Incidence of Hereditary Gastric Cancer May Be Much Higher than Reported
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Extraction of the DNA and Preparation of the Exome Library
2.2. DryLab
2.3. Statistical Analyses
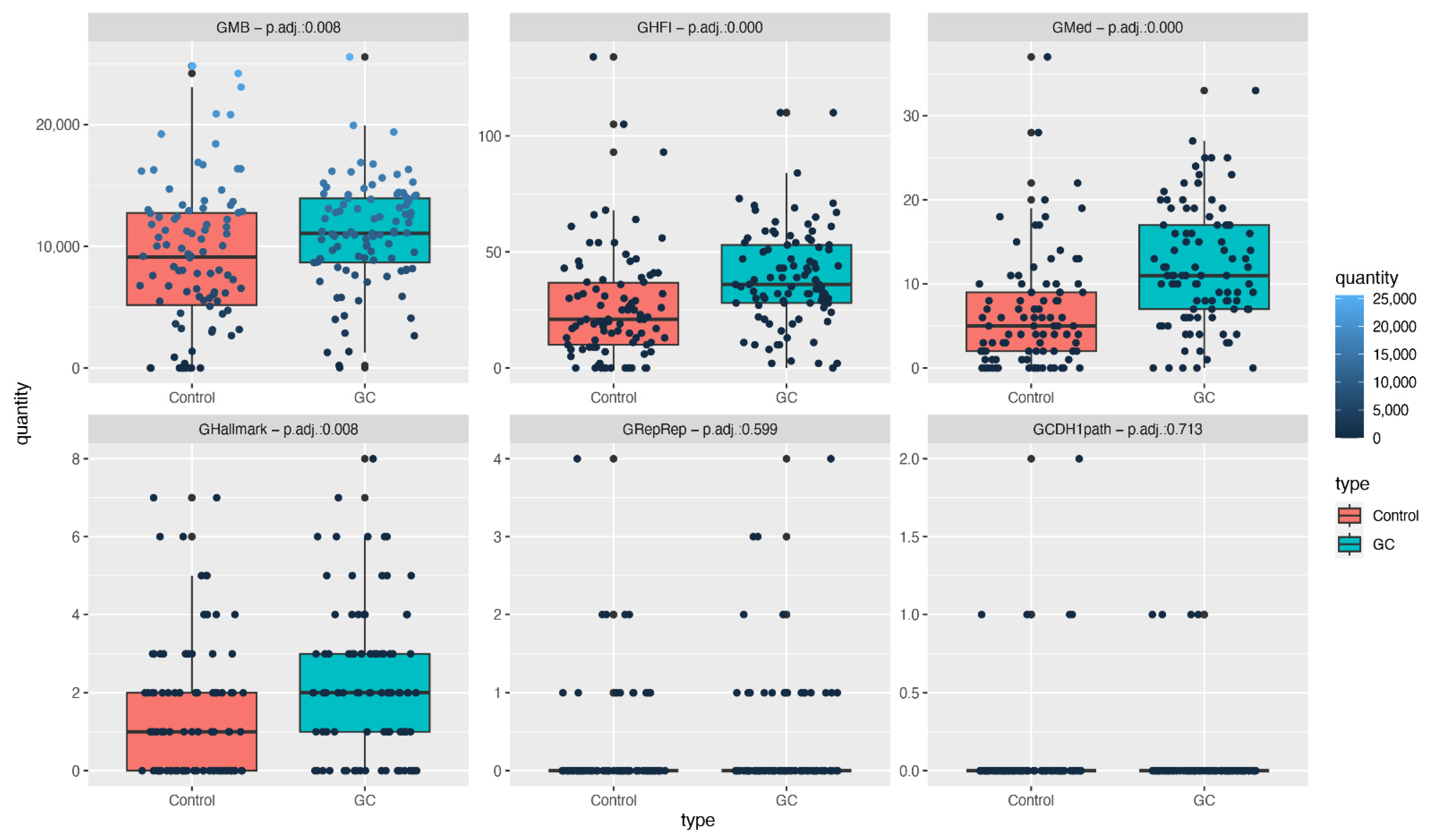
- GMB: the total number of germline mutations.
- GHFI: the number of non-synonymous mutations presenting high functional impact, according to Qing et al. [21].
- GMed: the number of non-synonymous mutations presenting high functional impact among genes related to clinical diseases [21].
- GRepRep: the number of non-synonymous mutations in genes involved in the replication or repair processes (GRepRep), listed in SF Table S2.
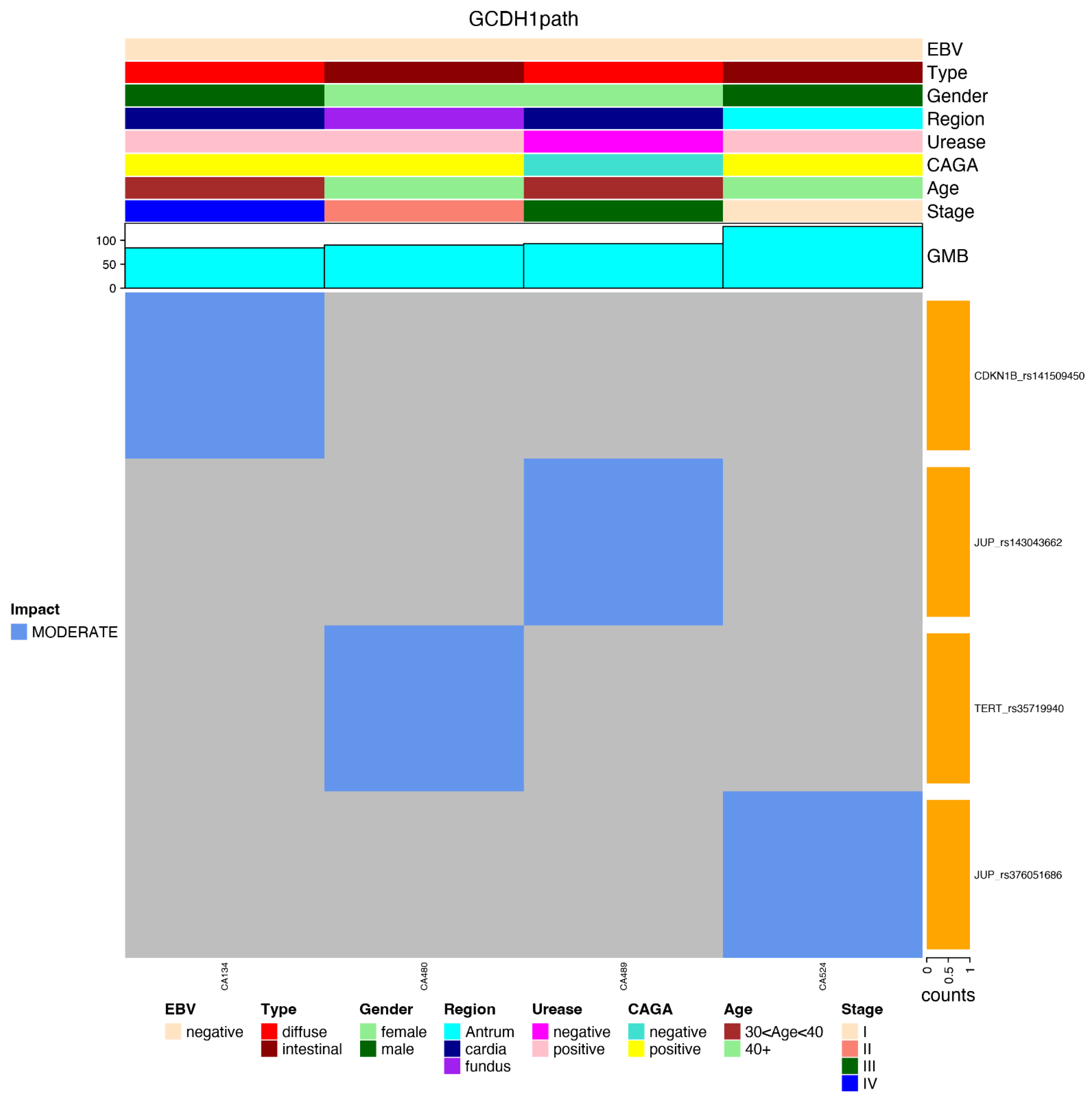
- GCDH1path: the number of germline mutations with high functional impact in genes of the CDH1 pathway (KEEG: N258_257_61) [23].
3. Results
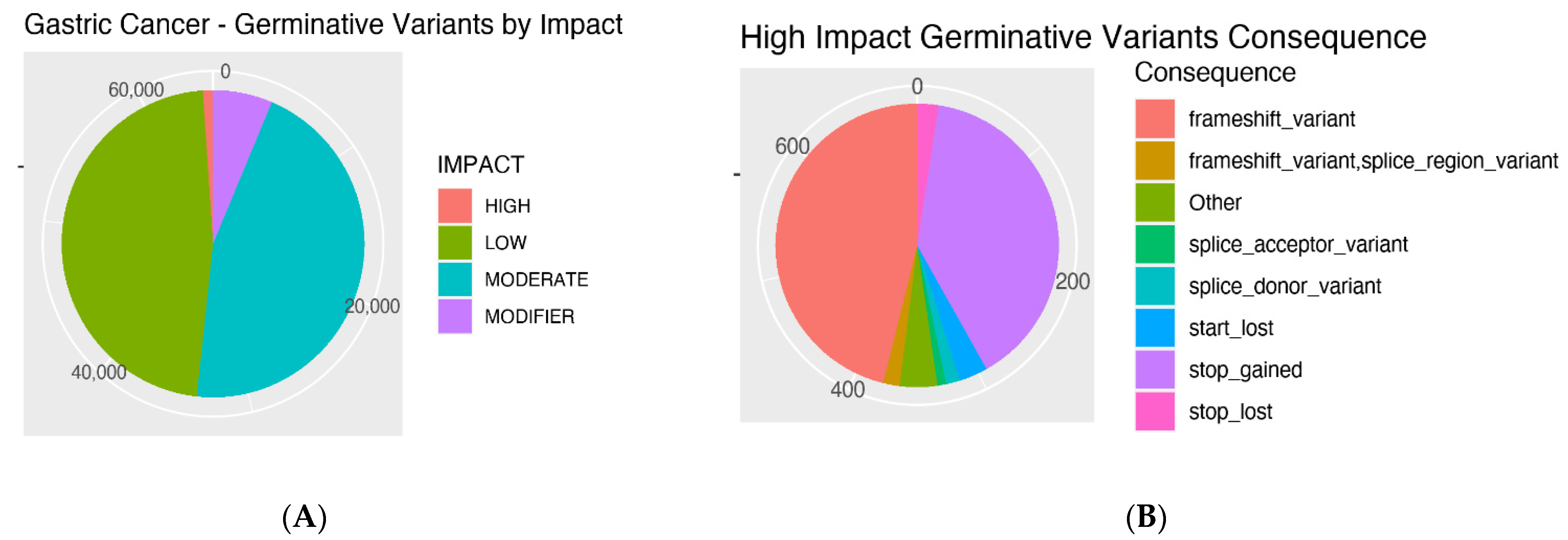
3.1. Mutations’ Impacts and Consequences
3.2. Number of Mutations
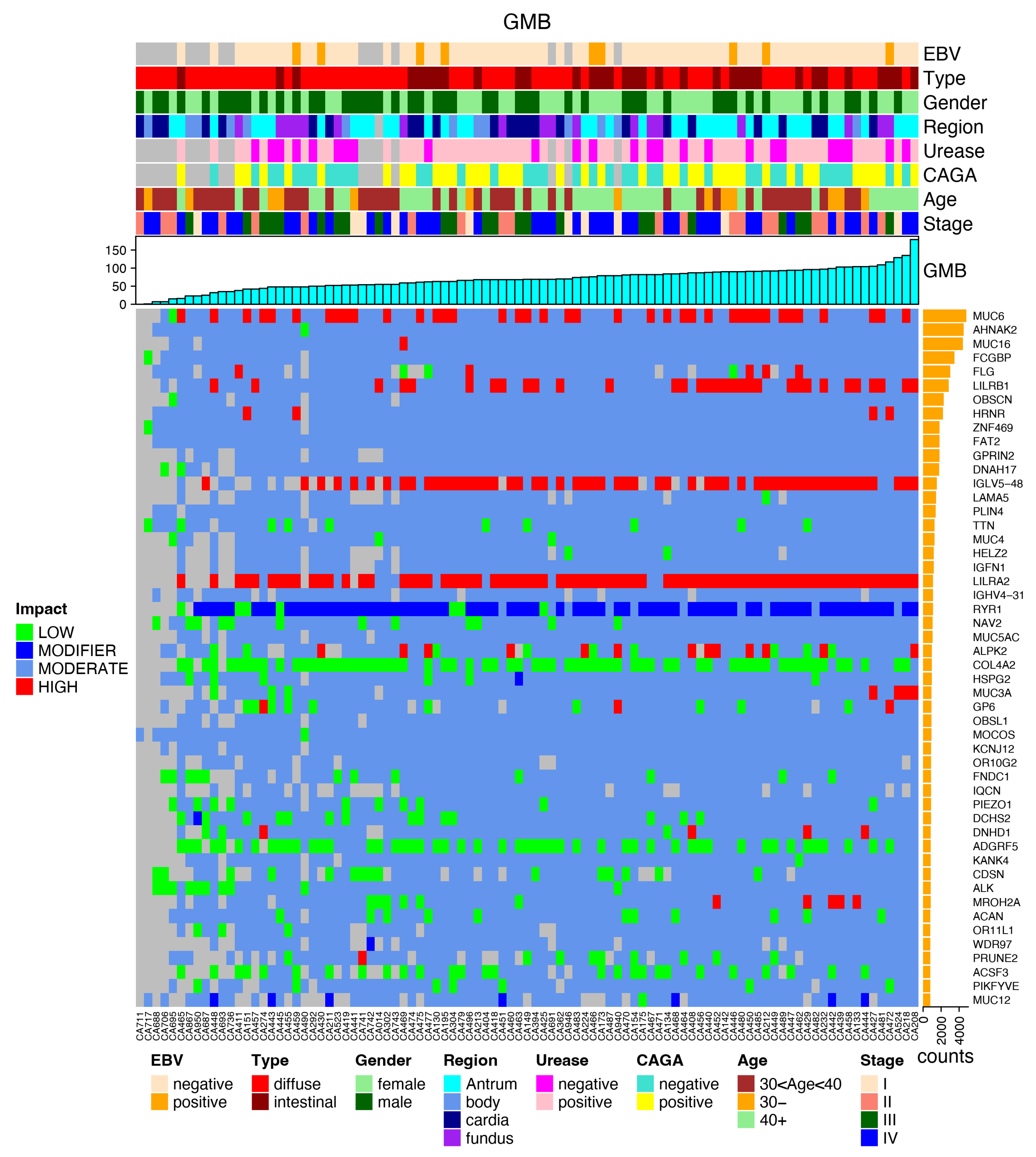
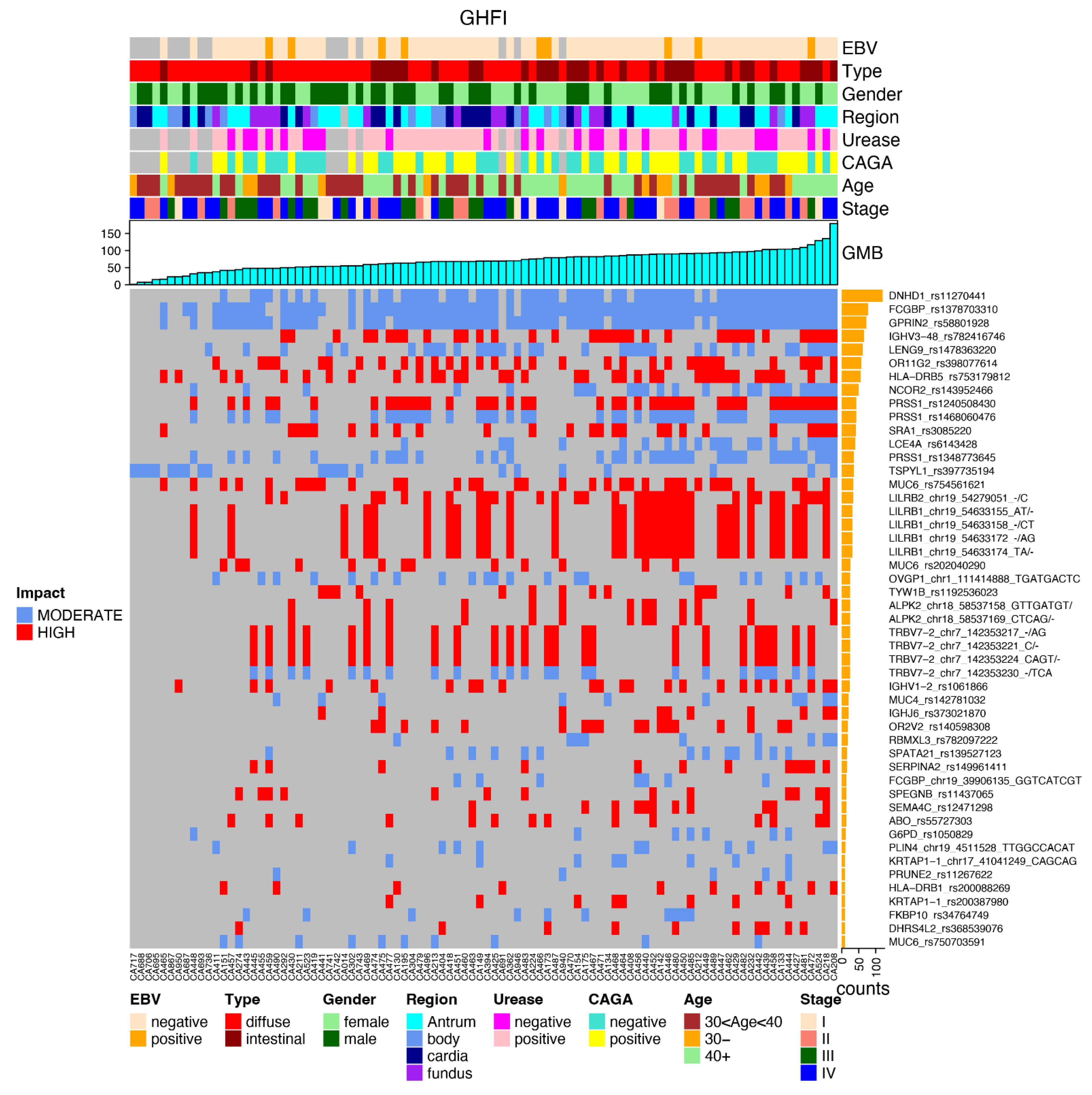
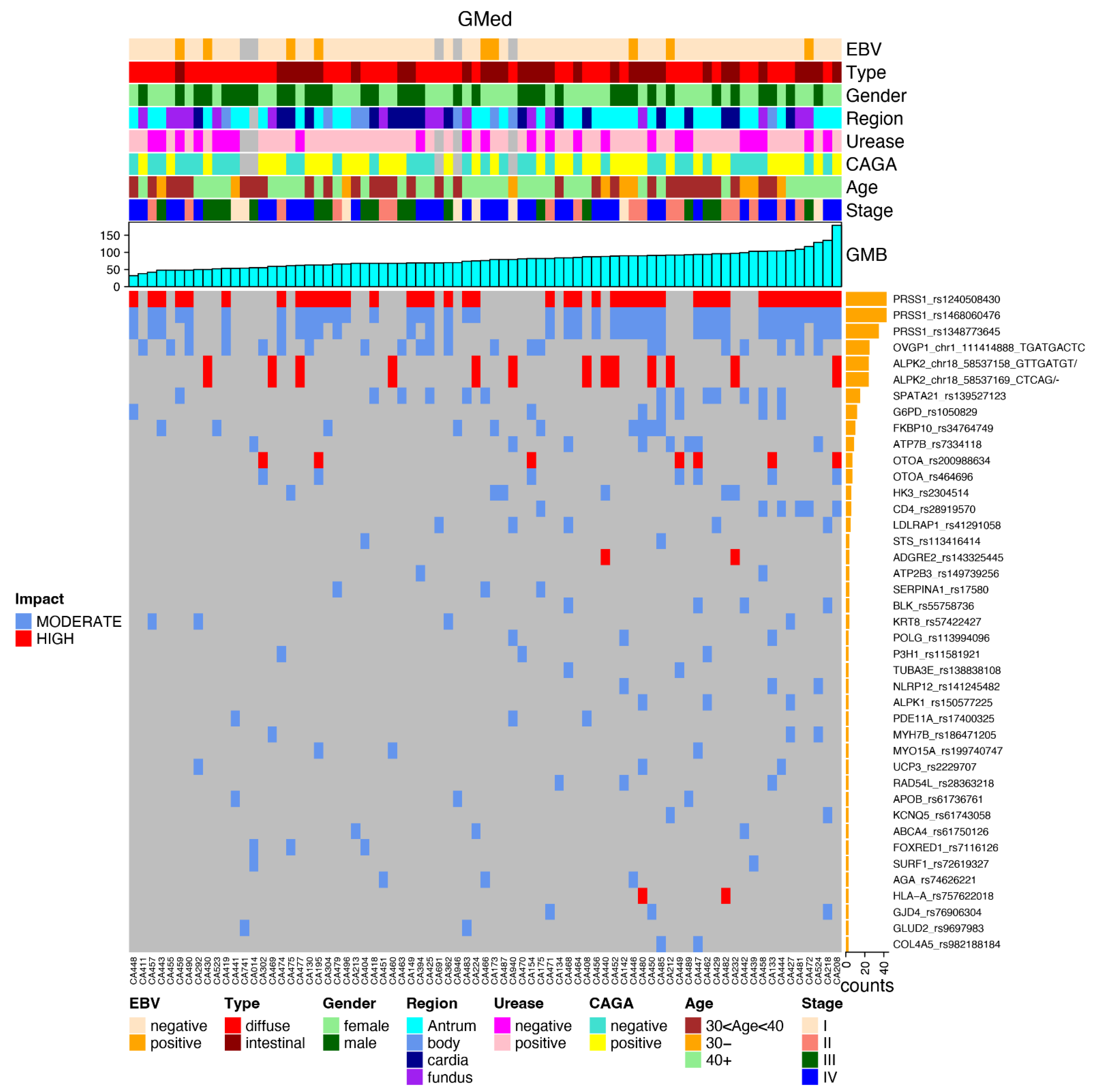
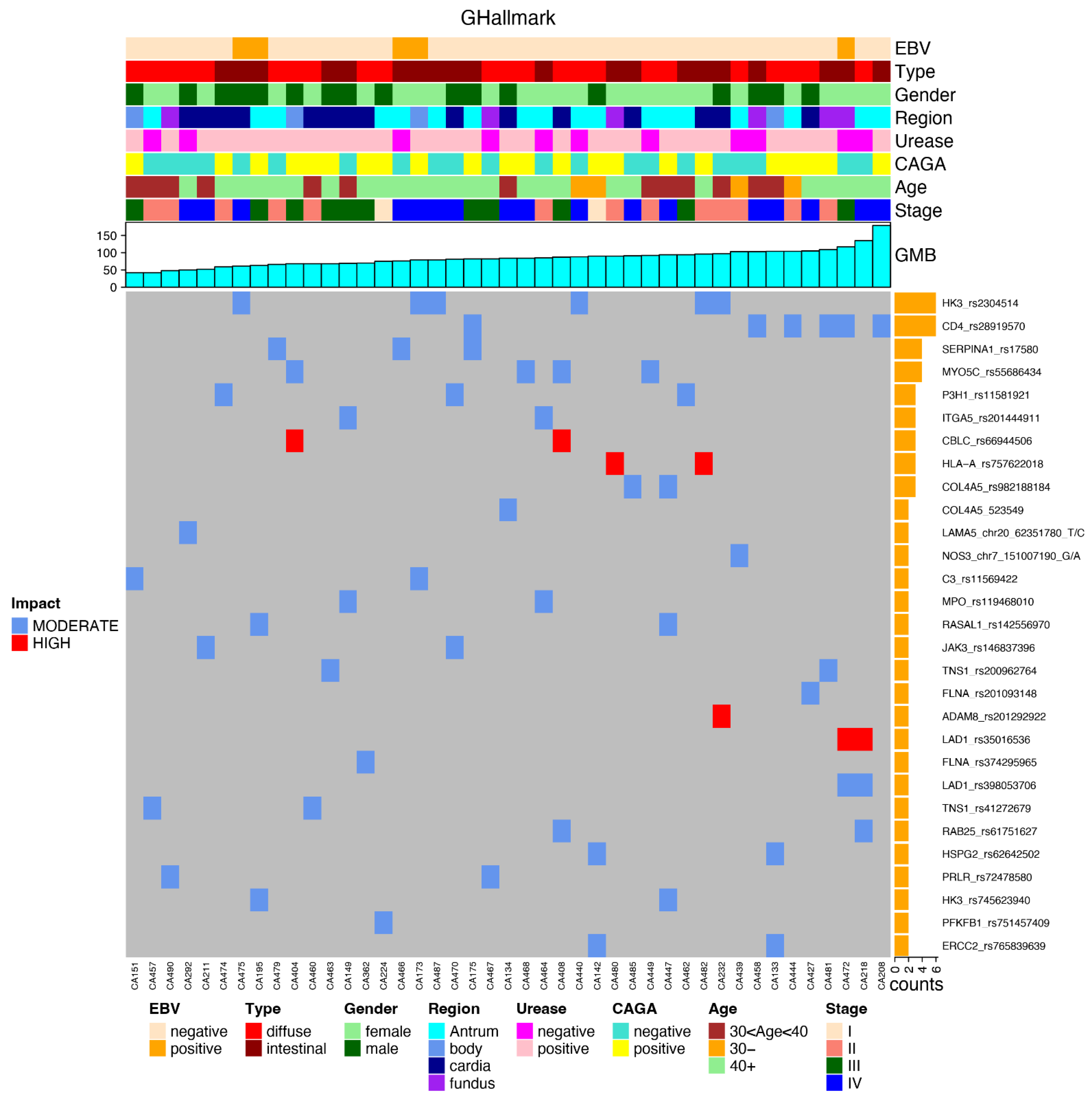
3.3. Most Mutated Genes and Correlations to Clinical Characteristics
3.4. Most Common Variants and Clinical Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ugai, T.; Sasamoto, N.; Lee, H.; Ando, M.; Song, M.; Tamimi, R.M.; Kawachi, I.; Campbell, P.T.; Giovannucci, E.L.; Weiderpass, E.; et al. Is early-onset cancer an emerging global epidemic? Current evidence and future implications. Nat. Rev. Clin. Oncol. 2022, 19, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, F. Familial and hereditary gastric cancer, an overview. Best Pract. Res. Clin. Gastroenterol. 2022, 58–59, 101800. [Google Scholar] [CrossRef] [PubMed]
- Van der Post, R.S.; Oliveira, C.; Guilford, P.; Carneiro, F. Hereditary gastric cancer: What’s new? Update 2013–2018. Fam. Cancer 2019, 18, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Pinheiro, H.; Figueiredo, J.; Seruca, R.; Carneiro, F. Familial gastric cancer: Genetic susceptibility, pathology, and implications for management. Lancet Oncol. 2015, 16, e60–e70. [Google Scholar] [CrossRef]
- Assumpção, P.; Araújo, T.; Khayat, A.; Ishak, G.; Santos, S.; Barra, W.; Acioli, J.F.; Rossi, B. Hereditary gastric cancer: Three rules to reduce missed diagnoses. World J. Gastroenterol. 2020, 26, 1382–1393. [Google Scholar] [CrossRef]
- Polom, K.; Marrelli, D.; D’Ignazio, A.; Roviello, F. Hereditary diffuse gastric cancer: How to look for and how to manage it. Updat. Surg. 2018, 70, 161–166. [Google Scholar] [CrossRef]
- Blair, V.R.; McLeod, M.; Carneiro, F.; Coit, D.G.; D’Addario, J.L.; van Dieren, J.M.; Harris, K.L.; Hoogerbrugge, N.; Oliveira, C.; van der Post, R.S.; et al. Hereditary diffuse gastric cancer: Updated clinical practice guidelines. Lancet Oncol. 2020, 21, e386–e397. [Google Scholar] [CrossRef]
- Yeoh, K.G.; Tan, P. Mapping the genomic diaspora of gastric cancer. Nat. Rev. Cancer 2021, 22, 71–84. [Google Scholar] [CrossRef]
- Porta-Pardo, E.; Valencia, A.; Godzik, A. Understanding oncogenicity of cancer driver genes and mutations in the cancer genomics era. FEBS Lett. 2020, 594, 4233–4246. [Google Scholar] [CrossRef]
- Tomasetti, C.; Li, L.; Vogelstein, B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 2017, 355, 1330–1334. [Google Scholar] [CrossRef]
- Kentsis, A. Why do young people get cancer? Pediatr. Blood Cancer 2020, 67, e28335. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, S. CDH1 (E-Cadherin) Mutation and Gastric Cancer: Genetics, Molecular Mechanisms and Guidelines for Management. Cancer Manag. Res. 2019, 11, 10477–10486. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.F.; Michalski, S.T.; Tondon, R.; Nehoray, B.; Ebrahimzadeh, J.; Hughes, S.K.; Soper, E.R.; Domchek, S.M.; Rustgi, A.K.; Pineda-Alvarez, D.; et al. Loss-of-function variants in CTNNA1 detected on multigene panel testing in individuals with gastric or breast cancer. Genet. Med. 2020, 22, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Hansford, S.; Kaurah, P.; Li-Chang, H.; Woo, M.; Senz, J.; Pinheiro, H.; Schrader, K.A.; Schaeffer, D.F.; Shumansky, K.; Zogopoulos, G.; et al. Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA Oncol. 2015, 1, 23–32. [Google Scholar] [CrossRef]
- Coudert, M.; Drouet, Y.; Delhomelle, H.; Svrcek, M.; Benusiglio, P.R.; Coulet, F.; Clark, D.F.; Katona, B.W.; van Hest, L.P.; van der Kolk, L.; et al. First estimates of diffuse gastric cancer risks for carriers of CTNNA1 germline pathogenic variants. J. Med. Genet. 2022, 59, 1189–1195. [Google Scholar] [CrossRef]
- Donner, I.; Kiviluoto, T.; Ristimäki, A.; Aaltonen, L.A.; Vahteristo, P. Exome sequencing reveals three novel candidate pre-disposition genes for diffuse gastric cancer. Fam. Cancer 2015, 14, 241–246. [Google Scholar] [CrossRef]
- Pocurull, A.; Herrera-Pariente, C.; Carballal, S.; Llach, J.; Sánchez, A.; Carot, L.; Botargues, J.M.; Cuatrecasas, M.; Ocaña, T.; Balaguer, F.; et al. Clinical, Molecular and Genetic Characteristics of Early Onset Gastric Cancer: Analysis of a Large Multicenter Study. Cancers 2021, 13, 3132. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Qing, T.; Mohsen, H.; Marczyk, M.; Ye, Y.; O’Meara, T.; Zhao, H.; Townsend, J.P.; Gerstein, M.; Hatzis, C.; Kluger, Y.; et al. Germline variant burden in cancer genes correlates with age at diagnosis and somatic mutation burden. Nat. Commun. 2020, 11, 2438. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, P.; Ranjbar, R. DNA microarray technology and bioinformatic web services. Acta Microbiol. Immunol. Hung. 2018, 66, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Li, J. Gastric Cancer in Young Adults: A Different Clinical Entity from Carcinogenesis to Prognosis. Gastroenterol. Res. Pract. 2020, 2020, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Chen, S.; Wu, W.; Kuo, Z.C.; Wei, Z.; Meng, S.; Chen, C.; Zhang, C.; He, Y. Gastric cancer in young patients: A separate entity with aggressive features and poor prognosis. J. Cancer Res. Clin. Oncol. 2020, 146, 2937–2947. [Google Scholar] [CrossRef]
- Kumar, S.; Long, J.M.; Ginsberg, G.G.; Katona, B. Role of endoscopy in the management of hereditary diffuse gastric cancer syndrome. World J. Gastroenterol. 2019, 25, 2878–2886. [Google Scholar] [CrossRef]
- Pantelis, D.; Hüneburg, R.; Adam, R.; Holzapfel, S.; Gevensleben, H.; Nattermann, J.; Strassburg, C.P.; Aretz, S.; Kalff, J.C. Prophy-lactic total gastrectomy in the management of hereditary tumor syndromes. Int. J. Color. Dis. 2016, 31, 1825–1833. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, M.; Feng, Y.; Bu, Z.; Li, Z.; Jia, S.; Ji, J. Germline mutations in hereditary diffuse gastric cancer. Chin. J. Cancer Res. 2018, 30, 122–130. [Google Scholar] [CrossRef]
- Stillman, M.D.; Kusche, N.; Toledano, S.; Hilfrank, K.J.; Yoon, C.; Gabre, J.T.; Rustgi, S.D.; Hur, C.; Kastrinos, S.; Ryeom, S.W.; et al. Short and long-term outcomes of prophylactic total gastrectomy in 54 consecutive individuals with germline pathogen-ic mutations in the CDH1 gene. J. Surg. Oncol. 2022, 126, 1413–1422. [Google Scholar] [CrossRef]
- Cosma, L.; Schlosser, S.; Tews, H.C.; Müller, M.; Kandulski, A. Hereditary Diffuse Gastric Cancer: Molecular Genetics, Biolog-ical Mechanisms and Current Therapeutic Approaches. Int. J. Mol. Sci. 2022, 23, 7821. [Google Scholar] [CrossRef]
- Saeki, N.; Ono, H.; Sakamoto, H.; Yoshida, T. Genetic factors related to gastric cancer susceptibility identified using a genome-wide association study. Cancer Sci. 2012, 104, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Zhu, M.; Ding, Y.; Yang, M.; Wang, M.; Li, G.; Ren, C.; Huang, T.; Yang, W.; He, B.; et al. Meta-analysis of genome-wide as-sociation studies and functional assays decipher susceptibility genes for gastric cancer in Chinese populations. Gut 2019, 69, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Park, B.; Yang, S.; Lee, J.; Woo, H.D.; Choi, I.J.; Kim, Y.W.; Ryu, K.W.; Kim, Y.; Kim, J. Genome-Wide Association of Genetic Varia-tion in the PSCA Gene with Gastric Cancer Susceptibility in a Korean Population. Cancer Res. Treat. 2018, 51, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Scherer, A.; Christensen, G.B. Concepts and Relevance of Genome-Wide Association Studies. Sci. Prog. 2016, 99, 59–67. [Google Scholar] [CrossRef]
- Qing, T.; Mohsen, H.; Cannataro, V.L.; Marczyk, M.; Rozenblit, M.; Foldi, J.; Murray, M.; Townsend, J.P.; Kluger, Y.; Gerstein, M.; et al. Cancer Relevance of Human Genes. J. Natl. Cancer Inst. 2022, 114, 988–995. [Google Scholar] [CrossRef]
- Knudson, A.G. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef]
- Chernoff, J. The two-hit theory hits. Mol. Biol. Cell 2021, 32, 1. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. The Path to Cancer–Three Strikes and You’re Out. N. Engl. J. Med. 2015, 373, 1895–1898. [Google Scholar] [CrossRef]
- Herrera-Pariente, C.; Capó-García, R.; Díaz-Gay, M.; Carballal, S.; Muñoz, J.; Llach, J.; Ariadna, S.; Bonjoch, L.; Ar-nau-Collell, C.; de Lima, Y.S.; et al. Identification of New Genes Involved in Germline Predisposition to Early-Onset Gastric Cancer. Int. J. Mol. Sci. 2021, 22, 1310. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, J.; Zhuang, Y.; Wu, H. Identification of the potential molecular mechanism and driving mutations in the pathogenesis of familial intestinal gastric cancer by whole exome sequencing. Oncol. Rep. 2018, 40, 2316–2324. [Google Scholar] [CrossRef] [PubMed]
- Shlien, A.; Campbell, B.B.; de Borja, R.; Alexandrov, L.B.; Merico, D.; Wedge, D.; Loo, P.V.; Tarpey, P.S.; Coupland, P.; Behjati, S.; et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat. Genet. 2015, 47, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Srinivasan, S.; Korkin, D. Enriching Human Interactome with Functional Mutations to Detect High-Impact Net-work Modules Underlying Complex Diseases. Genes 2019, 10, 933. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2015, 44, D862–D868. [Google Scholar] [CrossRef] [PubMed]
- Gall, T.M.H.; Frampton, A.E. Gene of the month: E-cadherin (CDH1). J. Clin. Pathol. 2013, 66, 928–932. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, J.; Chai, K.; Ying, X.; Zhou, B.P. The Role of Snail in EMT and Tumorigenesis. Curr. Cancer Drug Targets 2013, 13, 963–972. [Google Scholar] [CrossRef]
- Kraft, I.L.; Godley, L.A. Identifying potential germline variants from sequencing hematopoietic malignancies. Hematology 2020, 2020, 219–227. [Google Scholar] [CrossRef]
- Oak, N.; Cherniack, A.D.; Mashl, R.J.; Network, T.A.; Hirsch, F.R.; Ding, L.; Beroukhim, R.; Gümüş, Z.H.; Plon, S.E.; Huang, K. Ances-try-specific predisposing germline variants in cancer. Genome Med. 2020, 12, 51. [Google Scholar] [CrossRef]
- Ribeiro-Dos-Santos, A.M.; Vidal, A.F.; Vinasco-Sandoval, T.; Guerreiro, J.; Santos, S.; De Souza, S.J. Exome Sequencing of Native Populations from the Amazon Reveals Patterns on the Peopling of South America. Front. Genet. 2020, 11, 548507. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, D.; Huangfu, S.; Zhou, J.; Wang, W.; Liu, S.; Tang, H.; Pan, J.; Pan, Y. DNA Repair and Replication-Related Gene Signature Based on Tumor Mutation Burden Reveals Prognostic and Immunotherapy Response in Gastric Cancer. J. Oncol. 2022, 2022, 6469523. [Google Scholar] [CrossRef]


| GMB | GHFI | GMed | GHallmark | GRepRep | GCDH1path | ||
|---|---|---|---|---|---|---|---|
| Gender | adj.p | 0.12640 | 0.12640 | 0.47426 | 0.99146 | 0.12640 | 0.99146 |
| H. type | adj.p | 0.01328 | 0.09556 | 0.57447 | 0.10885 | 0.57447 | 0.57447 |
| Stage | adj.p | 0.89760 | 0.8976 | 0.89760 | 0.89760 | 0.03529 | 0.89760 |
| Age | adj.p | 0.32287 | 0.44947 | 0.7559 | 0.05163 | 0.51499 | 0.75590 |
| PCR.urease | adj.p | 0.88930 | 0.88930 | 0.88930 | 0.88930 | 0.88930 | 0.88930 |
| CAGA | adj.p | 0.99433 | 0.99433 | 0.99433 | 0.99433 | 0.99433 | 0.99433 |
| EBV | adj.p | 0.87695 | 0.87695 | 0.90029 | 0.87695 | 0.87695 | 0.87695 |
| Region | adj.p | 0.24500 | 0.16219 | 0.24500 | 0.24500 | 0.6612 | 0.66120 |
| EBV_urease | adj.p | 0.93363 | 0.93363 | 0.93363 | 0.93363 | 0.93363 | 0.93363 |
| EBV_CAGA | adj.p | 0.99597 | 0.99597 | 0.99597 | 0.99597 | 0.99597 | 0.99597 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Assumpção, P.B.; de Assumpção, P.P.; Moreira, F.C.; Ribeiro-dos-Santos, Â.; Vidal, A.F.; Magalhães, L.; Khayat, A.S.; Ribeiro-dos-Santos, A.M.; Cavalcante, G.C.; Pereira, A.L.; et al. Incidence of Hereditary Gastric Cancer May Be Much Higher than Reported. Cancers 2022, 14, 6125. https://doi.org/10.3390/cancers14246125
de Assumpção PB, de Assumpção PP, Moreira FC, Ribeiro-dos-Santos Â, Vidal AF, Magalhães L, Khayat AS, Ribeiro-dos-Santos AM, Cavalcante GC, Pereira AL, et al. Incidence of Hereditary Gastric Cancer May Be Much Higher than Reported. Cancers. 2022; 14(24):6125. https://doi.org/10.3390/cancers14246125
Chicago/Turabian Stylede Assumpção, Paula Baraúna, Paulo Pimentel de Assumpção, Fabiano Cordeiro Moreira, Ândrea Ribeiro-dos-Santos, Amanda F. Vidal, Leandro Magalhães, André Salim Khayat, André Maurício Ribeiro-dos-Santos, Giovanna C. Cavalcante, Adenilson Leão Pereira, and et al. 2022. "Incidence of Hereditary Gastric Cancer May Be Much Higher than Reported" Cancers 14, no. 24: 6125. https://doi.org/10.3390/cancers14246125
APA Stylede Assumpção, P. B., de Assumpção, P. P., Moreira, F. C., Ribeiro-dos-Santos, Â., Vidal, A. F., Magalhães, L., Khayat, A. S., Ribeiro-dos-Santos, A. M., Cavalcante, G. C., Pereira, A. L., Medeiros, I., de Souza, S. J., Burbano, R. M. R., de Souza, J. E. S., & Dos Santos, S. E. B. (2022). Incidence of Hereditary Gastric Cancer May Be Much Higher than Reported. Cancers, 14(24), 6125. https://doi.org/10.3390/cancers14246125







