Oral Microbiome in Nonsmoker Patients with Oral Cavity Squamous Cell Carcinoma, Defined by Metagenomic Shotgun Sequencing
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Recruitment of Human Subjects for OC-SCC Cases and Controls
2.2. Detection of Bacterial DNA Sequences in Mouthwash Samples of OC-SCC Patients and Control Patients Using MSS
3. Results
3.1. Patient and Tumor Characteristics
3.2. Alpha and Beta Diversity
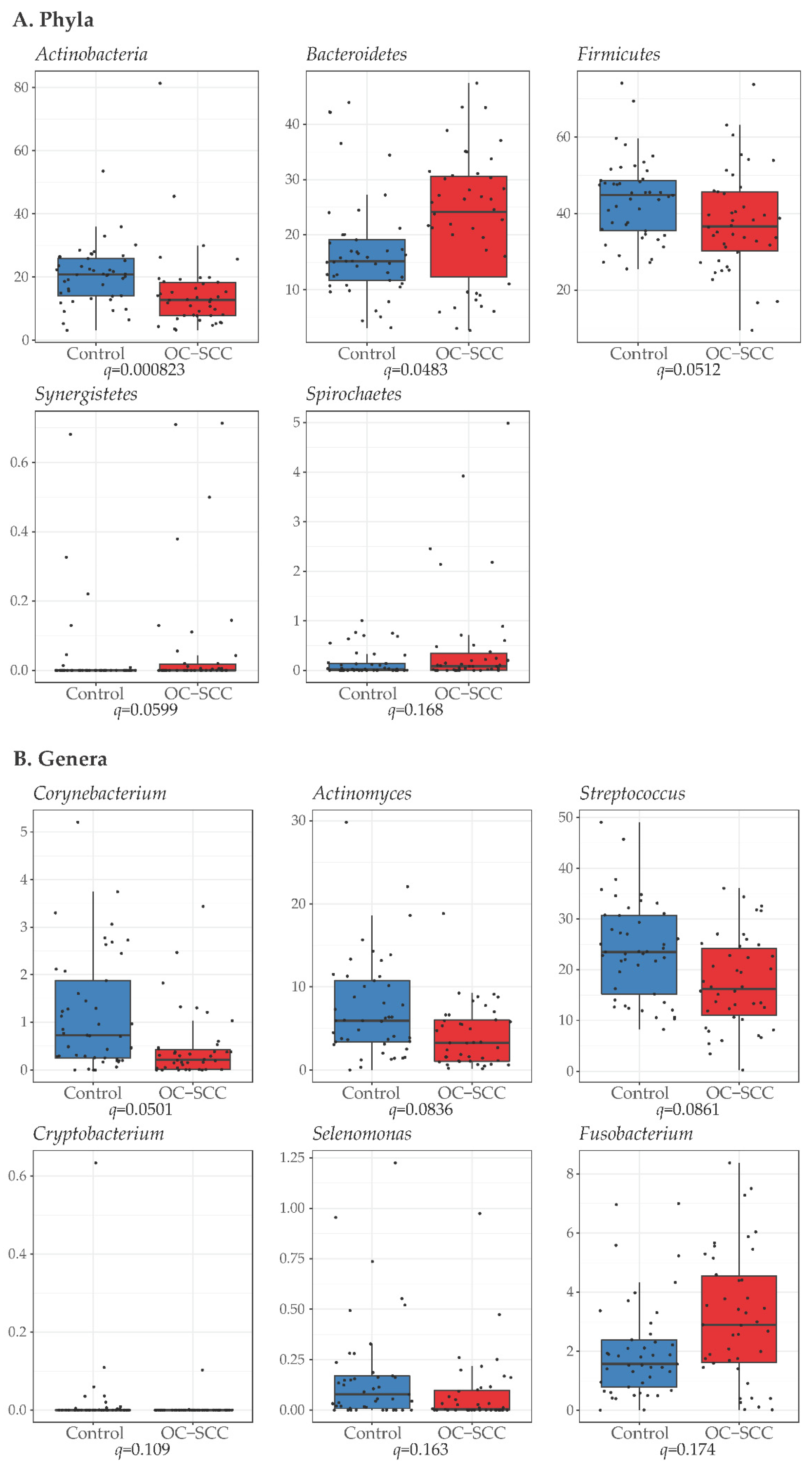
3.3. Differences in Bacteria Phyla, Genera and Species between Cases and Controls
3.4. Functional Prediction of Oral Microbiome Related to the Development of Oral Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Viale, P.H. The American Cancer Society’s Facts & Figures: 2020 Edition. J. Adv. Pract. Oncol. 2020, 11, 135–136. [Google Scholar] [PubMed]
- Scully, C.; Bagan, J. Oral squamous cell carcinoma overview. Oral Oncol. 2009, 45, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Scully, C.; Field, J.K.; Tanzawa, H. Genetic aberrations in oral or head and neck squamous cell carcinoma (SCCHN): 1. Carcinogen metabolism, DNA repair and cell cycle control. Oral Oncol. 2000, 36, 256–263. [Google Scholar] [CrossRef]
- Gondivkar, S.M.; Gondivkar, R.S.; Gadbail, A.R.; Chole, R.; Mankar, M.; Yuwanati, M. Chronic periodontitis and the risk of head and neck squamous cell carcinoma: Facts and figures. Exp. Oncol. 2013, 35, 163–167. [Google Scholar] [PubMed]
- Markman, M. Risk Factors for Oral Cancer. Available online: http://www.cancercenter.com/oral-cancer/risk-factors/ (accessed on 12 September 2022).
- Cancer Stat Facts: Oral Cavity and Pharynx Cancer. Available online: https://seer.cancer.gov/statfacts/html/oralcav.html (accessed on 12 September 2022).
- Wang, L.; Ganly, I. The oral microbiome and oral cancer. Clin. Lab. Med. 2014, 34, 711–719. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819867354. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]
- Saxena, R.; Prasoodanan, P.K.V.; Gupta, S.V.; Gupta, S.; Waiker, P.; Samaiya, A.; Sharma, A.K.; Sharma, V.K. Assessing the Effect of Smokeless Tobacco Consumption on Oral Microbiome in Healthy and Oral Cancer Patients. Front. Cell. Infect. Microbiol. 2022, 12, 841465. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, D.; Menon, R.K.; Wie, C.C.; Banerjee, M.; Panda, S.; Mandal, D.; Behera, P.K.; Roychoudhury, S.; Kheur, S.; Botelho, M.G.; et al. Differences in the bacteriome of swab, saliva, and tissue biopsies in oral cancer. Sci. Rep. 2021, 11, 1181. [Google Scholar] [CrossRef]
- Su, S.C.; Chang, L.C.; Huang, H.D.; Peng, C.Y.; Chuang, C.Y.; Chen, Y.T.; Lu, M.Y.; Chiu, Y.W.; Chen, P.Y.; Yang, S.F. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis 2021, 42, 127–135. [Google Scholar] [CrossRef]
- Yang, K.; Wang, Y.; Zhang, S.; Zhang, D.; Hu, L.; Zhao, T.; Zheng, H. Oral Microbiota Analysis of Tissue Pairs and Saliva Samples From Patients With Oral Squamous Cell Carcinoma—A Pilot Study. Front. Microbiol. 2021, 12, 719601. [Google Scholar] [CrossRef] [PubMed]
- Ganly, I.; Yang, L.; Giese, R.A.; Hao, Y.; Nossa, C.W.; Morris, L.G.T.; Rosenthal, M.; Migliacci, J.; Kelly, D.; Tseng, W.; et al. Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus. Int. J. Cancer 2019, 145, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Yeh, Y.M.; Yu, H.Y.; Chin, C.Y.; Hsu, C.W.; Liu, H.; Huang, P.J.; Hu, S.N.; Liao, C.T.; Chang, K.P.; et al. Oral Microbiota Community Dynamics Associated With Oral Squamous Cell Carcinoma Staging. Front. Microbiol. 2018, 9, 862. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chu, M.; Huang, Z.; Yang, X.; Ran, S.; Hu, B.; Zhang, C.; Liang, J. Variations in oral microbiota associated with oral cancer. Sci. Rep. 2017, 7, 11773. [Google Scholar] [CrossRef]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. mSphere 2021, 6, e01202-20. [Google Scholar] [CrossRef] [PubMed]
- Fouhy, F.; Clooney, A.G.; Stanton, C.; Claesson, M.J.; Cotter, P.D. 16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016, 16, 123. [Google Scholar] [CrossRef] [PubMed]
- Pei, A.Y.; Oberdorf, W.E.; Nossa, C.W.; Agarwal, A.; Chokshi, P.; Gerz, E.A.; Jin, Z.; Lee, P.; Yang, L.; Poles, M.; et al. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl. Environ. Microbiol. 2010, 76, 3886–3897. [Google Scholar] [CrossRef]
- Hassler, H.B.; Probert, B.; Moore, C.; Lawson, E.; Jackson, R.W.; Russell, B.T.; Richards, V.P. Phylogenies of the 16S rRNA gene and its hypervariable regions lack concordance with core genome phylogenies. Microbiome 2022, 10, 104. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Bowman, J.S.; Ducklow, H.W. Microbial Communities Can Be Described by Metabolic Structure: A General Framework and Application to a Seasonally Variable, Depth-Stratified Microbial Community from the Coastal West Antarctic Peninsula. PLoS ONE 2015, 10, e0135868. [Google Scholar] [CrossRef]
- Noll, K.M.; Lapierre, P.; Gogarten, J.P.; Nanavati, D.M. Evolution of mal ABC transporter operons in the Thermococcales and Thermotogales. BMC Evol. Biol. 2008, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.R.; Pacocha, S.; Pharino, C.; Klepac-Ceraj, V.; Hunt, D.E.; Benoit, J.; Sarma-Rupavtarm, R.; Distel, D.L.; Polz, M.F. Genotypic diversity within a natural coastal bacterioplankton population. Science 2005, 307, 1311–1313. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Makino, K.; Ohnishi, M.; Kurokawa, K.; Ishii, K.; Yokoyama, K.; Han, C.G.; Ohtsubo, E.; Nakayama, K.; Murata, T.; et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001, 8, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Kudva, I.T.; Evans, P.S.; Perna, N.T.; Barrett, T.J.; Ausubel, F.M.; Blattner, F.R.; Calderwood, S.B. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol. 2002, 184, 1873–1879. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ivanova, N.; Sorokin, A.; Anderson, I.; Galleron, N.; Candelon, B.; Kapatral, V.; Bhattacharyya, A.; Reznik, G.; Mikhailova, N.; Lapidus, A.; et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 2003, 423, 87–91. [Google Scholar] [CrossRef]
- Turnbull, P.C.B. Bacillus. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Pakbin, B.; Bruck, W.M.; Rossen, J.W.A. Virulence Factors of Enteric Pathogenic Escherichia coli: A Review. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef]
- Xu, W.; Chen, T.; Pei, Y.; Guo, H.; Li, Z.; Yang, Y.; Zhang, F.; Yu, J.; Li, X.; Yang, Y.; et al. Characterization of Shallow Whole-Metagenome Shotgun Sequencing as a High-Accuracy and Low-Cost Method by Complicated Mock Microbiomes. Front. Microbiol. 2021, 12, 678319. [Google Scholar] [CrossRef] [PubMed]
- Lamoureux, C.; Surgers, L.; Fihman, V.; Gricourt, G.; Demontant, V.; Trawinski, E.; N’Debi, M.; Gomart, C.; Royer, G.; Launay, N.; et al. Prospective Comparison Between Shotgun Metagenomics and Sanger Sequencing of the 16S rRNA Gene for the Etiological Diagnosis of Infections. Front. Microbiol. 2022, 13, 761873. [Google Scholar] [CrossRef]
- Peterson, D.; Bonham, K.S.; Rowland, S.; Pattanayak, C.W.; Consortium, R.; Klepac-Ceraj, V. Comparative Analysis of 16S rRNA Gene and Metagenome Sequencing in Pediatric Gut Microbiomes. Front. Microbiol. 2021, 12, 670336. [Google Scholar] [CrossRef]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef] [PubMed]
- Laudadio, I.; Fulci, V.; Palone, F.; Stronati, L.; Cucchiara, S.; Carissimi, C. Quantitative Assessment of Shotgun Metagenomics and 16S rDNA Amplicon Sequencing in the Study of Human Gut Microbiome. OMICS 2018, 22, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Hillmann, B.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Zhu, Q.; Gohl, D.M.; Beckman, K.B.; Knight, R.; Knights, D. Evaluating the Information Content of Shallow Shotgun Metagenomics. mSystems 2018, 3, e00069-18. [Google Scholar] [CrossRef] [PubMed]
- Ganly, I.; Pei, Z.; Hao, Y.; Ma, Y.; Rosenthal, M.; Wu, Z.; Migliacci, J.; Huang, B.; Katabi, N.; Tseng, W.; et al. Case control study comparing the HPV genome in patients with oral cavity squamous cell carcinoma to normal patients using metagenomic shotgun sequencing. Sci. Rep. 2021, 11, 3867. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef]
- Suzek, B.E.; Huang, H.; McGarvey, P.; Mazumder, R.; Wu, C.H. UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics 2007, 23, 1282–1288. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Pasolli, E.; Schiffer, L.; Manghi, P.; Renson, A.; Obenchain, V.; Truong, D.T.; Beghini, F.; Malik, F.; Ramos, M.; Dowd, J.B.; et al. Accessible, curated metagenomic data through ExperimentHub. Nat. Methods 2017, 14, 1023–1024. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, F.; Poco, S.E.; Ikeda, T.; Sato, M.; Kalfas, S.; Sundqvist, G.; Hoshino, E. Cryptobacterium curtum gen. nov., sp. nov., a new genus of gram-positive anaerobic rod isolated from human oral cavities. Int. J. Syst. Bacteriol. 1999, 49 Pt 3, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, H.; Sato, N.; Djais, A.; Hoshino, E. Degradation of arginine by Slackia exigua ATCC 700122 and Cryptobacterium curtum ATCC 700683. Oral. Microbiol. Immunol. 2006, 21, 381–384. [Google Scholar] [CrossRef]
- Gursoy, U.K.; Pollanen, M.; Kononen, E.; Uitto, V.J. Biofilm formation enhances the oxygen tolerance and invasiveness of Fusobacterium nucleatum in an oral mucosa culture model. J. Periodontol. 2010, 81, 1084–1091. [Google Scholar] [CrossRef]
- Hao, Y.; Karaoz, U.; Yang, L.; Yachimski, P.S.; Tseng, W.; Nossa, C.W.; Ye, W.; Tseng, M.; Poles, M.; Francois, F.; et al. Progressive dysbiosis of human orodigestive microbiota along the sequence of gastroesophageal reflux, Barrett’s esophagus and esophageal adenocarcinoma. Int. J. Cancer 2022, 151, 1703–1716. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yang, L.; Hao, Y.; Zhou, B.; Hu, J.; Yang, Y.; Bedi, S.; Sanichar, N.G.; Cheng, C.; Perez-Perez, G.; et al. Oral and gastric microbiome in relation to gastric intestinal metaplasia. Int. J. Cancer 2022, 150, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Binder Gallimidi, A.; Fischman, S.; Revach, B.; Bulvik, R.; Maliutina, A.; Rubinstein, A.M.; Nussbaum, G.; Elkin, M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 2015, 6, 22613–22623. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Baik, J.E.; Lagana, S.M.; Han, R.P.; Raab, W.J.; Sahoo, D.; Dalerba, P.; Wang, T.C.; Han, Y.W. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep. 2019, 20, e47638. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Zhang, Y.; Lu, Z.; Zhang, S.; Pan, Y. Fusobacterium nucleatum Caused DNA Damage and Promoted Cell Proliferation by the Ku70/p53 Pathway in Oral Cancer Cells. DNA Cell Biol. 2020, 39, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, P.; Guo, Y.; Liang, X.; Li, Y.; Ding, S. Helicobacter pylori-Induced DNA Damage Is a Potential Driver for Human Gastric Cancer AGS Cells. DNA Cell Biol. 2019, 38, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; He, F.; Kwang, J.; Engelward, B.P.; Chow, V.T. Pneumococcal Pneumolysin Induces DNA Damage and Cell Cycle Arrest. Sci. Rep. 2016, 6, 22972. [Google Scholar] [CrossRef]
- McIlvanna, E.; Linden, G.J.; Craig, S.G.; Lundy, F.T.; James, J.A. Fusobacterium nucleatum and oral cancer: A critical review. BMC Cancer 2021, 21, 1212. [Google Scholar] [CrossRef]
- Leon-Del-Rio, A. Biotin in metabolism, gene expression, and human disease. J. Inherit. Metab. Dis. 2019, 42, 647–654. [Google Scholar] [CrossRef]
- Yang, H.T.; Chao, P.C.; Yin, M.C. Riboflavin at high doses enhances lung cancer cell proliferation, invasion, and migration. J. Food Sci. 2013, 78, H343–H349. [Google Scholar] [CrossRef]
- Kaur, P.; Nagar, S.; Bhagwat, M.; Uddin, M.; Zhu, Y.; Vancurova, I.; Vancura, A. Activated heme synthesis regulates glycolysis and oxidative metabolism in breast and ovarian cancer cells. PLoS ONE 2021, 16, e0260400. [Google Scholar] [CrossRef]
- Zhou, X.; Kandalai, S.; Hossain, F.; Zheng, Q. Tumor microbiome metabolism: A game changer in cancer development and therapy. Front Oncol. 2022, 12, 933407. [Google Scholar] [CrossRef]


| Characteristics | OC-SCC (n = 42) | Controls (n = 45) | p Value |
|---|---|---|---|
| Sex (%) | |||
| Male | 19 (45%) | 24 (53%) | 0.5 |
| Female | 23 (55%) | 21 (47%) | |
| Age (mean ± SD) | 0.7 | ||
| 63 ± 13 | 63 ± 11 | ||
| Race (%) | |||
| White | 34 (81%) | 37 (82%) | 0.9 |
| Others | 8 (19%) | 8 (18%) | |
| Alcohol drinking (%) | 0.3 | ||
| Never/social drinking | 12 (29%) | 15 (33%) | |
| Quit | 3 (7.1%) | 0 (0%) | |
| Active | 27 (64%) | 30 (67%) | |
| Social/mild | 20 (71%) | 21 (70%) | |
| Moderate | 4 (14%) | 5 (17%) | >0.9 |
| Heavy | 4 (14%) | 4 (13%) | |
| Smoking (%) | >0.9 | ||
| Never | 22 (52%) | 24 (53%) | |
| Quit | 20 (48%) | 21 (47%) |
| Characteristic | No (%) |
|---|---|
| Tumor subsite | |
| Tongue | 24 (57%) |
| Floor of mouth | 5 (12%) |
| Upper gum | 3 (7.2%) |
| Lower gum | 6 (14%) |
| Buccal | 2 (4.8%) |
| Retromolar trigone | 2 (4.8%) |
| Lip | 0 |
| Treatment | |
| Surgery alone | 24 (57%) |
| Surgery + postop radiation | 18 (43%) |
| Tumor size (mm) | |
| 1–10 | 11 (26%) |
| 11–20 | 14 (33%) |
| 21–30 | 8 (19%) |
| 31–40 | 5 (12%) |
| 41–50 | 4 (9.5%) |
| Pathology T stage | |
| T1 | 21 (51%) |
| T2 | 11 (27%) |
| T3 | 2 (4.9%) |
| T4 | 7 (17%) |
| Pathology N stage | |
| N0/Nx | 27 (64%) |
| N+ | 15 (35.7%) |
| Overall pathological stage | |
| 1 | 20 (47%) |
| 2 | 4 (9.3%) |
| 3 | 6 (14%) |
| 4 | 12 (28%) |
| Tumor grade | |
| Well differentiated | 10 (24%) |
| Moderately differentiated | 30 (71%) |
| Poorly differentiated | 2 (4.8%) |
| Class | Pathway | Control * | Cancer * | Fold Change | q Value |
|---|---|---|---|---|---|
| Vitamin | PWY-6519: 8-amino-7-oxononanoate biosynthesis I | 11 | 17 | 1.530 | 0.028 |
| BIOTIN-BIOSYNTHESIS-PWY: biotin biosynthesis I | 12 | 18 | 1.491 | 0.028 | |
| PWY-7539: 6-hydroxymethyl-dihydropterin diphosphate biosynthesis III | 28 | 37 | 1.293 | 0.057 | |
| RIBOSYN2-PWY: flavin biosynthesis I bacteria and plants | 29 | 36 | 1.243 | 0.023 | |
| THISYNARA-PWY: superpathway of thiamin diphosphate biosynthesis III | 29 | 34 | 1.202 | 0.059 | |
| PWY-6168: flavin biosynthesis III | 30 | 35 | 1.194 | 0.059 | |
| PWY-6147: 6-hydroxymethyl-dihydropterin diphosphate biosynthesis I | 65 | 76 | 1.166 | 0.031 | |
| PWY-6897: thiamin salvage II | 39 | 43 | 1.112 | 0.094 | |
| PWY-3841: folate transformations II | 66 | 58 | 0.874 | 0.059 | |
| 1CMET2-PWY: N10-formyl-tetrahydrofolate biosynthesis | 57 | 50 | 0.873 | 0.059 | |
| Heme | HEME-BIOSYNTHESIS-II: heme biosynthesis I aerobic | 23 | 30 | 1.322 | 0.028 |
| PWY-5918: heme biosynthesis from glutamate | 33 | 40 | 1.216 | 0.059 | |
| Nucleotide | PWY-7228: guanosine nucleotides de novo biosynthesis I | 85 | 80 | 0.947 | 0.077 |
| PWY-6126: adenosine nucleotides de novo biosynthesis II | 98 | 92 | 0.946 | 0.098 | |
| PWY-841: purine nucleotides de novo biosynthesis I | 71 | 66 | 0.938 | 0.094 | |
| PWY-6125: guanosine nucleotides de novo biosynthesis II | 81 | 76 | 0.938 | 0.077 | |
| PWY-7208: pyrimidine nucleobases salvage | 79 | 73 | 0.923 | 0.059 | |
| PWY-7197: pyrimidine deoxyribonucleotide phosphorylation | 66 | 61 | 0.920 | 0.077 | |
| PWY-7220: adenosine deoxyribonucleotides de novo biosynthesis II | 72 | 66 | 0.917 | 0.059 | |
| PWY-7222: guanosine deoxyribonucleotides de novo biosynthesis II | 72 | 66 | 0.917 | 0.059 | |
| PWY0-1296: purine ribonucleosides degradation | 67 | 57 | 0.850 | 0.057 | |
| tRNA | TRNA-CHARGING-PWY: tRNA charging | 34 | 39 | 1.152 | 0.059 |
| Amino acid | ILEUSYN-PWY: L-isoleucine biosynthesis I from threonine | 93 | 87 | 0.937 | 0.094 |
| VALSYN-PWY: L-valine biosynthesis | 93 | 87 | 0.937 | 0.094 | |
| PWY-2941: L-lysine biosynthesis II | 59 | 54 | 0.914 | 0.090 | |
| PWY-6936: seleno-amino acid biosynthesis | 76 | 69 | 0.907 | 0.059 | |
| PWY0-781: aspartate superpathway | 35 | 32 | 0.895 | 0.065 | |
| P4-PWY: L-lysine L-threonine and L-methionine biosynthesis I | 42 | 37 | 0.895 | 0.094 | |
| PWY-5347: L-methionine biosynthesis transsulfuration | 52 | 43 | 0.831 | 0.023 | |
| Sugar | DTDPRHAMSYN-PWY: dTDP-L-rhamnose biosynthesis I | 43 | 49 | 1.148 | 0.094 |
| CALVIN-PWY: Calvin-Benson-Bassham cycle | 49 | 55 | 1.120 | 0.038 | |
| Fatty acid | PWY-5973: cis-vaccenate biosynthesis | 54 | 58 | 1.087 | 0.094 |
| Fermentation | ANAEROFRUCAT-PWY: homolactic fermentation | 38 | 41 | 1.096 | 0.059 |
| PWY-7111: pyruvate fermentation to isobutanol engineered | 94 | 87 | 0.932 | 0.077 | |
| PWY-7383: anaerobic energy metabolism invertebrates cytosol | 27 | 22 | 0.849 | 0.059 | |
| Cell wall | PWY0-1586: peptidoglycan maturation | 20 | 17 | 0.848 | 0.063 |
| PWY-6471: peptidoglycan biosynthesis IV Enterococcus faecium | 21 | 16 | 0.742 | 0.028 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganly, I.; Hao, Y.; Rosenthal, M.; Wang, H.; Migliacci, J.; Huang, B.; Katabi, N.; Brown, S.; Tang, Y.-W.; Pei, Z.; et al. Oral Microbiome in Nonsmoker Patients with Oral Cavity Squamous Cell Carcinoma, Defined by Metagenomic Shotgun Sequencing. Cancers 2022, 14, 6096. https://doi.org/10.3390/cancers14246096
Ganly I, Hao Y, Rosenthal M, Wang H, Migliacci J, Huang B, Katabi N, Brown S, Tang Y-W, Pei Z, et al. Oral Microbiome in Nonsmoker Patients with Oral Cavity Squamous Cell Carcinoma, Defined by Metagenomic Shotgun Sequencing. Cancers. 2022; 14(24):6096. https://doi.org/10.3390/cancers14246096
Chicago/Turabian StyleGanly, Ian, Yuhan Hao, Matthew Rosenthal, Hongmei Wang, Jocelyn Migliacci, Bin Huang, Nora Katabi, Stuart Brown, Yi-Wei Tang, Zhiheng Pei, and et al. 2022. "Oral Microbiome in Nonsmoker Patients with Oral Cavity Squamous Cell Carcinoma, Defined by Metagenomic Shotgun Sequencing" Cancers 14, no. 24: 6096. https://doi.org/10.3390/cancers14246096
APA StyleGanly, I., Hao, Y., Rosenthal, M., Wang, H., Migliacci, J., Huang, B., Katabi, N., Brown, S., Tang, Y.-W., Pei, Z., & Yang, L. (2022). Oral Microbiome in Nonsmoker Patients with Oral Cavity Squamous Cell Carcinoma, Defined by Metagenomic Shotgun Sequencing. Cancers, 14(24), 6096. https://doi.org/10.3390/cancers14246096








