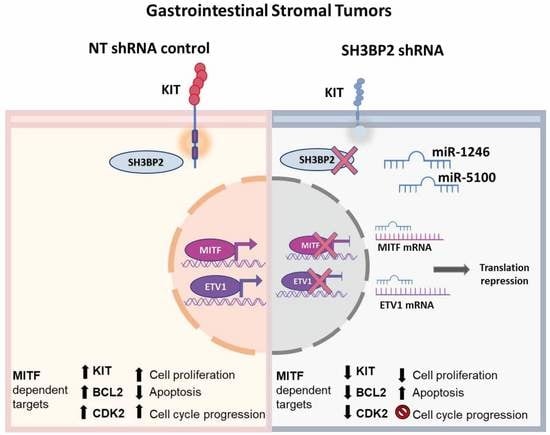
SH3BP2 Silencing Increases miRNAs Targeting ETV1 and Microphthalmia-Associated Transcription Factor, Decreasing the Proliferation of Gastrointestinal Stromal Tumors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibodies and Reagents
2.2. Cell Culture
2.3. RNA Extraction, Retrotranscription, and PCR Assays
2.4. MicroRNA Array Profiling
2.5. Lentiviral Transduction
2.6. Cell Viability, Proliferation, and Caspases 3/7 Activity Assays
2.7. Western Blotting
2.8. Cell Cycle Analysis by Flow Cytometry
2.9. MITF Overexpression
2.10. LNA Anti-miRNA Treatment
2.11. Statistical Data Analysis
3. Results
3.1. SH3BP2 Silencing Reduces ETV1 Levels in GIST Cell Lines
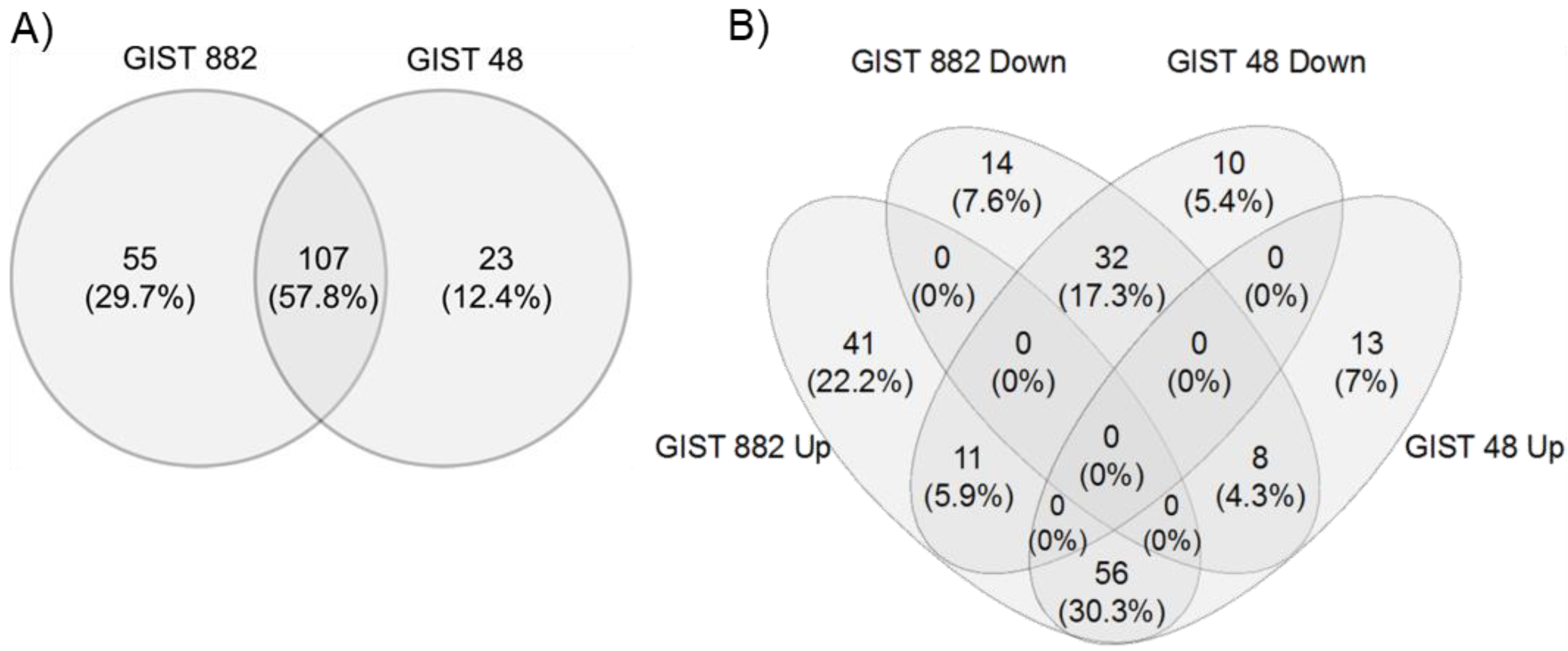
3.2. miRNA Profiling of SH3BP2-Silenced GIST Cells
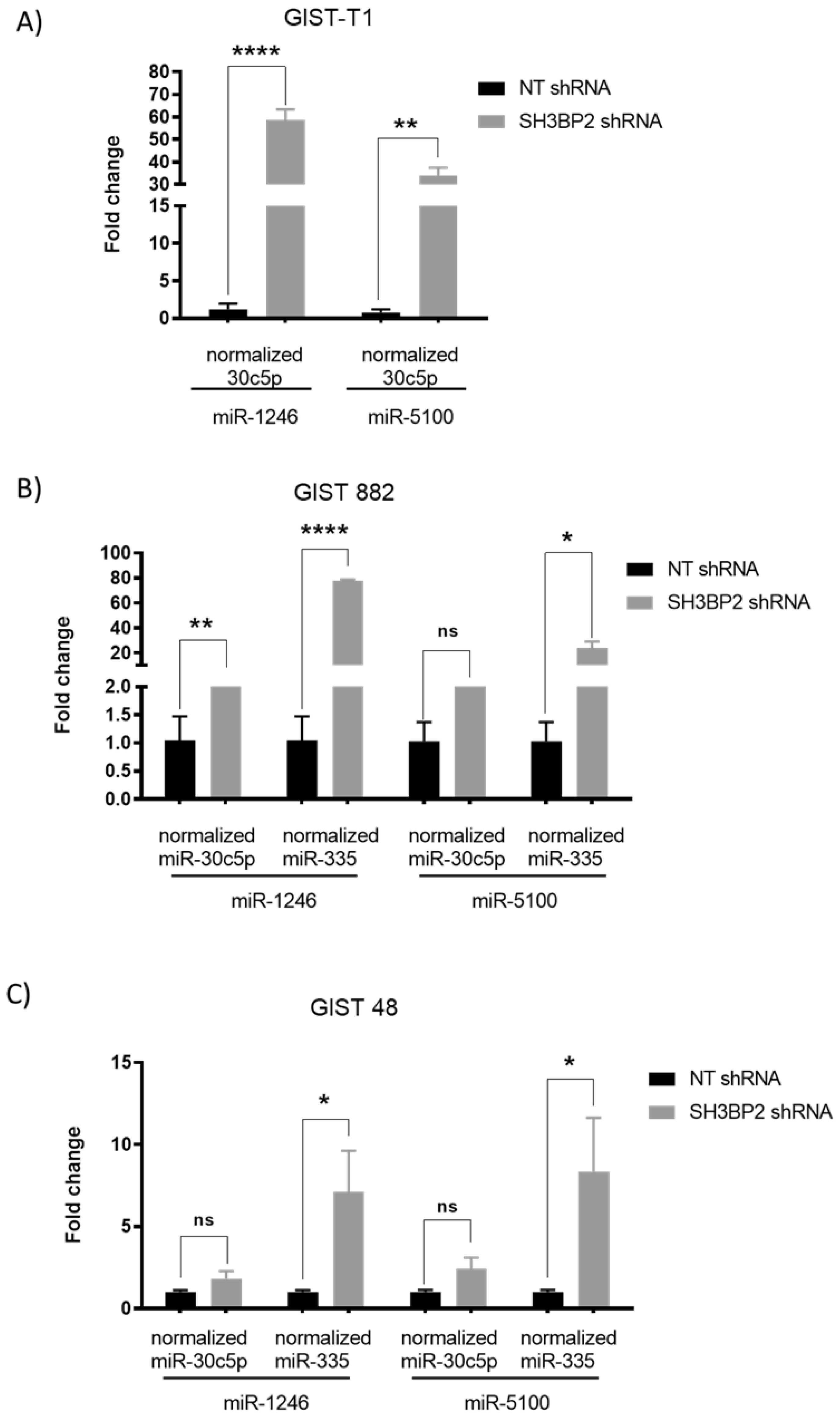
3.3. Validation of Up-Regulated miRNAs That Target MITF and ETV1in GIST Cell Lines
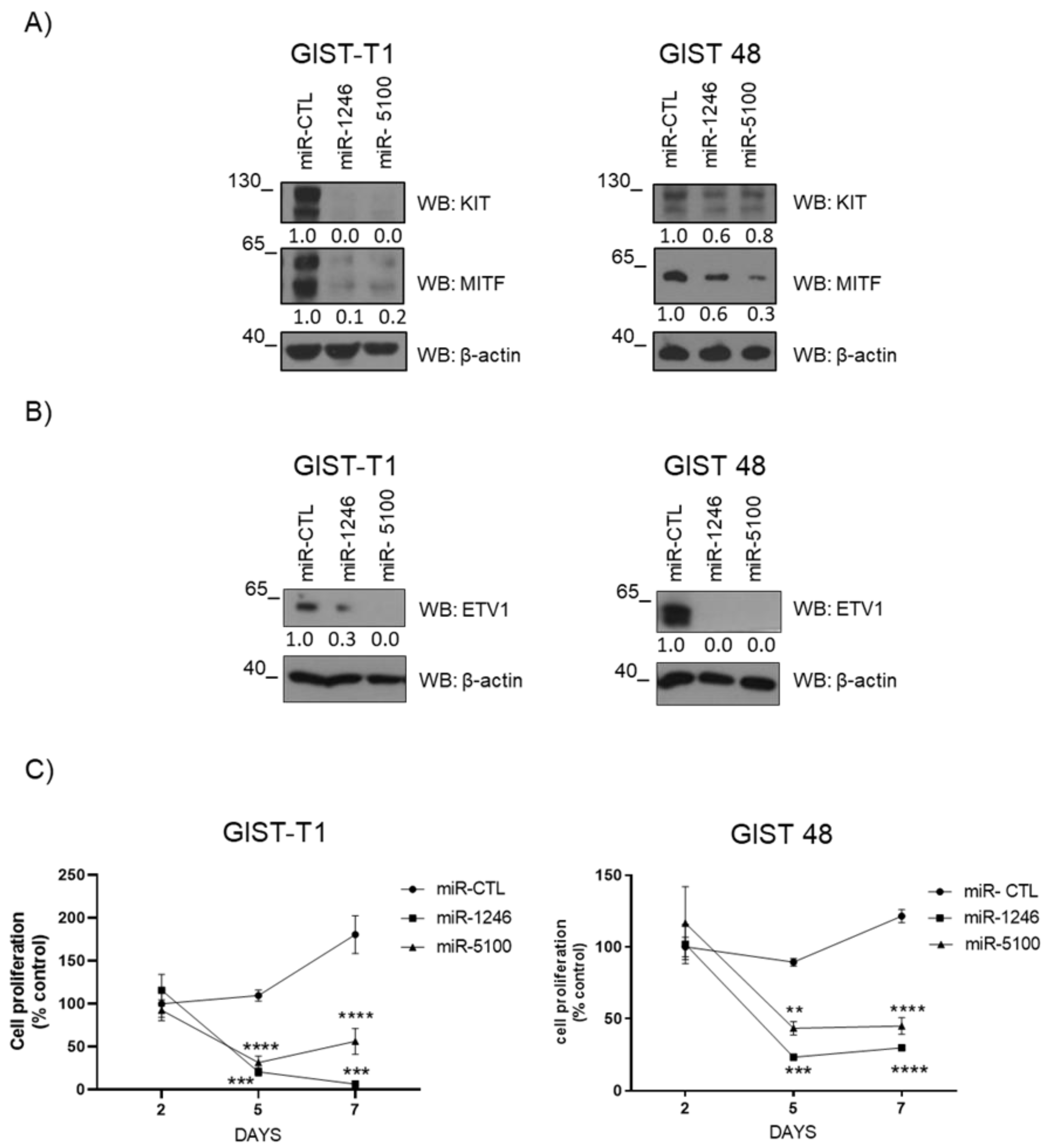
3.4. MiR-1246 and miR-5100 Target ETV1 and MITF, and Overexpression Significantly Affects Cell Proliferation
3.5. MiR-1246 and miR-5100 Promote Apoptosis by Caspases 3/7 in GIST Cells
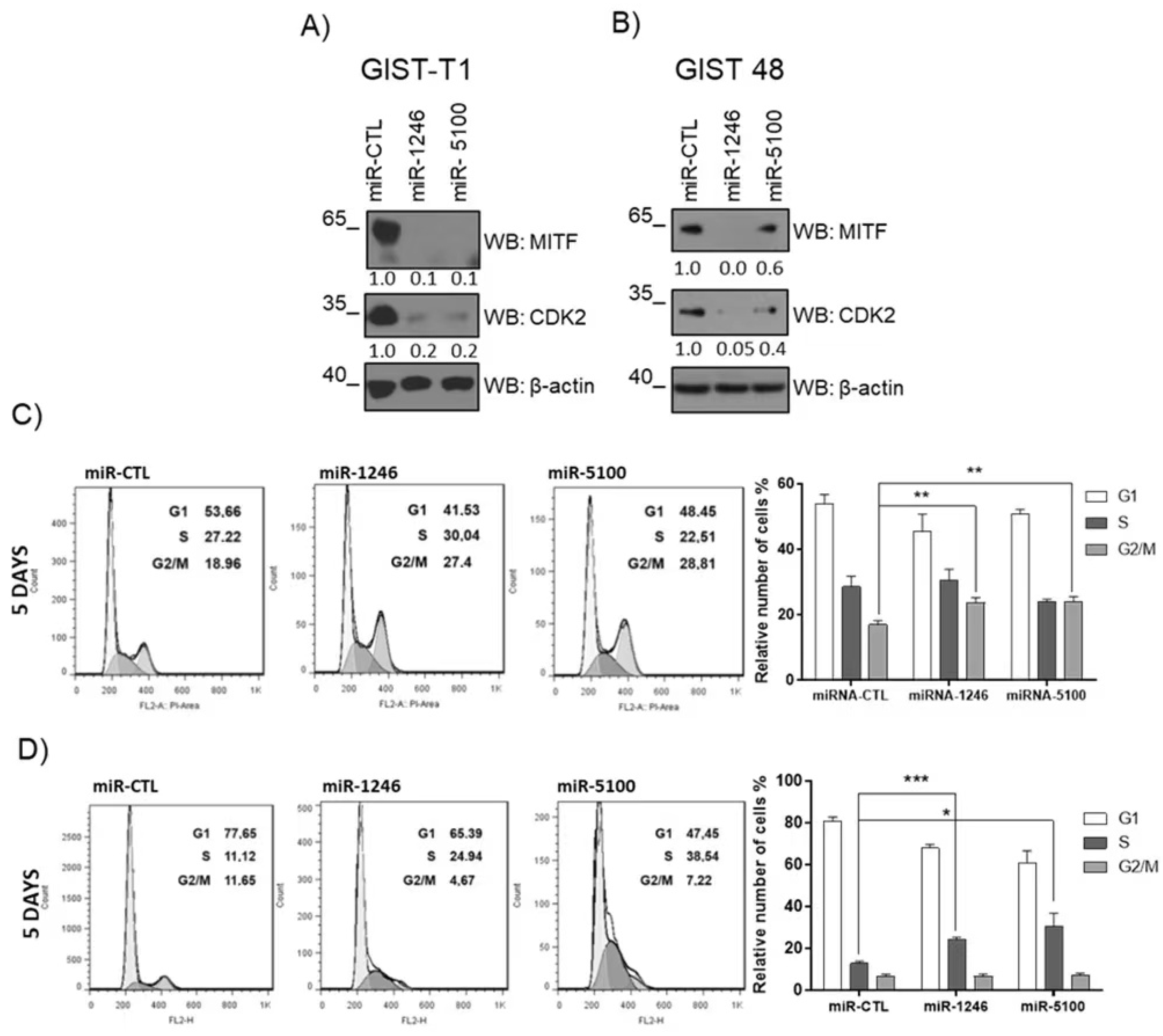
3.6. MiR-1246 and miR-5100 Affect Cell Cycle Progression
3.7. MITF Overexpression Significantly Restores Cell Proliferation
3.8. LNA Treatment (Anti-miR-1246 Anti-miR-5100) Was Not Adequate to Revert the Apoptotic Phenotype in GIST-T1 SH3BP2 Silenced Cell
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kelly, C.M.; Gutierrez Sainz, L.; Chi, P. The management of metastatic GIST: Current standard and investigational therapeutics. J. Hematol. Oncol. 2021, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Beham, A.W.; Schaefer, I.-M.; Schüler, P.; Cameron, S.; Michael Ghadimi, B. Gastrointestinal stromal tumors. Int. J. Color. Dis. 2012, 27, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Wada, R.; Arai, H.; Kure, S.; Peng, W.X.; Naito, Z. “Wild type” GIST: Clinicopathological features and clinical practice. Pathol. Int. 2016, 66, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Ainsua-Enrich, E.; Serrano-Candelas, E.; Álvarez-Errico, D.; Picado, C.; Sayós, J.; Rivera, J.; Martín, M. The Adaptor 3BP2 Is Required for KIT Receptor Expression and Human Mast Cell Survival. J. Immunol. 2015, 194, 4309–4318. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Candelas, E.; Ainsua-Enrich, E.; Navinés-Ferrer, A.; Rodrigues, P.; García-Valverde, A.; Bazzocco, S.; Macaya, I.; Arribas, J.; Serrano, C.; Sayós, J.; et al. Silencing of adaptor protein SH3BP2 reduces KIT/PDGFRA receptors expression and impairs gastrointestinal stromal tumors growth. Mol. Oncol. 2018, 12, 1383–1397. [Google Scholar] [CrossRef] [PubMed]
- Hoek, K.S.; Schlegel, N.C.; Eichhoff, O.M.; Widmer, D.S.; Praetorius, C.; Einarsson, S.O.; Valgeirsdottir, S.; Bergsteinsdottir, K.; Schepsky, A.; Dummer, R.; et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008, 21, 665–676. [Google Scholar] [CrossRef]
- Du, J.; Widlund, H.R.; Horstmann, M.A.; Ramaswamy, S.; Ross, K.; Huber, W.E.; Nishimura, E.K.; Golub, T.R.; Fisher, D.E. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 2004, 6, 565–576. [Google Scholar] [CrossRef]
- Levy, C.; Khaled, M.; Fisher, D.E. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef]
- Morii, E.; Ogihara, H.; Kim, D.-K.K.; Ito, A.; Oboki, K.; Lee, Y.-M.M.; Jippo, T.; Nomura, S.; Maeyama, K.; Lamoreux, M.L.; et al. Importance of leucine zipper domain of mitranscription factor (MITF) for differentiation of mast cells demonstrated using mice/mice mutant mice of which MITF lacks the zipper domain. Blood 2001, 97, 2038–2044. [Google Scholar] [CrossRef]
- Tsujimura, T.; Morii, E.; Nozaki, M.; Hashimoto, K.; Moriyama, Y.; Takebayashi, K.; Kondo, T.; Kanakura, Y.; Kitamura, Y. Involvement of Transcription Factor Encoded by the mi Locus in the Expression of c-kit Receptor Tyrosine Kinase in Cultured Mast Cells of Mice. Blood 1996, 88, 1225–1233. [Google Scholar] [CrossRef]
- Proaño-Pérez, E.; Serrano-Candelas, E.; García-Valverde, A.; Rosell, J.; Gómez-Peregrina, D.; Navinés-Ferrer, A.; Guerrero, M.; Serrano, C.; Martín, M.; Elizabeth, P.P.; et al. The microphthalmia-associated transcription factor is involved in gastrointestinal stromal tumor growth. Cancer Gene Ther. 2022, 115–117. [Google Scholar] [CrossRef]
- Lee, Y.-N.; Brandal, S.; Noel, P.; Wentzel, E.; Mendell, J.T.; McDevitt, M.A.; Kapur, R.; Carter, M.; Metcalfe, D.D.; Takemoto, C.M. KIT signaling regulates MITF expression through miRNAs in normal and malignant mast cell proliferation. Blood 2011, 117, 3629–3640. [Google Scholar] [CrossRef] [PubMed]
- Ran, L.; Sirota, I.; Cao, Z.; Murphy, D.; Chen, Y.; Shukla, S.; Xie, Y.; Kaufmann, M.C.; Gao, D.; Zhu, S.; et al. Combined Inhibition of MAP Kinase and KIT Signaling Synergistically Destabilizes ETV1 and Suppresses GIST Tumor Growth. Cancer Discov. 2015, 5, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Chi, P.; Chen, Y.; Zhang, L.; Guo, X.; Wongvipat, J.; Shamu, T.; Fletcher, J.A.; Dewell, S.; Maki, R.G.; Zheng, D.; et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature 2010, 467, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Silver, J.; Oshlack, A.; Holmes, M.; Diyagama, D.; Holloway, A.; Smyth, G.K. A comparison of background correction methods for two-colour microarrays. Bioinformatics 2007, 23, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal Violet Assay for Determining Viability of Cultured Cells. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot087379. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Errico, D.; Oliver-Vila, I.; Ainsua-Enrich, E.; Gilfillan, A.M.; Picado, C.; Sayós, J.; Martín, M. CD84 Negatively Regulates IgE High-Affinity Receptor Signaling in Human Mast Cells. J. Immunol. 2011, 187, 5577–5586. [Google Scholar] [CrossRef]
- Pozarowski, P.; Darzynkiewicz, Z. Analysis of Cell Cycle by Flow Cytometry. In Checkpoint Controls and Cancer; Humana Press: Totowa, NJ, USA, 2004; Volume 281, pp. 301–312. [Google Scholar]
- Zhang, Y.; Gu, M.-L.; Zhou, X.-X.; Ma, H.; Yao, H.-P.; Ji, F. Altered expression of ETV1 and its contribution to tumorigenic phenotypes in gastrointestinal stromal tumors. Oncol. Rep. 2014, 32, 927–934. [Google Scholar] [CrossRef][Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Hsu, J.; Chiu, C.-M.; Hsu, S.-D.; Huang, W.-Y.; Chien, C.-H.; Lee, T.-Y.; Huang, H.-D. miRTar: An integrated system for identifying miRNA-target interactions in human. BMC Bioinform. 2011, 12, 300. [Google Scholar] [CrossRef]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk—Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.-C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1—Integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef]
- Zhang, H.M.; Li, H.; Wang, G.X.; Wang, J.; Xiang, Y.; Huang, Y.; Shen, C.; Dai, Z.T.; Li, J.P.; Zhang, T.C.; et al. MKL1/miR-5100/CAAP1 loop regulates autophagy and apoptosis in gastric cancer cells. Neoplasia 2020, 22, 220–230. [Google Scholar] [CrossRef]
- Fang, Y.; Gao, F.; Hao, J.; Liu, Z. microRNA-1246 mediates lipopolysaccharide-induced pulmonary endothelial cell apoptosis and acute lung injury by targeting angiotensin-converting enzyme 2. Am. J. Transl. Res. 2017, 9, 1287–1296. [Google Scholar]
- Kawakami, A.; Fisher, D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. 2017, 97, 649–656. [Google Scholar] [CrossRef]
- Poveda, A.; García del Muro, X.; López-Guerrero, J.A.; Cubedo, R.; Martínez, V.; Romero, I.; Serrano, C.; Valverde, C.; Martín-Broto, J. GEIS guidelines for gastrointestinal sarcomas (GIST). Cancer Treat. Rev. 2017, 55, 107–119. [Google Scholar] [CrossRef]
- Bhagirath, D.; Yang, T.L.; Bucay, N.; Sekhon, K.; Majid, S.; Shahryari, V.; Dahiya, R.; Tanaka, Y.; Saini, S. microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res. 2018, 78, 1833–1844. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Z.; Zhu, X.; Chen, L.; Ma, Y.; Wang, J.; Yang, X.; Liu, Z. Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int. J. Clin. Oncol. 2019, 25, 89–99. [Google Scholar] [CrossRef]
- Zhang, Y.; Liao, J.M.; Zeng, S.X.; Lu, H. p53 downregulates Down syndrome-associated DYRK1A through miR-1246. EMBO Rep. 2011, 12, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Z.; Hu, J.; Wan, X.; Huang, W.; Zhang, H.; Jiang, N. MiR-1246 regulates the PI3K/AKT signaling pathway by targeting PIK3AP1 and inhibits thyroid cancer cell proliferation and tumor growth. Mol. Cell. Biochem. 2022, 477, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Suo, T.; Chen, G.; Huang, Y.; Zhao, K.; Wang, T.; Hu, K. miRNA-1246 suppresses acute lung injury-induced inflammation and apoptosis via the NF-κB and Wnt/β-catenin signal pathways. Biomed. Pharmacother. 2018, 108, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.T.; Fan, W.X. MiRNA-1246 suppresses the proliferation and migration of renal cell carcinoma through targeting CXCR4. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5979–5987. [Google Scholar] [CrossRef]
- Kim, J.; Ahn, J.; Lee, M. Upregulation of MicroRNA-1246 Is Associated with BRAF Inhibitor Resistance in Melanoma Cells with Mutant BRAF. Cancer Res. Treat. 2017, 49, 947–959. [Google Scholar] [CrossRef]
- Sun, Z.; Meng, C.; Wang, S.; Zhou, N.; Guan, M.; Bai, C.; Lu, S.; Han, Q.; Zhao, R.C. MicroRNA-1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC Cancer 2014, 14, 616. [Google Scholar] [CrossRef]
- Chijiiwa, Y.; Moriyama, T.; Ohuchida, K.; Nabae, T.; Ohtsuka, T.; Miyasaka, Y.; Fujita, H.; Maeyama, R.; Manabe, T.; Abe, A.; et al. Overexpression of microRNA-5100 decreases the aggressive phenotype of pancreatic cancer cells by targeting PODXL. Int. J. Oncol. 2016, 48, 1688–1700. [Google Scholar] [CrossRef]
- Huang, H.; Jiang, Y.; Wang, Y.; Chen, T.; Yang, L.; He, H.; Lin, Z.; Liu, T.; Yang, T.; Kamp, D.W.; et al. miR-5100 promotes tumor growth in lung cancer by targeting Rab6. Cancer Lett. 2015, 362, 15–24. [Google Scholar] [CrossRef]
- Long, D.; Lee, R.; Williams, P.; Chan, C.Y.; Ambros, V.; Ding, Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007, 14, 287–294. [Google Scholar] [CrossRef]
- McGill, G.G.; Horstmann, M.; Widlund, H.R.; Du, J.; Motyckova, G.; Nishimura, E.K.; Lin, Y.L.; Ramaswamy, S.; Avery, W.; Ding, H.F.; et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 2002, 109, 707–718. [Google Scholar] [CrossRef]
- Haq, R.; Yokoyama, S.; Hawryluk, E.B.; Jönsson, G.B.; Frederick, D.T.; McHenry, K.; Porter, D.; Tran, T.N.; Love, K.T.; Langer, R.; et al. BCL2A1 is a lineage-specific anti-apoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc. Natl. Acad. Sci. USA 2013, 110, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- Hartman, M.L.; Czyz, M. Pro-Survival Role of MITF in Melanoma. J. Investig. Dermatol. 2015, 135, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Changchien, C.R.; Wu, M.-C.; Tasi, W.-S.; Tang, R.; Chiang, J.-M.; Chen, J.-S.; Huang, S.-F.; Wang, J.-Y.; Yeh, C.-Y. Evaluation of prognosis for malignant rectal gastrointestinal stromal tumor by clinical parameters and immunohistochemical staining. Dis. Colon Rectum 2004, 47, 1922–1929. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Mitra, J.; van den Heuvel, S.; Enders, G.H. S and G2 Phase Roles for Cdk2 Revealed by Inducible Expression of a Dominant-Negative Mutant in Human Cells. Mol. Cell. Biol. 2001, 21, 2755–2766. [Google Scholar] [CrossRef]
- Bačević, K.; Lossaint, G.; Achour, T.N.; Georget, V.; Fisher, D.; Dulić, V. Cdk2 strengthens the intra-S checkpoint and counteracts cell cycle exit induced by DNA damage. Sci. Rep. 2017, 7, 13429. [Google Scholar] [CrossRef]
- Goding, C.R.; Arnheiter, H. MITF—The first 25 years. Genes Dev. 2019, 33, 983–1007. [Google Scholar] [CrossRef]
- Zarei, M.; Giannikou, K.; Du, H.; Liu, H.-J.; Duarte, M.; Johnson, S.; Nassar, A.H.; Widlund, H.R.; Henske, E.P.; Long, H.W.; et al. MITF is a driver oncogene and potential therapeutic target in kidney angiomyolipoma tumors through transcriptional regulation of CYR61. Oncogene 2021, 40, 112–126. [Google Scholar] [CrossRef]
- Kim, N.; Kim, S.; Lee, M.W.; Jeon, H.J.; Ryu, H.; Kim, J.M.; Lee, H.J. MITF Promotes Cell Growth, Migration and Invasion in Clear Cell Renal Cell Carcinoma by Activating the RhoA/YAP Signal Pathway. Cancers 2021, 13, 2920. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Proaño-Pérez, E.; Serrano-Candelas, E.; Mancia, C.; Navinés-Ferrer, A.; Guerrero, M.; Martin, M. SH3BP2 Silencing Increases miRNAs Targeting ETV1 and Microphthalmia-Associated Transcription Factor, Decreasing the Proliferation of Gastrointestinal Stromal Tumors. Cancers 2022, 14, 6198. https://doi.org/10.3390/cancers14246198
Proaño-Pérez E, Serrano-Candelas E, Mancia C, Navinés-Ferrer A, Guerrero M, Martin M. SH3BP2 Silencing Increases miRNAs Targeting ETV1 and Microphthalmia-Associated Transcription Factor, Decreasing the Proliferation of Gastrointestinal Stromal Tumors. Cancers. 2022; 14(24):6198. https://doi.org/10.3390/cancers14246198
Chicago/Turabian StyleProaño-Pérez, Elizabeth, Eva Serrano-Candelas, Cindy Mancia, Arnau Navinés-Ferrer, Mario Guerrero, and Margarita Martin. 2022. "SH3BP2 Silencing Increases miRNAs Targeting ETV1 and Microphthalmia-Associated Transcription Factor, Decreasing the Proliferation of Gastrointestinal Stromal Tumors" Cancers 14, no. 24: 6198. https://doi.org/10.3390/cancers14246198
APA StyleProaño-Pérez, E., Serrano-Candelas, E., Mancia, C., Navinés-Ferrer, A., Guerrero, M., & Martin, M. (2022). SH3BP2 Silencing Increases miRNAs Targeting ETV1 and Microphthalmia-Associated Transcription Factor, Decreasing the Proliferation of Gastrointestinal Stromal Tumors. Cancers, 14(24), 6198. https://doi.org/10.3390/cancers14246198