; Kanazawa, A.; Hidaka, H.; et al. FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology 2013, 57, 1407–1415. [Google Scholar] [CrossRef] [Green Version]
- Takeda, H.; Nishikawa, H.; Osaki, Y.; Tsuchiya, K.; Joko, K.; Ogawa, C.; Taniguchi, H.; Orito, E.; Uchida, Y.; Izumi, N.; et al. Clinical features associated with radiological response to sorafenib in unresectable hepatocellular carcinoma: A large multicenter study in Japan. Liver Int. Off. J. Int. Assoc. Study Liver 2015, 35, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Kaibori, M.; Sakai, K.; Ishizaki, M.; Matsushima, H.; De Velasco, M.A.; Matsui, K.; Iida, H.; Kitade, H.; Kwon, A.H.; Nagano, H.; et al. Increased FGF19 copy number is frequently detected in hepatocellular carcinoma with a complete response after sorafenib treatment. Oncotarget 2016, 7, 49091–49098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudo, M. Recent Advances in Systemic Therapy for Hepatocellular Carcinoma in an Aging Society: 2020 Update. Liver Cancer 2020, 9, 640–662. [Google Scholar] [CrossRef]
- Llovet, J.M.; Pena, C.E.; Lathia, C.D.; Shan, M.; Meinhardt, G.; Bruix, J.; Group, S.I.S. Plasma biomarkers as predictors of outcome in patients with advanced hepatocellular carcinoma. Clin. Cancer Res. 2012, 18, 2290–2300. [Google Scholar] [CrossRef] [Green Version]
- Kudo, M. Biomarkers and Personalized Sorafenib Therapy. Liver Cancer 2014, 3, 399–404. [Google Scholar] [CrossRef]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.C.; Llovet, J.M. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015, 149, 1226–1239.e1224. [Google Scholar] [CrossRef] [Green Version]
- Totoki, Y.; Tatsuno, K.; Covington, K.R.; Ueda, H.; Creighton, C.J.; Kato, M.; Tsuji, S.; Donehower, L.A.; Slagle, B.L.; Nakamura, H.; et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014, 46, 1267–1273. [Google Scholar] [CrossRef]
- Fujimoto, A.; Furuta, M.; Shiraishi, Y.; Gotoh, K.; Kawakami, Y.; Arihiro, K.; Nakamura, T.; Ueno, M.; Ariizumi, S.; Nguyen, H.H.; et al. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat. Commun. 2015, 6, 6120. [Google Scholar] [CrossRef]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Deng, Q.; Wang, Q.; Li, K.Y.; Dai, J.H.; Li, N.; Zhu, Z.D.; Zhou, B.; Liu, X.Y.; Liu, R.F.; et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat. Genet. 2012, 44, 1117–1121. [Google Scholar] [CrossRef]
- Schulze, K.; Imbeaud, S.; Letouze, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Totoki, Y.; Tatsuno, K.; Yamamoto, S.; Arai, Y.; Hosoda, F.; Ishikawa, S.; Tsutsumi, S.; Sonoda, K.; Totsuka, H.; Shirakihara, T.; et al. High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 2011, 43, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Marusawa, H.; Endo, Y.; Chiba, T. Inflammation-mediated genomic instability: Roles of activation-induced cytidine deaminase in carcinogenesis. Cancer Sci. 2012, 103, 1201–1206. [Google Scholar] [CrossRef]
- Levrero, M.; Zucman-Rossi, J. Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol. 2016, 64, S84–S101. [Google Scholar] [CrossRef]
- Brunner, S.F.; Roberts, N.D.; Wylie, L.A.; Moore, L.; Aitken, S.J.; Davies, S.E.; Sanders, M.A.; Ellis, P.; Alder, C.; Hooks, Y.; et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature 2019, 574, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Lu, T.; Jia, Y.; Luo, X.; Gopal, P.; Li, L.; Odewole, M.; Renteria, V.; Singal, A.G.; Jang, Y.; et al. Somatic Mutations Increase Hepatic Clonal Fitness and Regeneration in Chronic Liver Disease. Cell 2019, 177, 608–621.e612. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.K.; Takeda, H.; Takai, A.; Matsumoto, T.; Kakiuchi, N.; Yokoyama, A.; Yoshida, K.; Kaido, T.; Uemoto, S.; Minamiguchi, S.; et al. Comprehensive analysis of genetic aberrations linked to tumorigenesis in regenerative nodules of liver cirrhosis. J. Gastroenterol. 2019, 54, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Midorikawa, Y.; Yamamoto, S.; Tsuji, S.; Kamimura, N.; Ishikawa, S.; Igarashi, H.; Makuuchi, M.; Kokudo, N.; Sugimura, H.; Aburatani, H. Allelic imbalances and homozygous deletion on 8p23.2 for stepwise progression of hepatocarcinogenesis. Hepatology 2009, 49, 513–522. [Google Scholar] [CrossRef]
- Midorikawa, Y.; Yamamoto, S.; Tatsuno, K.; Renard-Guillet, C.; Tsuji, S.; Hayashi, A.; Ueda, H.; Fukuda, S.; Fujita, T.; Katoh, H.; et al. Accumulation of Molecular Aberrations Distinctive to Hepatocellular Carcinoma Progression. Cancer Res. 2020, 80, 3810–3819. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Takai, A.; Kumagai, K.; Iguchi, E.; Arasawa, S.; Eso, Y.; Shimizu, T.; Ueda, Y.; Taura, K.; Uemoto, S.; et al. Multiregional whole-genome sequencing of hepatocellular carcinoma with nodule-in-nodule appearance reveals stepwise cancer evolution. J. Pathol. 2020, 252, 398–410. [Google Scholar] [CrossRef]
- Nault, J.C.; Calderaro, J.; Di Tommaso, L.; Balabaud, C.; Zafrani, E.S.; Bioulac-Sage, P.; Roncalli, M.; Zucman-Rossi, J. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology 2014, 60, 1983–1992. [Google Scholar] [CrossRef]
- Hudson, T.J.; Anderson, W.; Artez, A.; Barker, A.D.; Bell, C.; Bernabe, R.R.; Bhan, M.K.; Calvo, F.; Eerola, I.; Gerhard, D.S.; et al. International network of cancer genome projects. Nature 2010, 464, 993–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consortium, I.T.P.-C.A.o.W.G. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341.e1323. [CrossRef] [Green Version]
- Nault, J.C.; Datta, S.; Imbeaud, S.; Franconi, A.; Mallet, M.; Couchy, G.; Letouze, E.; Pilati, C.; Verret, B.; Blanc, J.F.; et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 2015, 47, 1187–1193. [Google Scholar] [CrossRef]
- Fujimoto, A.; Totoki, Y.; Abe, T.; Boroevich, K.A.; Hosoda, F.; Nguyen, H.H.; Aoki, M.; Hosono, N.; Kubo, M.; Miya, F.; et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012, 44, 760–764. [Google Scholar] [CrossRef]
- Nault, J.C.; Mallet, M.; Pilati, C.; Calderaro, J.; Bioulac-Sage, P.; Laurent, C.; Laurent, A.; Cherqui, D.; Balabaud, C.; Zucman-Rossi, J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 2013, 4, 2218. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Furuta, M.; Ueno, M.; Fujimoto, A.; Hayami, S.; Yasukawa, S.; Kojima, F.; Arihiro, K.; Kawakami, Y.; Wardell, C.P.; Shiraishi, Y.; et al. Whole genome sequencing discriminates hepatocellular carcinoma with intrahepatic metastasis from multi-centric tumors. J. Hepatol. 2016, 66, 363–373. [Google Scholar] [CrossRef]
- Shibata, T.; Aburatani, H. Exploration of liver cancer genomes. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yoo, J.E.; Cho, J.Y.; Oh, B.K.; Yoon, Y.S.; Han, H.S.; Lee, H.S.; Jang, J.J.; Jeong, S.H.; Kim, J.W.; et al. Telomere length, TERT and shelterin complex proteins in hepatocellular carcinomas expressing “stemness”-related markers. J. Hepatol. 2013, 59, 746–752. [Google Scholar] [CrossRef]
- Castelo-Branco, P.; Choufani, S.; Mack, S.; Gallagher, D.; Zhang, C.; Lipman, T.; Zhukova, N.; Walker, E.J.; Martin, D.; Merino, D.; et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: An integrative genomic and molecular study. Lancet Oncol. 2013, 14, 534–542. [Google Scholar] [CrossRef]
- Lee, D.D.; Leao, R.; Komosa, M.; Gallo, M.; Zhang, C.H.; Lipman, T.; Remke, M.; Heidari, A.; Nunes, N.M.; Apolonio, J.D.; et al. DNA hypermethylation within TERT promoter upregulates TERT expression in cancer. J. Clin. Investig. 2019, 129, 223–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Weng, X.; Ye, J.; He, L.; Zhou, D.; Liu, Y. Promoter hypermethylation of TERT is associated with hepatocellular carcinoma in the Han Chinese population. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 600–609. [Google Scholar] [CrossRef]
- Satoh, S.; Daigo, Y.; Furukawa, Y.; Kato, T.; Miwa, N.; Nishiwaki, T.; Kawasoe, T.; Ishiguro, H.; Fujita, M.; Tokino, T.; et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat. Genet. 2000, 24, 245–250. [Google Scholar] [CrossRef] [PubMed]
- De La Coste, A.; Romagnolo, B.; Billuart, P.; Renard, C.A.; Buendia, M.A.; Soubrane, O.; Fabre, M.; Chelly, J.; Beldjord, C.; Kahn, A.; et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 1998, 95, 8847–8851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abitbol, S.; Dahmani, R.; Coulouarn, C.; Ragazzon, B.; Mlecnik, B.; Senni, N.; Savall, M.; Bossard, P.; Sohier, P.; Drouet, V.; et al. AXIN deficiency in human and mouse hepatocytes induces hepatocellular carcinoma in the absence of β-catenin activation. J. Hepatol. 2018, 68, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Désert, R.; Rohart, F.; Canal, F.; Sicard, M.; Desille, M.; Renaud, S.; Turlin, B.; Bellaud, P.; Perret, C.; Clément, B.; et al. Human hepatocellular carcinomas with a periportal phenotype have the lowest potential for early recurrence after curative resection. Hepatology 2017, 66, 1502–1518. [Google Scholar] [CrossRef]
- Boyault, S.; Rickman, D.S.; de Reyniès, A.; Balabaud, C.; Rebouissou, S.; Jeannot, E.; Hérault, A.; Saric, J.; Belghiti, J.; Franco, D.; et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007, 45, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Désert, R.; Nieto, N.; Musso, O. Dimensions of hepatocellular carcinoma phenotypic diversity. World J. Gastroenterol. 2018, 24, 4536–4547. [Google Scholar] [CrossRef]
- Bayard, Q.; Meunier, L.; Peneau, C.; Renault, V.; Shinde, J.; Nault, J.C.; Mami, I.; Couchy, G.; Amaddeo, G.; Tubacher, E.; et al. Cyclin A2/E1 activation defines a hepatocellular carcinoma subclass with a rearrangement signature of replication stress. Nat. Commun. Commun. 2018, 9, 5235. [Google Scholar] [CrossRef] [Green Version]
- Hoshida, Y.; Nijman, S.M.; Kobayashi, M.; Chan, J.A.; Brunet, J.P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurent-Puig, P.; Legoix, P.; Bluteau, O.; Belghiti, J.; Franco, D.; Binot, F.; Monges, G.; Thomas, G.; Bioulac-Sage, P.; Zucman-Rossi, J. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology 2001, 120, 1763–1773. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.P.; Schwank, J.; Staib, F.; Wang, X.W.; Harris, C.C. TP53 mutations and hepatocellular carcinoma: Insights into the etiology and pathogenesis of liver cancer. Oncogene 2007, 26, 2166–2176. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.; Li, M.; Parsons, D.W.; Zhang, X.; Wesseling, J.; Kristel, P.; Schmidt, M.K.; Markowitz, S.; Yan, H.; Bigner, D.; et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum. Mutat. 2012, 33, 100–103. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.; Zhang, Q.; Du, Y.; Liu, W.; Li, Z.; Hou, X.; Cao, G. Somatic mutations, viral integration and epigenetic modification in the evolution of hepatitis B virus-induced hepatocellular carcinoma. Curr. Genom. 2014, 15, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Sung, W.K.; Zheng, H.; Li, S.; Chen, R.; Liu, X.; Li, Y.; Lee, N.P.; Lee, W.H.; Ariyaratne, P.N.; Tennakoon, C.; et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012, 44, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 2021, 7, 6. [Google Scholar] [CrossRef]
- Chiang, D.Y.; Villanueva, A.; Hoshida, Y.; Peix, J.; Newell, P.; Minguez, B.; LeBlanc, A.C.; Donovan, D.J.; Thung, S.N.; Sole, M.; et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008, 68, 6779–6788. [Google Scholar] [CrossRef] [Green Version]
- Shuai, S.; Suzuki, H.; Diaz-Navarro, A.; Nadeu, F.; Kumar, S.A.; Gutierrez-Fernandez, A.; Delgado, J.; Pinyol, M.; López-Otín, C.; Puente, X.S.; et al. The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature 2019, 574, 712–716. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Zhai, W.; Lim, T.K.; Zhang, T.; Phang, S.T.; Tiang, Z.; Guan, P.; Ng, M.H.; Lim, J.Q.; Yao, F.; Li, Z.; et al. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat. Commun. 2017, 8, 4565. [Google Scholar] [CrossRef] [Green Version]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.C.; Mayakonda, A.; Dinh, H.Q.; Huang, P.; Lin, L.; Liu, X.; Ding, L.W.; Wang, J.; Berman, B.; Song, E.; et al. Genomic and epigenomic heterogeneity of hepatocellular carcinoma. Cancer Res. 2017, 77, 2255–2265. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Lee, I.H.; Cho, H.J.; Park, C.K.; Jung, Y.S.; Kim, Y.; Nam, S.H.; Kim, B.S.; Johnson, M.D.; Kong, D.S.; et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell 2015, 28, 318–328. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.K.; Wang, J.; Sa, J.K.; Ladewig, E.; Lee, H.O.; Lee, I.H.; Kang, H.J.; Rosenbloom, D.S.; Camara, P.G.; Liu, Z.; et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat. Genet. 2017; 49, 594–599. [Google Scholar] [CrossRef]
- Makohon-Moore, A.P.; Zhang, M.; Reiter, J.G.; Bozic, I.; Allen, B.; Kundu, D.; Chatterjee, K.; Wong, F.; Jiao, Y.; Kohutek, Z.A.; et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 2017, 49, 358–366. [Google Scholar] [CrossRef]
- Kumar, A.; Coleman, I.; Morrissey, C.; Zhang, X.; True, L.D.; Gulati, R.; Etzioni, R.; Bolouri, H.; Montgomery, B.; White, T.; et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat. Med. 2016, 22, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Uchi, R.; Takahashi, Y.; Niida, A.; Shimamura, T.; Hirata, H.; Sugimachi, K.; Sawada, G.; Iwaya, T.; Kurashige, J.; Shinden, Y.; et al. Integrated Multiregional Analysis Proposing a New Model of Colorectal Cancer Evolution. PLoS Genet. 2016, 12, e1005778. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.; Li, R.; Guo, H.; Guo, L.; Su, Z.; Ni, X.; Qi, L.; Zhang, T.; Li, Q.; Zhang, Z.; et al. Variable Intra-Tumor Genomic Heterogeneity of Multiple Lesions in Patients With Hepatocellular Carcinoma. Gastroenterology 2016, 150, 998–1008. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Wu, L.; Lin, J.; Han, L.; Bian, J.; Wu, Y.; Robson, S.C.; Xue, L.; Ge, Y.; Sang, X.; et al. Whole-exome sequencing reveals the origin and evolution of hepato-cholangiocarcinoma. Nat. Commun. 2018, 9, 894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lou, Y.; Yang, J.; Wang, J.; Feng, J.; Zhao, Y.; Wang, L.; Huang, X.; Fu, Q.; Ye, M.; et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut 2019, 68, 2019–2031. [Google Scholar] [CrossRef]
- Jeng, K.S.; Chang, C.F.; Jeng, W.J.; Sheen, I.S.; Jeng, C.J. Heterogeneity of hepatocellular carcinoma contributes to cancer progression. Crit. Rev. Oncol. Hematol. 2015, 94, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Guo, H.; Cao, C.; Li, X.; Hu, B.; Zhu, P.; Wu, X.; Wen, L.; Tang, F.; Huang, Y.; et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016, 26, 304–319. [Google Scholar] [CrossRef] [PubMed]
- Aihara, T.; Noguchi, S.; Sasaki, Y.; Nakano, H.; Imaoka, S. Clonal analysis of regenerative nodules in hepatitis C virus-induced liver cirrhosis. Gastroenterology 1994, 107, 1805–1811. [Google Scholar] [CrossRef]
- Gong, L.; Li, Y.H.; Su, Q.; Chu, X.; Zhang, W. Clonality of nodular lesions in liver cirrhosis and chromosomal abnormalities in monoclonal nodules of altered hepatocytes. Histopathology 2010, 56, 589–599. [Google Scholar] [CrossRef]
- Lin, W.R.; Lim, S.N.; McDonald, S.A.; Graham, T.; Wright, V.L.; Peplow, C.L.; Humphries, A.; Kocher, H.M.; Wright, N.A.; Dhillon, A.P.; et al. The histogenesis of regenerative nodules in human liver cirrhosis. Hepatology 2010, 51, 1017–1026. [Google Scholar] [CrossRef]
- Ochiai, T.; Urata, Y.; Yamano, T.; Yamagishi, H.; Ashihara, T. Clonal expansion in evolution of chronic hepatitis to hepatocellular carcinoma as seen at an X-chromosome locus. Hepatology 2000, 31, 615–621. [Google Scholar] [CrossRef]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoyama, A.; Kakiuchi, N.; Yoshizato, T.; Nannya, Y.; Suzuki, H.; Takeuchi, Y.; Shiozawa, Y.; Sato, Y.; Aoki, K.; Kim, S.K.; et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 2019, 565, 312–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuma, M.; Sakamoto, M.; Yamazaki, K.; Ohta, T.; Ohki, M.; Asaka, M.; Hirohashi, S. Expression profiling in multistage hepatocarcinogenesis: Identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology 2003, 37, 198–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nault, J.C.; Martin, Y.; Caruso, S.; Hirsch, T.Z.; Bayard, Q.; Calderaro, J.; Charpy, C.; Copie-Bergman, C.; Ziol, M.; Bioulac-Sage, P.; et al. Clinical Impact of Genomic Diversity From Early to Advanced Hepatocellular Carcinoma. Hepatology 2020, 71, 164–182. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).