Epigenetic Therapies and Biomarkers in Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
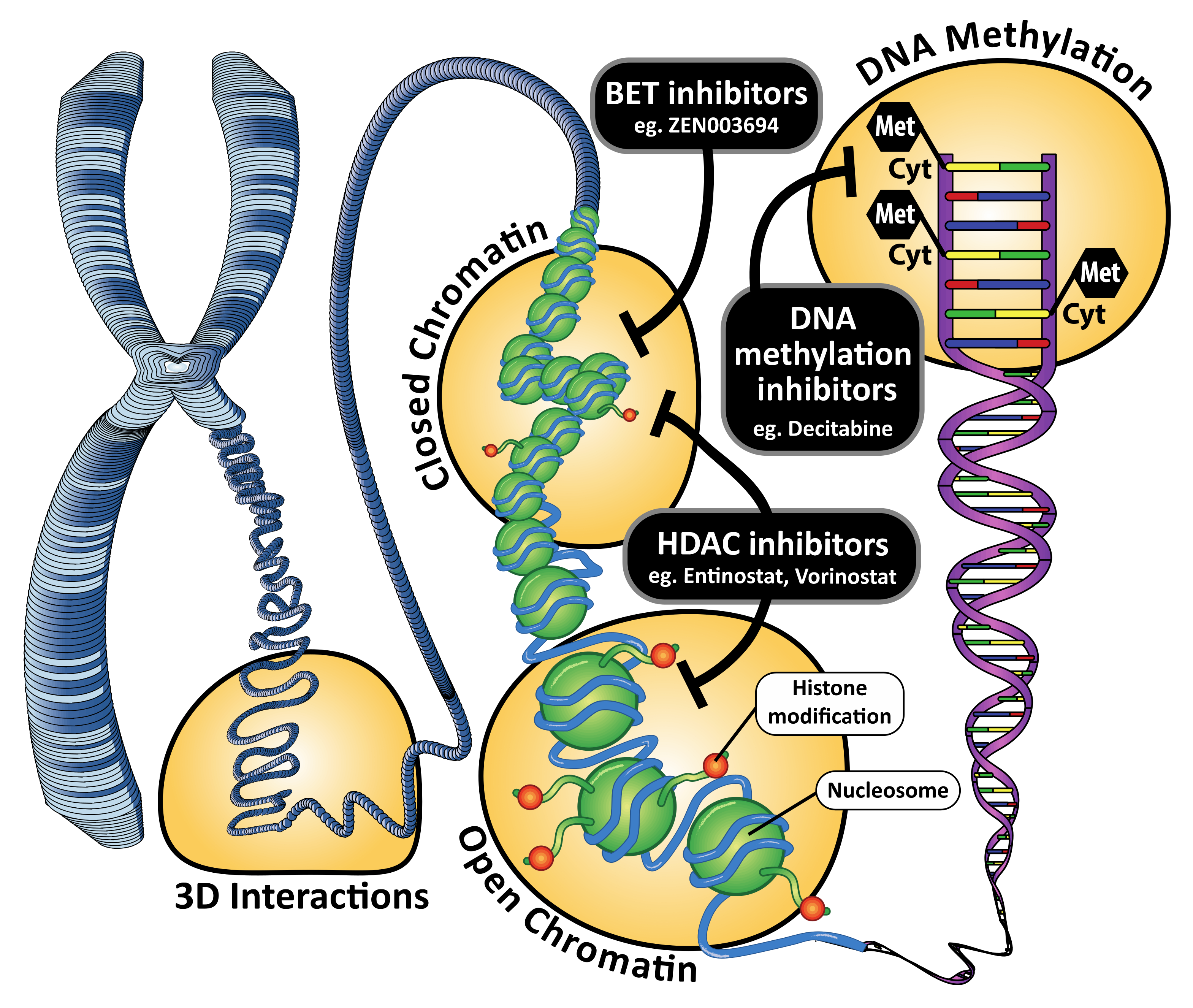
2. DNA Methylation and DNA Methyltransferase Inhibitors
3. Histone Modifications and Histone Deacetylase Inhibitors
4. Chromatin Remodeling, Super-Enhancers and Bromodomain and Extra-Terminal Motif Inhibitors
5. Promising Epigenetic Therapy Combinations
6. DNA Methylation Biomarkers in Breast Cancer
7. Discussion and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Shi, Y.; Dong, M.; Hong, X.; Zhang, W.; Feng, J.; Zhu, J.; Yu, L.; Ke, X.; Huang, H.; Shen, Z.; et al. Results from a multicenter, open-label, pivotal phase II study of chidamide in relapsed or refractory peripheral T-cell lymphoma. Ann. Oncol. 2015, 26, 1766–1771. [Google Scholar] [CrossRef]
- Silverman, L.R.; Mufti, G.J. Methylation inhibitor therapy in the treatment of myelodysplastic syndrome. Nat. Clin. Pr. Oncol. 2005, 2 (Suppl. 1), S12–S23. [Google Scholar] [CrossRef]
- Silverman, L.R.; Demakos, E.P.; Peterson, B.L.; Kornblith, A.B.; Holland, J.C.; Odchimar-Reissig, R.; Stone, R.M.; Nelson, D.; Powell, B.L.; DeCastro, C.M.; et al. Randomized Controlled Trial of Azacitidine in Patients with the Myelodysplastic Syndrome: A Study of the Cancer and Leukemia Group B. J. Clin. Oncol. 2002, 20, 2429–2440. [Google Scholar] [CrossRef]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Finelli, C.; Giagounidis, A.; Schoch, R.; Gattermann, N.; Sanz, G.; List, A.; et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009, 10, 223–232. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Griffiths, E.A.; Steensma, D.P.; Roboz, G.J.; Wells, R.; McCloskey, J.; Odenike, O.; DeZern, A.E.; Yee, K.; Busque, L.; et al. Oral cedazuridine/decitabine for MDS and CMML: A phase 2 pharmacokinetic/pharmacodynamic randomized crossover study. Blood 2020, 136, 674–683. [Google Scholar] [CrossRef]
- Morel, D.; Jeffery, D.; Aspeslagh, S.; Almouzni, G.; Postel-Vinay, S. Combining epigenetic drugs with other therapies for solid tumours—Past lessons and future promise. Nat. Rev. Clin. Oncol. 2020, 17, 91–107. [Google Scholar] [CrossRef]
- Garcia-Martinez, L.; Zhang, Y.; Nakata, Y.; Chan, H.L.; Morey, L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021, 12, 1786. [Google Scholar] [CrossRef]
- Luu, T.H.; Morgan, R.J.; Leong, L.; Lim, D.; McNamara, M.; Portnow, J.; Frankel, P.; Smith, D.; Doroshow, J.H.; Gandara, D.R.; et al. A Phase II Trial of Vorinostat (Suberoylanilide Hydroxamic Acid) in Metastatic Breast Cancer: A California Cancer Consortium Study. Clin. Cancer Res. 2008, 14, 7138–7142. [Google Scholar] [CrossRef]
- Piha-Paul, S.A.; Sachdev, J.C.; Barve, M.; Lorusso, P.; Szmulewitz, R.; Patel, S.P.; Lara, P.N., Jr.; Chen, X.; Hu, B.; Freise, K.J.; et al. First-in-Human Study of Mivebresib (ABBV-075), an Oral Pan-Inhibitor of Bromodomain and Extra Terminal Proteins, in Patients with Relapsed/Refractory Solid Tumors. Clin. Cancer Res. 2019, 25, 6309–6319. [Google Scholar] [CrossRef]
- Ramaswamy, B.; Fiskus, W.; Cohen, B.; Pellegrino, C.; Hershman, D.L.; Chuang, E.; Luu, T.; Somlo, G.; Goetz, M.; Swaby, R.; et al. Phase I–II study of vorinostat plus paclitaxel and bevacizumab in metastatic breast cancer: Evidence for vorinostat-induced tubulin acetylation and Hsp90 inhibition in vivo. Breast Cancer Res. Treat. 2012, 132, 1063–1072. [Google Scholar] [CrossRef]
- Candelaria, M.; Gallardo-Rincón, D.; Arce, C.; Cetina, L.; Aguilar-Ponce, J.; Arrieta, O.; González-Fierro, A.; Chávez-Blanco, A.; de la Cruz-Hernández, E.; Camargo, M.; et al. A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann. Oncol. 2007, 18, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Appleton, K.; Mackay, H.J.; Judson, I.; Plumb, J.A.; McCormick, C.; Strathdee, G.; Lee, C.; Barrett, S.; Reade, S.; Jadayel, D.; et al. Phase I and Pharmacodynamic Trial of the DNA Methyltransferase Inhibitor Decitabine and Carboplatin in Solid Tumors. J. Clin. Oncol. 2007, 25, 4603–4609. [Google Scholar] [CrossRef]
- Munster, P.N.; Marchion, D.; Thomas, S.; Egorin, M.; Minton, S.; Springett, G.; Lee, J.-H.; Simon, G.; Chiappori, A.; Sullivan, D.; et al. Phase I trial of vorinostat and doxorubicin in solid tumours: Histone deacetylase 2 expression as a predictive marker. Br. J. Cancer 2009, 101, 1044–1050. [Google Scholar] [CrossRef]
- Jiang, Z.; Li, W.; Hu, X.; Zhang, Q.; Sun, T.; Cui, S.; Wang, S.; Ouyang, Q.; Yin, Y.; Geng, C.; et al. Tucidinostat plus exemestane for postmenopausal patients with advanced, hormone receptor-positive breast cancer (ACE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019, 20, 806–815. [Google Scholar] [CrossRef]
- Yardley, D.A.; Ismail-Khan, R.R.; Melichar, B.; Lichinitser, M.; Munster, P.N.; Klein, P.M.; Cruickshank, S.; Miller, K.D.; Lee, M.J.; Trepel, J.B. Randomized Phase II, Double-Blind, Placebo-Controlled Study of Exemestane with or Without Entinostat in Postmenopausal Women with Locally Recurrent or Metastatic Estrogen Receptor-Positive Breast Cancer Progressing on Treatment With a Nonsteroidal Aromatase Inhibitor. J. Clin. Oncol. 2013, 31, 2128–2135. [Google Scholar] [CrossRef]
- Yeruva, S.L.H.; Zhao, F.; Miller, K.D.; Tevaarwerk, A.J.; Wagner, L.I.; Gray, R.J.; Sparano, J.A.; Connolly, R.M. E2112: Randomized Phase III Trial of Endocrine Therapy Plus Entinostat or Placebo in Hormone Receptor—Positive Advanced Breast Cancer. A Trial of the ECOG-ACRIN Cancer Research Group. J. Clin. Oncol. 2021, 39, 3171–3181. [Google Scholar]
- Munster, P.N.; Thurn, K.T.; Thomas, S.; Raha, P.; Lacevic, M.; Miller, A.; Melisko, M.; Ismail-Khan, R.; Rugo, H.; Moasser, M.; et al. A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br. J. Cancer 2011, 104, 1828–1835. [Google Scholar] [CrossRef]
- Bauman, J.; Verschraegen, C.; Belinsky, S.; Muller, C.; Rutledge, T.; Fekrazad, M.; Ravindranathan, M.; Lee, S.-J.; Jones, D. A phase I study of 5-azacytidine and erlotinib in advanced solid tumor malignancies. Cancer Chemother. Pharmacol. 2011, 69, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Garrido-Laguna, I.; Naing, A.; Fu, S.; Falchook, G.S.; Piha-Paul, S.; Wheler, J.J.; Hong, D.S.; Tsimberidou, A.M.; Subbiah, V.; et al. Phase I dose-escalation study of the mTOR inhibitor sirolimus and the HDAC inhibitor vorinostat in patients with advanced malignancy. Oncotarget 2016, 7, 67521–67531. [Google Scholar] [CrossRef]
- Goldstein, L.J.; Zhao, F.; Wang, M.; Swaby, R.F.; Sparano, J.A.; Meropol, N.J.; Bhalla, K.N.; Pellegrino, C.M.; Alpaugh, R.K.; Falkson, C.I.; et al. A Phase I/II study of suberoylanilide hydroxamic acid (SAHA) in combination with trastuzumab (Herceptin) in patients with advanced metastatic and/or local chest wall recurrent HER2-amplified breast cancer: A trial of the ECOG-ACRIN Cancer Research Group (E1104). Breast Cancer Res. Treat. 2017, 165, 375–382. [Google Scholar] [CrossRef]
- Skvortsova, K.; Stirzaker, C.; Taberlay, P. The DNA methylation landscape in cancer. Essays Biochem. 2019, 63, 797–811. [Google Scholar] [CrossRef]
- Ottaviano, Y.L.; Issa, J.-P.; Parl, F.F.; Smith, H.S.; Baylin, S.B.; Davidson, N. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994, 54, 2552–5255. [Google Scholar] [PubMed]
- Dobrovic, A.; Simpfendorfer, D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997, 57, 3347–3350. [Google Scholar] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Baylin, S.; Bestor, T.H. Altered methylation patterns in cancer cell genomes: Cause or consequence? Cancer Cell 2002, 1, 299–305. [Google Scholar] [CrossRef]
- Hon, G.C.; Hawkins, R.D.; Caballero, O.L.; Lo, C.; Lister, R.; Pelizzola, M.; Valsesia, A.; Ye, Z.; Kuan, S.; Edsall, L.E.; et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2011, 22, 246–258. [Google Scholar] [CrossRef]
- Brinkman, A.B.; Nik-Zainal, S.; Simmer, F.; Rodríguez-González, F.G.; Smid, M.; Alexandrov, L.B.; Butler, A.; Martin, S.; Davies, H.; Glodzik, D.; et al. Partially methylated domains are hypervariable in breast cancer and fuel widespread CpG island hypermethylation. Nat. Commun. 2019, 10, 1749. [Google Scholar] [CrossRef]
- Batra, R.N.; Lifshitz, A.; Vidakovic, A.T.; Chin, S.-F.; Sati-Batra, A.; Sammut, S.-J.; Provenzano, E.; Ali, H.R.; Dariush, A.; Bruna, A.; et al. DNA methylation landscapes of 1538 breast cancers reveal a replication-linked clock, epigenomic instability and cis-regulation. Nat. Commun. 2021, 12, 5406. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Sabato, S.; Hellman, A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013, 14, R21. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef]
- Bulger, M.; Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 2011, 144, 327–339. [Google Scholar] [CrossRef]
- Shlyueva, D.; Stampfel, G.; Stark, A. Transcriptional enhancers: From properties to genome-wide predictions. Nat. Rev. Genet. 2014, 15, 272–286. [Google Scholar] [CrossRef]
- Locke, W.J.; Zotenko, E.; Stirzaker, C.; Robinson, M.D.; Hinshelwood, R.; Stone, A.; Reddel, R.R.; Huschtscha, L.; Clark, S.J. Coordinated epigenetic remodelling of transcriptional networks occurs during early breast carcinogenesis. Clin. Epigenetics 2015, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, T.; Oslo Breast Cancer Research Consortium (OSBREAC); Tekpli, X.; Mathelier, A.; Wang, S.; Nebdal, D.; Dhakal, H.P.; Sahlberg, K.K.; Schlichting, E.; Børresen-Dale, A.-L.; et al. DNA methylation at enhancers identifies distinct breast cancer lineages. Nat. Commun. 2017, 8, 1379. [Google Scholar] [CrossRef]
- Fleischer, T.; Frigessi, A.; Johnson, K.C.; Edvardsen, H.; Touleimat, N.; Klajic, J.; Riis, M.L.; Haakensen, V.D.; Wärnberg, F.; Naume, B.; et al. Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol. 2014, 15, 435. [Google Scholar] [PubMed]
- Broome, R.; Chernukhin, I.; Jamieson, S.; Kishore, K.; Papachristou, E.K.; Mao, S.-Q.; Tejedo, C.G.; Mahtey, A.; Theodorou, V.; Groen, A.J.; et al. TET2 is a component of the estrogen receptor complex and controls 5mC to 5hmC conversion at estrogen receptor cis-regulatory regions. Cell Rep. 2021, 34, 108776. [Google Scholar] [CrossRef]
- Lin, X.; Li, J.; Yin, G.; Zhao, Q.; Elias, D.; Lykkesfeldt, A.; Stenvang, J.; Brünner, N.; Wang, J.; Yang, H.; et al. Integrative analyses of gene expression and DNA methylation profiles in breast cancer cell line models of tamoxifen-resistance indicate a potential role of cells with stem-like properties. Breast Cancer Res. 2013, 15, R119. [Google Scholar] [CrossRef] [PubMed]
- Stone, A.; Zotenko, E.; Locke, W.J.; Korbie, D.; Millar, E.K.A.; Pidsley, R.; Stirzaker, C.; Graham, P.; Trau, M.; Musgrove, E.A.; et al. DNA methylation of oestrogen-regulated enhancers defines endocrine sensitivity in breast cancer. Nat. Commun. 2015, 6, 7758. [Google Scholar] [CrossRef]
- Achinger-Kawecka, J.; Valdes-Mora, F.; Luu, P.-L.; Giles, K.A.; Caldon, C.E.; Qu, W.; Nair, S.; Soto, S.; Locke, W.J.; Yeo-Teh, N.S.; et al. Epigenetic reprogramming at estrogen-receptor binding sites alters 3D chromatin landscape in endocrine-resistant breast cancer. Nat. Commun. 2020, 11, 320. [Google Scholar] [CrossRef]
- Zhou, Y.; Gerrard, D.L.; Wang, J.; Li, T.; Yang, Y.; Fritz, A.J.; Rajendran, M.; Fu, X.; Stein, G.; Schiff, R.; et al. Temporal dynamic reorganization of 3D chromatin architecture in hormone-induced breast cancer and endocrine resistance. Nat. Commun. 2019, 10, 1522. [Google Scholar] [CrossRef]
- Hickey, T.E.; Selth, L.A.; Chia, K.M.; Laven-Law, G.; Milioli, H.H.; Roden, D.; Jindal, S.; Hui, M.; Finlay-Schultz, J.; Ebrahimie, E.; et al. The androgen receptor is a tumor suppressor in estrogen receptor–positive breast cancer. Nat. Med. 2021, 27, 310–320. [Google Scholar] [CrossRef]
- Bates, S.E. Epigenetic Therapies for Cancer. N. Engl. J. Med. 2020, 383, 650–663. [Google Scholar] [CrossRef]
- Wong, K.K. DNMT1: A key drug target in triple-negative breast cancer. Semin. Cancer Biol. 2020, 72, 198–213. [Google Scholar] [CrossRef]
- Stirzaker, C.; Zotenko, E.; Song, J.Z.; Qu, W.; Nair, S.S.; Locke, W.; Stone, A.; Armstong, N.J.; Robinson, M.D.; Dobrovic, A.; et al. Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat. Commun. 2015, 6, 5899. [Google Scholar] [CrossRef]
- Mehdipour, P.; Marhon, S.A.; Ettayebi, I.; Chakravarthy, A.; Hosseini, A.; Wang, Y.; De Castro, F.A.; Yau, H.L.; Ishak, C.; Abelson, S.; et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 2020, 588, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef]
- Deblois, G.; Tonekaboni, S.A.M.; Grillo, G.; Martinez, C.; Kao, Y.I.; Tai, F.; Ettayebi, I.; Fortier, A.-M.; Savage, P.; Fedor, A.N.; et al. Epigenetic Switch–Induced Viral Mimicry Evasion in Chemotherapy-Resistant Breast Cancer. Cancer Discov. 2020, 10, 1312–1329. [Google Scholar] [CrossRef] [PubMed]
- Janin, M.; Esteller, M. Epigenetic Awakening of Viral Mimicry in Cancer. Cancer Discov. 2020, 10, 1258–1260. [Google Scholar] [CrossRef]
- Dedeurwaerder, S.; Desmedt, C.; Calonne, E.; Singhal, S.K.; Haibe-Kains, B.; Defrance, M.; Michiels, S.; Volkmar, M.; Deplus, R.; Luciani, J.; et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 2011, 3, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Tan, Y.J.; Falasca, M.; Oon, C.E. Cancer-Associated Fibroblasts: Epigenetic Regulation and Therapeutic Intervention in Breast Cancer. Cancers 2020, 12, 2949. [Google Scholar] [CrossRef]
- Xiong, H.; Qiu, H.; Zhuang, L.; Xiong, H.; Jiang, R.; Chen, Y. Effects of 5-Aza-CdR on the proliferation of human breast cancer cell line MCF-7 and on the expression of Apaf-1 gene. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2009, 29, 498–502. [Google Scholar] [CrossRef]
- Khan, G.N.; Kim, E.J.; Shin, T.S.; Lee, S.H. Azacytidine-induced Chemosensitivity to Doxorubicin in Human Breast Cancer MCF7 Cells. Anticancer Res. 2017, 37, 2355–2364. [Google Scholar] [CrossRef]
- Tsai, H.-C.; Li, H.; Van Neste, L.; Cai, Y.; Robert, C.; Rassool, F.V.; Shin, J.J.; Harbom, K.M.; Beaty, R.; Pappou, E.; et al. Transient Low Doses of DNA-Demethylating Agents Exert Durable Antitumor Effects on Hematological and Epithelial Tumor Cells. Cancer Cell 2012, 21, 430–446. [Google Scholar] [CrossRef] [PubMed]
- Achinger-Kawecka, J.; Stirzaker, C.; Chia, K.M.; Portman, N.; Campbell, E.; Du, Q.; Clark, S. Epigenetic therapy suppresses endocrine-resistant breast tumour growth by re-wiring ER-mediated 3D chromatin interactions. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dahn, M.L.; Cruickshank, B.M.; Jackson, A.J.; Dean, C.; Holloway, R.W.; Hall, S.R.; Coyle, K.M.; Maillet, H.; Waisman, D.M.; Goralski, K.B.; et al. Decitabine Response in Breast Cancer Requires Efficient Drug Processing and Is Not Limited by Multidrug Resistance. Mol. Cancer Ther. 2020, 19, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Butler, C.; Sprowls, S.; Szalai, G.; Arsiwala, T.; Saralkar, P.; Straight, B.; Hatcher, S.; Tyree, E.; Yost, M.; Kohler, W.J.; et al. Hypomethylating Agent Azacitidine Is Effective in Treating Brain Metastasis Triple-Negative Breast Cancer Through Regulation of DNA Methylation of Keratin 18 Gene. Transl. Oncol. 2020, 13, 100775. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Qin, B.; Moyer, A.M.; Nowsheen, S.; Liu, T.; Qin, S.; Zhuang, Y.; Liu, D.; Lu, S.W.; Kalari, K.R.; et al. DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J. Clin. Investig. 2018, 128, 2376–2388. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Pardee, A.B. Suberoylanilide Hydroxamic Acid as a Potential Therapeutic Agent for Human Breast Cancer Treatment. Mol. Med. 2000, 6, 849–866. [Google Scholar] [CrossRef]
- Robertson, F.M.; Woodward, W.; Pickei, R.; Ye, Z.; Bornmann, W.; Pal, A.; Peng, Z.; Hall, C.S.; Cristofanilli, M.; Woodward, W.A.; et al. Suberoylanilide hydroxamic acid blocks self-renewal and homotypic aggregation of inflammatory breast cancer spheroids. Cancer 2010, 116 (Suppl. 11), 2760–2767. [Google Scholar] [CrossRef] [PubMed]
- Aztopal, N.; Erkisa, M.; Erturk, E.; Ulukaya, E.; Tokullugil, A.H.; Ari, F. Valproic acid, a histone deacetylase inhibitor, induces apoptosis in breast cancer stem cells. Chem. Interact. 2018, 280, 51–58. [Google Scholar] [CrossRef]
- Thomas, S.; Thurn, K.T.; Biçaku, E.; Marchion, D.C.; Münster, P.N. Addition of a histone deacetylase inhibitor redirects tamoxifen-treated breast cancer cells into apoptosis, which is opposed by the induction of autophagy. Breast Cancer Res. Treat. 2011, 130, 437–447. [Google Scholar] [CrossRef]
- Sabnis, G.J.; Goloubeva, O.G.; Kazi, A.A.; Shah, P.; Brodie, A.H. HDAC Inhibitor Entinostat Restores Responsiveness of Letrozole-Resistant MCF-7Ca Xenografts to Aromatase Inhibitors through Modulation of Her-2. Mol. Cancer Ther. 2013, 12, 2804–2816. [Google Scholar] [CrossRef] [PubMed]
- Sabnis, G.J.; Goloubeva, O.; Chumsri, S.; Nguyen, N.; Sukumar, S.; Brodie, A.M.H. Functional Activation of the Estrogen Receptor-α and Aromatase by the HDAC Inhibitor Entinostat Sensitizes ER-Negative Tumors to Letrozole. Cancer Res. 2011, 71, 1893–1903. [Google Scholar] [CrossRef]
- Fuino, L.; Bali, P.; Wittmann, S.; Donapaty, S.; Guo, F.; Yamaguchi, H.; Wang, H.-G.; Atadja, P.; Bhalla, K. Histone deacetylase inhibitor LAQ824 down-regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine, and epothilone B. Mol. Cancer Ther. 2003, 2, 971. [Google Scholar]
- Miller, K.; Wang, M.; Gralow, J.; Dickler, M.; Cobleigh, M.; Perez, E.A.; Shenkier, T.; Cella, D.; Davidson, N.E. Paclitaxel plus Bevacizumab versus Paclitaxel Alone for Metastatic Breast Cancer. N. Engl. J. Med. 2007, 357, 2666–2676. [Google Scholar] [CrossRef]
- Plass, C.; Pfister, S.; Lindroth, A.; Bogatyrova, O.; Claus, R.; Lichter, P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 2013, 14, 765–780. [Google Scholar] [CrossRef] [PubMed]
- Skulte, K.; Phan, L.; Clark, S.J.; Taberlay, P.C. Chromatin remodeler mutations in human cancers: Epigenetic implications. Epigenomics 2014, 6, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Belandia, B.; Orford, R.L.; Hurst, H.C.; Parker, M.G. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 2002, 21, 4094–4103. [Google Scholar] [CrossRef]
- García-Pedrero, J.M.; Kiskinis, E.; Parker, M.G.; Belandia, B. The SWI/SNF Chromatin Remodeling Subunit BAF57 Is a Critical Regulator of Estrogen Receptor Function in Breast Cancer Cells. J. Biol. Chem. 2006, 281, 22656–22664. [Google Scholar] [CrossRef] [PubMed]
- Razavi, P.; Chang, M.T.; Xu, G.; Bandlamudi, C.; Ross, D.S.; Vasan, N.; Cai, Y.; Bielski, C.M.; Donoghue, M.T.A.; Jonsson, P.; et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018, 34, 427–438.e6. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Chhangawala, S.; Cocco, E.; Razavi, P.; Cai, Y.; Otto, J.; Ferrando, L.; Selenica, P.; Ladewig, E.; Chan, C.; et al. ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat. Genet. 2020, 52, 198–207. [Google Scholar] [CrossRef]
- Nagarajan, S.; Rao, S.V.; Sutton, J.; Cheeseman, D.; Dunn, S.; Papachristou, E.K.; Prada, J.-E.G.; Couturier, D.-L.; Kumar, S.; Kishore, K.; et al. ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat. Genet. 2020, 52, 187–197. [Google Scholar] [CrossRef]
- Shorstova, T.; Foulkes, W.D.; Witcher, M. Achieving clinical success with BET inhibitors as anti-cancer agents. Br. J. Cancer 2021, 124, 1478–1490. [Google Scholar] [CrossRef]
- Da Motta, L.L.; Ledaki, I.; Purshouse, K.; Haider, S.; De Bastiani, M.A.; Baban, D.; Morotti, M.; Steers, G.; Wigfield, S.; Bridges, E.; et al. The BET inhibitor JQ1 selectively impairs tumour response to hypoxia and downregulates CA9 and angiogenesis in triple negative breast cancer. Oncogene 2017, 36, 122–132. [Google Scholar] [CrossRef]
- Vázquez, R.; Riveiro, M.E.; Astorgues-Xerri, L.; Odore, E.; Rezai, K.; Erba, E.; Panini, N.; Rinaldi, A.; Kwee, I.; Beltrame, L.; et al. The bromodomain inhibitor OTX015 (MK-8628) exerts anti-tumor activity in triple-negative breast cancer models as single agent and in combination with everolimus. Oncotarget 2016, 8, 7598–7613. [Google Scholar] [CrossRef]
- Shu, S.; Lin, C.Y.; He, H.H.; Witwicki, R.M.; Tabassum, D.P.; Roberts, J.M.; Janiszewska, M.; Huh, S.J.; Liang, Y.; Ryan, J.; et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 2016, 529, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Zhang, Z.; Shea, M.J.; Creighton, C.J.; Coarfa, C.; Hilsenbeck, S.G.; Lanz, R.; He, B.; Wang, L.; Fu, X.; et al. An epigenomic approach to therapy for tamoxifen-resistant breast cancer. Cell Res. 2014, 24, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Perez-Salvia, M.; Simó-Riudalbas, L.; Llinàs-Arias, P.; Roa, L.; Setien, F.; Soler, M.; De Moura, M.C.; Bradner, J.E.; Gonzalez-Suarez, E.; Moutinho, C.; et al. Bromodomain inhibition shows antitumoral activity in mice and human luminal breast cancer. Oncotarget 2017, 8, 51621–51629. [Google Scholar] [CrossRef]
- Yan, X.; Zhao, Y.; Liu, Y.; Yang, Q.; Dong, L.; Wu, Z.; Nie, J.; Chen, D.; Bai, M.; Ti, D.; et al. Case Report: Low-Dose Decitabine Plus Anti-PD-1 Inhibitor Camrelizumab for Previously Treated Advanced Metastatic Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 558572. [Google Scholar] [CrossRef]
- Weng, R.R.; Lu, H.-H.; Lin, C.-T.; Fan, C.-C.; Lin, R.-S.; Huang, T.-C.; Lin, S.-Y.; Huang, Y.-J.; Juan, Y.-H.; Wu, Y.-C.; et al. Epigenetic modulation of immune synaptic-cytoskeletal networks potentiates γδ T cell-mediated cytotoxicity in lung cancer. Nat. Commun. 2021, 12, 2163. [Google Scholar] [CrossRef]
- Yu, G.; Wu, Y.; Wang, W.; Xu, J.; Lv, X.; Cao, X.; Wan, T. Low-dose decitabine enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by re-modulating the tumor microenvironment. Cell. Mol. Immunol. 2018, 16, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.C.-Y.; Chiang, S.-F.; Chen, W.T.-L.; Chen, T.-W.; Hu, C.-H.; Yang, P.-C.; Ke, T.-W.; Chao, K.S.C. Decitabine Augments Chemotherapy-Induced PD-L1 Upregulation for PD-L1 Blockade in Colorectal Cancer. Cancers 2020, 12, 462. [Google Scholar] [CrossRef]
- Cortes, J.; Cescon, D.W.; Rugo, H.S.; Nowecki, Z.; Im, S.-A.; Yusof, M.M.; Gallardo, C.; Lipatov, O.; Barrios, C.H.; Holgado, E.; et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020, 396, 1817–1828. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.-A.; Wright, G.S.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chiappinelli, K.B.; Guzzetta, A.A.; Easwaran, H.; Yen, R.-W.C.; Vatapalli, R.; Topper, M.J.; Luo, J.; Connolly, R.M.; Azad, N.S.; et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014, 5, 587–598. [Google Scholar] [CrossRef]
- Christmas, B.J.; Rafie, C.I.; Hopkins, A.C.; Scott, B.A.; Ma, H.S.; Cruz, K.A.; Woolman, S.; Armstrong, T.D.; Connolly, R.M.; Azad, N.A.; et al. Entinostat Converts Immune-Resistant Breast and Pancreatic Cancers into Checkpoint-Responsive Tumors by Reprogramming Tumor-Infiltrating MDSCs. Cancer Immunol. Res. 2018, 6, 1561–1577. [Google Scholar] [CrossRef]
- Tripathy, D.; Im, S.-A.; Colleoni, M.; Franke, F.; Bardia, A.; Harbeck, N.; Hurvitz, S.A.; Chow, L.; Sohn, J.; Lee, K.S.; et al. Ribociclib plus endocrine therapy for premenopausal women with hormone-receptor-positive, advanced breast cancer (MONALEESA-7): A randomised phase 3 trial. Lancet Oncol. 2018, 19, 904–915. [Google Scholar] [CrossRef]
- Goetz, M.P.; Toi, M.; Campone, M.; Sohn, J.; Paluch-Shimon, S.; Huober, J.; Park, I.H.; Trédan, O.; Chen, S.-C.; Manso, L.; et al. MONARCH 3: Abemaciclib as Initial Therapy for Advanced Breast Cancer. J. Clin. Oncol. 2017, 35, 3638–3646. [Google Scholar] [CrossRef]
- Finn, R.S.; Martin, M.; Rugo, H.S.; Jones, S.; Im, S.-A.; Gelmon, K.; Harbeck, N.; Lipatov, O.N.; Walshe, J.M.; Moulder, S.; et al. Palbociclib and Letrozole in Advanced Breast Cancer. N. Engl. J. Med. 2016, 375, 1925–1936. [Google Scholar] [CrossRef]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.-S.; Sonke, G.S.; Paluch-Shimon, S.; Campone, M.; Blackwell, K.L.; Andre, F.; Winer, E.P.; et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N. Engl. J. Med. 2016, 375, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Stefansson, O.A.; Esteller, M. Epigenetic Modifications in Breast Cancer and Their Role in Personalized Medicine. Am. J. Pathol. 2013, 183, 1052–1063. [Google Scholar] [CrossRef]
- Drew, Y.; Mulligan, E.A.; Vong, W.-T.; Thomas, H.D.; Kahn, S.; Kyle, S.; Mukhopadhyay, A.; Los, G.; Hostomsky, Z.; Plummer, E.R.; et al. Therapeutic Potential of Poly(ADP-ribose) Polymerase Inhibitor AG014699 in Human Cancers With Mutated or Methylated BRCA1 or BRCA2. JNCI J. Natl. Cancer Inst. 2010, 103, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Marijon, H.; Lee, D.H.; Ding, L.-W.; Sun, H.; Gery, S.; de Gramont, A.; Koeffler, H.P. Co-targeting poly(ADP-ribose) polymerase (PARP) and histone deacetylase (HDAC) in triple-negative breast cancer: Higher synergism in BRCA mutated cells. Biomed. Pharmacother. 2018, 99, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Min, A.; Im, S.-A.; Kim, D.K.; Song, S.-H.; Kim, H.-J.; Lee, K.-H.; Kim, T.-Y.; Han, S.-W.; Oh, D.-Y.; O’Connor, M.J.; et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res. 2015, 17, 33. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, Y.; Shan, W.; Hu, Z.; Yuan, J.; Pi, J.; Wang, Y.; Fan, L.; Tang, Z.; Li, C.; et al. Repression of BET activity sensitizes homologous recombination–proficient cancers to PARP inhibition. Sci. Transl. Med. 2017, 9, eaal1645. [Google Scholar] [CrossRef]
- Mio, C.; Gerratana, L.; Bolis, M.; Caponnetto, F.; Zanello, A.; Barbina, M.; Di Loreto, C.; Garattini, E.; Damante, G.; Puglisi, F. BET proteins regulate homologous recombination-mediated DNA repair: BRCAness and implications for cancer therapy. Int. J. Cancer 2019, 144, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef]
- Stirzaker, C.; Taberlay, P.; Statham, A.L.; Clark, S.J. Mining cancer methylomes: Prospects and challenges. Trends Genet. 2014, 30, 75–84. [Google Scholar] [CrossRef]
- Alix-Panabières, C. The future of liquid biopsy. Nature 2020, 579, S9. [Google Scholar] [CrossRef]
- Coombes, R.C.; Page, K.; Salari, R.; Hastings, R.K.; Armstrong, A.C.; Ahmed, S.; Ali, S.; Cleator, S.J.; Kenny, L.M.; Stebbing, J.; et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin. Cancer Res. 2019, 25, 4255–4263. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Fortina, P.; Dawson, S.-J.; Rosenfeld, N.; Caldas, C.; Weiss, L.; Hufnagl, C.; Greil, R. Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N. Engl. J. Med. 2013, 369, 93–94. [Google Scholar] [CrossRef] [PubMed]
- Lam, D.; Luu, P.-L.; Song, J.Z.; Qu, W.; Risbridger, G.P.; Lawrence, M.G.; Lu, J.; Trau, M.; Korbie, D.; Clark, S.J.; et al. Comprehensive evaluation of targeted multiplex bisulphite PCR sequencing for validation of DNA methylation biomarker panels. Clin. Epigenetics 2020, 12, 90. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, S.E.; Laird, P.W. Sensitive Detection of DNA Methylation. Ann. N. Y. Acad. Sci. 2003, 983, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Shan, M.; Yin, H.; Li, J.; Li, X.; Wang, D.; Su, Y.; Niu, M.; Zhong, Z.; Wang, J.; Zhang, X.; et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget 2016, 7, 18485–18494. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, D.; Yin, Y.; Yang, T.; You, Z.; Li, D.; Chen, Y.; Jiang, Y.; Xu, S.; Geng, J.; et al. Circulating cell-free DNA-based methylation patterns for breast cancer diagnosis. NPJ Breast Cancer 2021, 7, 106. [Google Scholar] [CrossRef]
- Klein, E.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Sharma, G.; Mirza, S.; Parshad, R.; Gupta, S.D.; Ralhan, R. DNA methylation of circulating DNA: A marker for monitoring efficacy of neoadjuvant chemotherapy in breast cancer patients. Tumor Biol. 2012, 33, 1837–1843. [Google Scholar] [CrossRef]
- Zurita, M.; Lara, P.C.; del Moral, R.; Torres, B.; Linares-Fernández, J.L.; Arrabal, S.R.; de Almodóvar, J.M.R. Hypermethylated 14-3-3-sigma and ESR1 gene promoters in serum as candidate biomarkers for the diagnosis and treatment efficacy of breast cancer metastasis. BMC Cancer 2010, 10, 217. [Google Scholar] [CrossRef]
- Fiegl, H.; Jones, A.; Hauser-Kronberger, C.; Hutarew, G.; Reitsamer, R.; Jones, R.L.; Dowsett, M.; Mueller-Holzner, E.; Windbichler, G.; Daxenbichler, G.; et al. Methylated NEUROD1 Promoter is a Marker for Chemosensitivity in Breast Cancer. Clin. Cancer Res. 2008, 14, 3494–3502. [Google Scholar] [CrossRef][Green Version]
- Fiegl, H.; Millinger, S.; Mueller-Holzner, E.; Marth, C.; Ensinger, C.; Berger, A.; Klocker, H.; Goebel, G.; Widschwendter, M. Circulating Tumor-Specific DNA: A Marker for Monitoring Efficacy of Adjuvant Therapy in Cancer Patients. Cancer Res. 2005, 65, 1141–1145. [Google Scholar] [CrossRef]
- Moss, J.; Zick, A.; Grinshpun, A.; Carmon, E.; Maoz, M.; Ochana, B.; Abraham, O.; Arieli, O.; Germansky, L.; Meir, K.; et al. Circulating breast-derived DNA allows universal detection and monitoring of localized breast cancer. Ann. Oncol. 2020, 31, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, K.; Fackler, M.S.; Zhang, Z.; Bujanda, Z.L.; Jeter, S.C.; Sokoll, L.J.; Garrett-Mayer, E.; Cope, L.M.; Umbricht, C.B.; Euhus, D.M.; et al. Monitoring of Serum DNA Methylation as an Early Independent Marker of Response and Survival in Metastatic Breast Cancer: TBCRC 005 Prospective Biomarker Study. J. Clin. Oncol. 2017, 35, 751–758. [Google Scholar] [CrossRef]
- Fackler, M.J.; Bujanda, Z.L.; Umbricht, C.; Teo, W.W.; Cho, S.; Zhang, Z.; Visvanathan, K.; Jeter, S.; Argani, P.; Wang, C.; et al. Novel Methylated Biomarkers and a Robust Assay to Detect Circulating Tumor DNA in Metastatic Breast Cancer. Cancer Res. 2014, 74, 2160–2170. [Google Scholar] [CrossRef]
- De Ruijter, T.C.; van der Heide, F.; Smits, K.M.; Aarts, M.J.; van Engeland, M.; Heijnen, V.C. Prognostic DNA methylation markers for hormone receptor breast cancer: A systematic review. Breast Cancer Res. 2020, 22, 13. [Google Scholar] [CrossRef] [PubMed]
- Fontes-Sousa, M.; Amorim, M.; Salta, S.; Palma De Sousa, S.; Henrique, R.; Jerónimo, C. Predicting resistance to endocrine therapy in breast cancer: It’s time for epigenetic biomarkers (Review). Oncol. Rep. 2019, 41, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Martens, J.W.; Nimmrich, I.; Koenig, T.; Look, M.P.; Harbeck, N.; Model, F.; Kluth, A.; Vries, J.B.-D.; Sieuwerts, A.M.; Portengen, H.; et al. Association of DNA Methylation of Phosphoserine Aminotransferase with Response to Endocrine Therapy in Patients with Recurrent Breast Cancer. Cancer Res. 2005, 65, 4101–4117. [Google Scholar] [CrossRef]
- Dodaran, M.S.; Borgoni, S.; Sofyalı, E.; Verschure, P.J.; Wiemann, S.; Moerland, P.D.; van Kampen, A.H. Candidate methylation sites associated with endocrine therapy resistance in ER+/HER2- breast cancer. BMC Cancer 2020, 20, 676. [Google Scholar]
- Fackler, M.J.; Cho, S.; Cope, L.; Gabrielson, E.; Visvanathan, K.; Wilsbach, K.; Meir-Levi, D.; Lynch, C.F.; Marks, J.; Geradts, J.; et al. DNA methylation markers predict recurrence-free interval in triple-negative breast cancer. NPJ Breast Cancer 2020, 6, 3. [Google Scholar] [CrossRef]
- Pineda, B.; Diaz-Lagares, A.; Pérez-Fidalgo, J.A.; Burgués, O.; González-Barrallo, I.; Crujeiras, A.B.; Sandoval, J.; Esteller, M.; Lluch, A.; Eroles, P. A two-gene epigenetic signature for the prediction of response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Clin. Epigenetics 2019, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Tost, J.; Gut, I.G. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007, 2, 2265–2275. [Google Scholar] [CrossRef] [PubMed]
- Tost, J.; Gut, I.G. Analysis of Gene-Specific DNA Methylation Patterns by Pyrosequencing® Technology. Methods Mol. Biol. 2007, 373, 89–102. [Google Scholar] [CrossRef]
- Brena, R.M.; Auer, H.; Kornacker, K.; Plass, C. Quantification of DNA methylation in electrofluidics chips (Bio-COBRA). Nat. Protoc. 2006, 1, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Ehrich, M.; Nelson, M.R.; Stanssens, P.; Zabeau, M.; Liloglou, T.; Xinarianos, G.; Cantor, C.R.; Field, J.; Boom, D.V.D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. USA 2005, 102, 15785–15790. [Google Scholar] [CrossRef]
- Eads, C.A.; Danenberg, K.D.; Kawakami, K.; Saltz, L.; Blake, C.; Shibata, D.; Danenberg, P.V.; Laird, P.W. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000, 28, 8.e32. [Google Scholar] [CrossRef]
- Herman, J.G.; Graff, J.R.; Myohanen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef] [PubMed]
- Montavon, C.; Gloss, B.S.; Warton, K.; Barton, C.A.; Statham, A.L.; Scurry, J.P.; Tabor, B.; Nguyen, T.V.; Qu, W.; Samimi, G.; et al. Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol. Oncol. 2012, 124, 582–588. [Google Scholar] [CrossRef]
- Devaney, J.; Stirzaker, C.; Qu, W.; Song, J.Z.; Statham, A.L.; Patterson, K.I.; Horvath, L.G.; Tabor, B.; Coolen, M.W.; Hulf, T.; et al. Epigenetic Deregulation Across Chromosome 2q14.2 Differentiates Normal from Prostate Cancer and Provides a Regional Panel of Novel DNA Methylation Cancer Biomarkers. Cancer Epidemiol. Biomark. Prev. 2011, 20, 148–159. [Google Scholar] [CrossRef]
- Rand, K.N.; Ho, T.; Qu, W.; Mitchell, S.M.; White, R.; Clark, S.; Molloy, P.L. Headloop suppression PCR and its application to selective amplification of methylated DNA sequences. Nucleic Acids Res. 2005, 33, e127. [Google Scholar] [CrossRef] [PubMed]
- Weisenberger, D.J.; Trinh, B.N.; Campan, M.; Sharma, S.; Long, T.I.; Ananthnarayan, S.; Liang, G.; Esteva, F.J.; Hortobagyi, G.N.; McCormick, F.; et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res. 2008, 36, 4689–4698. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Korbie, D.; Lin, E.; Wall, D.; Nair, S.S.; Stirzaker, C.; Clark, S.J.; Trau, M. Multiplex bisulfite PCR resequencing of clinical FFPE DNA. Clin. Epigenetics 2015, 7, 28. [Google Scholar] [CrossRef]
- Legendre, C.; Gooden, G.C.; Johnson, K.; Martinez, R.A.; Liang, W.S.; Salhia, B. Whole-genome bisulfite sequencing of cell-free DNA identifies signature associated with metastatic breast cancer. Clin. Epigenetics 2015, 7, 100. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Lüönd, F.; Tiede, S.; Christofori, G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br. J. Cancer 2021, 125, 164–175. [Google Scholar] [CrossRef]
- Ektefaie, Y.; Yuan, W.; Dillon, D.A.; Lin, N.U.; Golden, J.A.; Kohane, I.S.; Yu, K.-H. Integrative multiomics-histopathology analysis for breast cancer classification. NPJ Breast Cancer 2021, 7, 147. [Google Scholar] [CrossRef]
| Therapy | Phase | Patient Population | Breast Cancer Patients (n) | ORR (%) | OS (months) | PFS (months) | AE’s ≥ Grade 3–4 | Biomarkers | Ref |
|---|---|---|---|---|---|---|---|---|---|
| Epigenetic Therapies Alone | |||||||||
| Vorinostat | II | Advancedbreast cancer | 14 | 0 | NR | NR | Diarrhoea (7%) Nausea (7%) Dehydration (7%) | NA | [8] |
| Mivebresib | I | Solid organ tumours | 8/72 | 0 | NR | 1.8 |
Thrombocytopenia (35%) Anaemia (6%) | NA | [9] |
| Epigenetic Therapies + Cytotoxic Agents | |||||||||
| Breast | |||||||||
| Vorinostat + paclitaxel + bevacizumab | I/II | Advancedbreast cancer | 44 | 55 | 29.4 | 11.9 | Neutropenia (27%) Anaemia (6%) Thrombocytopenia (2%) Diarrhoea (6%) Fatigue (19%) | HSP90 lysine 69-acetylation Acetyltubulin Acetylhistone H3 Acetylhistone H4 | [10] |
| All Comer Trials | |||||||||
| Hydralazine + magnesium valproate + chemotherapy | II | Solid organ tumours | 3/17 | 24 | NR | NR | Thrombocytopenia (47%) Anaemia (41%) Neutropenia (23%) Hypoalbuminaemia (23%) Infection (23%) | Histone Deacetylation DNA demethylation | [11] |
| Decitabine + Carboplatin | I | Solid organ tumours | 5/35 | 3% | NR | NR | Neutropenia (46%) Leucopenia (43%) Anaemia (6%) Thrombocytopenia (3%) Fatigue (9%) Mucositis (3%) | DNA methylation (dose dependent decrease) MAGE1 CpG Demethylatio | [12] |
| Vorinostat + Doxorubicin | I | Solid organ tumours | 5/32 | 8% | NR | NR | Neutropenia (25%) Thrombocytopenia (12%) Mucositis (3%) | Histone H3 H4 acetylation HDAC2 expression | [13] |
| Epigenetic Therapies + Endocrine Therapy | |||||||||
| Breast | |||||||||
| Exemestane +/− tucidinostat | III (2:1) | Advanced ER+ breast cancer | 365 | 18% vs. 9% (p = 0.025) | NR | 7.4 vs. 3.8 (p = 0.033) | Neutropenia (51% vs. 3%) Thrombocytopenia (27% vs. 3%) Leucopenia (19% vs. 3%) | NR | [14] |
| Exemestane +/− entinostat | II | Advanced ER+ breast cancer | 135 | NA | 28.1 vs. 19.8 m (HR 0.59; p = 0.036) | 4.3 vs. 2.3 | Fatigue (13%) Neutropenia (15%) Thrombocytopenia Higher rates of treatment discontinuation in the entinostat group (11% vs. 2%) | Protein lysine hyperacetylation associated with prolonged PFS in entinostat arm | [15] |
| Exemestane +/− entinostat | III (1:1) | Advanced ER+ breast cancer | 608 | 5.8% vs. 5.6% | 23.4 vs. 21.7 (HR 0.99; p = 0.94) | 3.3 vs. 3.1 (HR 0.87; p = 0.030) | Neutropenia (20%) Hypophosphatemia (14%) Anaemia (8%) Leukopenia (6%) Fatigue (4%) Diarrhoea (4%) Thrombocytopenia (3%) | Higher increase in lysine acetylation in PMBCs in the entinostat arm | [16] |
| Vorinostat + tamoxifen | II | Advanced ER+ breast cancer | 43 | ORR 19% | 29 | 10.3 | Thrombocytopenia 9% Neutropenia 16% Fatigue 16% VTE 7% | HDAC2 expression Histone hyperaceytlation | [17] |
| Epigenetic Therapies + Targeted therapy | |||||||||
| All Comers | |||||||||
| 5-azacytidine + erlotinib | I | Solid organ tumours | 1/30 | 7% | 7.5 | 2 | Neutropenia (27%) Neuropathy (3%) Anaemia (3%) Infection (7%) | NA | [18] |
| Vorinostat + sirolimus | I | Solid organ tumours | 1/70 | 3% | 10.3 | 2 | Thrombocytopenia (31%) Neutropenia (8%) Anaemia (7%) Fatigue (3%) | NA | [19] |
| Breast | |||||||||
| SAHA + trastuzumab | I/II | HER2-positive and negative metastatic breast cancer | 15 | 7% | 9.3 | 1.5 | Thrombocytopenia 6% | NA | [20] |
| Epigenetic Target | Breast Cancer Subtype | Phase of Trial | Interventions | Status | Clinical Trials Reference |
|---|---|---|---|---|---|
| DNMT | Advanced HR+, HER2– Progressed on AI. | 2 | Fulvestrant + Azacitadine | Terminated | NCT02374099 |
| Locally advanced, resectable HER2– | 2 | Pembrolizumab + Decitabine followed by neoadjuvant chemotherapy | Recruiting | NCT02957968 | |
| Advanced HER2– | 1b | Paclitaxel + Decitabine | Unknown | NCT03282825 | |
| Advanced TNBC | 2 | Carboplatin + Decitabine | Recruiting | NCT03295552 | |
| Advanced, any subtype | 1b/2 | Nab-paclitaxel + Azacitadine | Completed | NCT00748553 | |
| DNMT + HDAC | Advanced HER2– | 2 | Azacitadine + Entinostat | Active, not recruiting | NCT01349959 |
| HDAC | Advanced HR+ | 2 | Exemestane +/− Entinostat | Active, not recruiting | NCT02115282 |
| Advanced HR+ | 3 | Exemestane +/− Entinostat | Active, not recruiting | NCT03538171 | |
| Advanced HR+ | 2 | Exemestane +/− Entinostat | Active, not recruiting | NCT03291886 | |
| Advanced HR+, HER2– | 1b/2 | Atezolizumab + Entinostat vs. Fulvestrant | Recruiting | NCT03280563 | |
| Advanced HR+ | 1 | Nivolumab + Ipilimumab + Entinostat | Active, not recruiting | NCT02453620 | |
| Advanced HR+, PD1 > 10% | 2 | Tamoxifen + Pembrolizumab +/− Vorinostat | Active, not recruiting | NCT04190056 | |
| Advanced HER2–and Stage I-III HER2–, with residual disease following neoadjuvant chemotherapy | 1 | Capecitabine + Entinostat | Recruiting | NCT03473639 | |
| Advanced HER2+ | 1 | Lapatinib + Trastuzumab + Entinostat | Completed | NCT01434303 | |
| Early stage TNBC | 2 | Neoadjuvant Anastrozole + Entinostat | Terminated | NCT01234532 | |
| Advanced TNBC | 1 | Ribociclib + Belinostat | Recruiting | NCT04315233 | |
| Advanced TNBC | 1/2 | Cisplatin + Nivolumab + Romidepsin | Suspended | NCT02393794 | |
| BET | Advanced HR+ | 1 | Fulvestrant + Molibresib/GSK525762 | Active, not recruiting | NCT02964507 |
| Advanced HR+ | 1 | Fulvestrant or Exemestane + Alobresib | Completed | NCT02392611 | |
| Advanced TNBC, BRCA wildtype | 2 | Talazoparib + ZEN003694 | Active, not recruiting | NCT03901469 |
| Epigenetic Biomarkers | Genes with Poor Outcomes | Genes with Improved Outcomes | Methods of Assessment |
|---|---|---|---|
| Prognostic Biomarkers | Hypermethylation of BRCA, PITX2, CDH1, RARB, PCDH10, PGR, GSTP1, RASSF1, PTEN, PTGER4, CDK10, HOXC10, ID4, NAT1 | Hypermethylation of ESRI and CYP1B1 | Methylight Digital Droplet MSP Bisulphite pyrosequencing Combined bisulphite restriction analysis (COBRA) EpiTYPER MethyLight Methylation-Specific PCR Headloop-MSP Digital bisulphite genomic sequencing Methylation-sensitive high resolution melting Targeted multiplex bisulphite amplicon sequencing. Whole-genome bisulphite sequencing MeDIP-seq |
| Predictive Biomarkers | Hypermethylation of DAXX, MSI2, NCOR2, RXRA, C8orf46, GATA3, ITPK1, ESR1 and GET4 genes | Hypermethylation of PSAT1 promoter hypermethylation, FERD3L, TRIP10 BRCA1 Hypomethylation of ESR1-responsive enhancer elements |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, L.J.; Achinger-Kawecka, J.; Portman, N.; Clark, S.; Stirzaker, C.; Lim, E. Epigenetic Therapies and Biomarkers in Breast Cancer. Cancers 2022, 14, 474. https://doi.org/10.3390/cancers14030474
Brown LJ, Achinger-Kawecka J, Portman N, Clark S, Stirzaker C, Lim E. Epigenetic Therapies and Biomarkers in Breast Cancer. Cancers. 2022; 14(3):474. https://doi.org/10.3390/cancers14030474
Chicago/Turabian StyleBrown, Lauren Julia, Joanna Achinger-Kawecka, Neil Portman, Susan Clark, Clare Stirzaker, and Elgene Lim. 2022. "Epigenetic Therapies and Biomarkers in Breast Cancer" Cancers 14, no. 3: 474. https://doi.org/10.3390/cancers14030474
APA StyleBrown, L. J., Achinger-Kawecka, J., Portman, N., Clark, S., Stirzaker, C., & Lim, E. (2022). Epigenetic Therapies and Biomarkers in Breast Cancer. Cancers, 14(3), 474. https://doi.org/10.3390/cancers14030474






