GD2 Expression in Medulloblastoma and Neuroblastoma for Personalized Immunotherapy: A Matter of Subtype
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissues
2.2. Materials for Lipid Analysis
2.3. Lipid Extraction
2.4. TLC and Immune Overlay Analysis
2.5. LC-MS2 Analysis of GD2
2.6. Gene Expression Analysis of Tumor Samples
2.7. RNA-Seq Data
2.8. RNA-Seq Data Preparation and Principal Component Analysis
2.9. Differential Gene Expression Analysis
2.10. Two-Gene Signature for GD2 Quantification
3. Results
3.1. Developpement of a LC-MS2 Assay for the Detection of N-glycolyl GM3 and Quantification of GD2
3.2. Neuroblastoma Have the Highest Expression of GD2 and Also Express Complex Gangliosides but Not N-glycolyl GM3
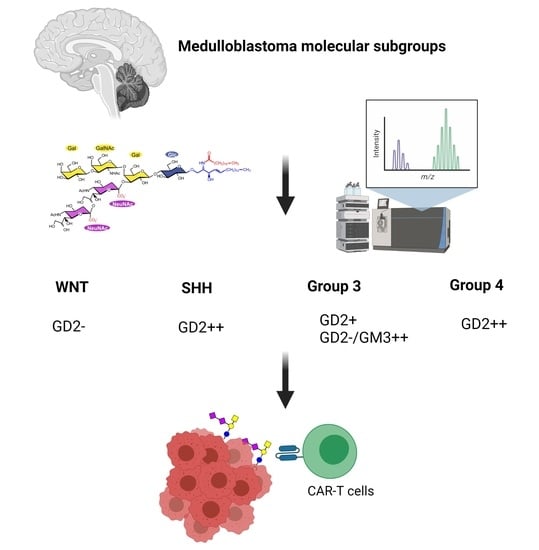
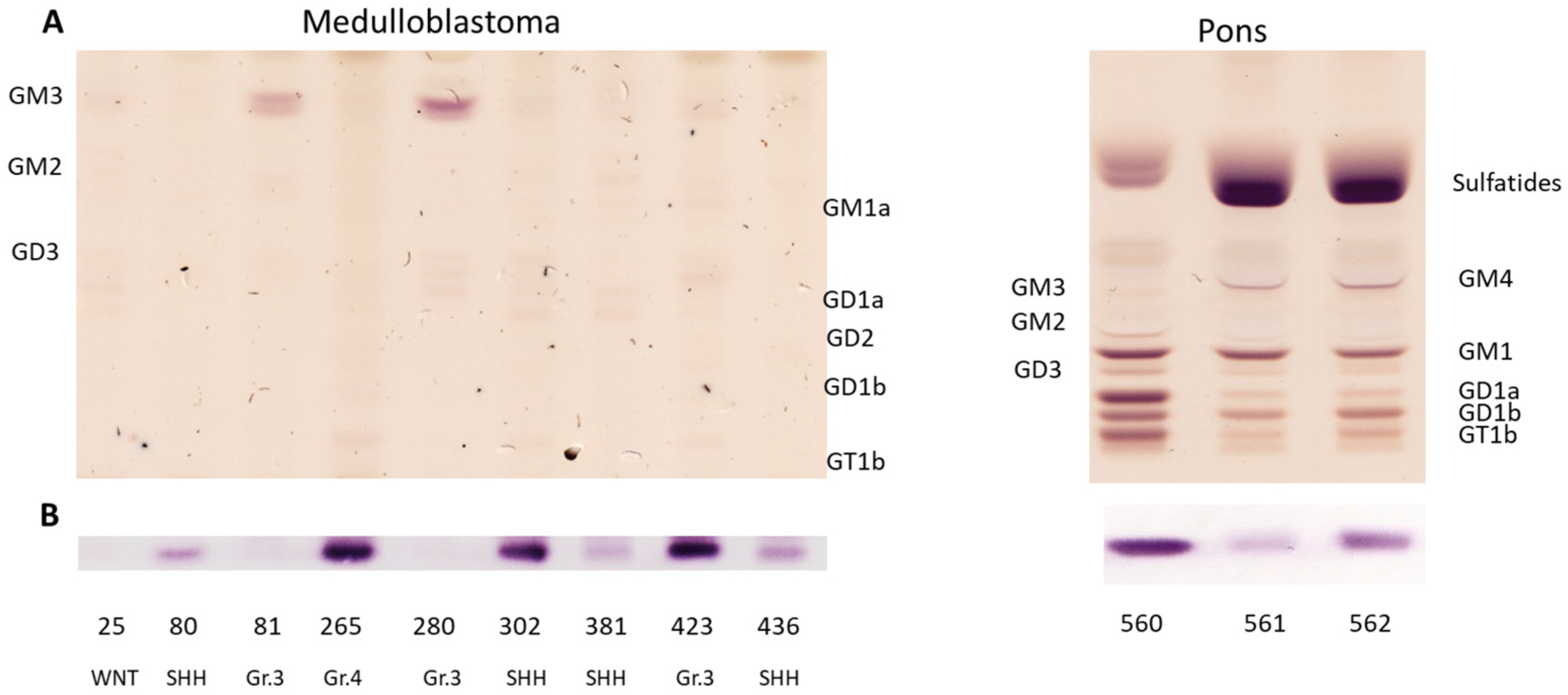
3.3. Medulloblastoma Mainly Express the Gangliosides GD2 or GM3
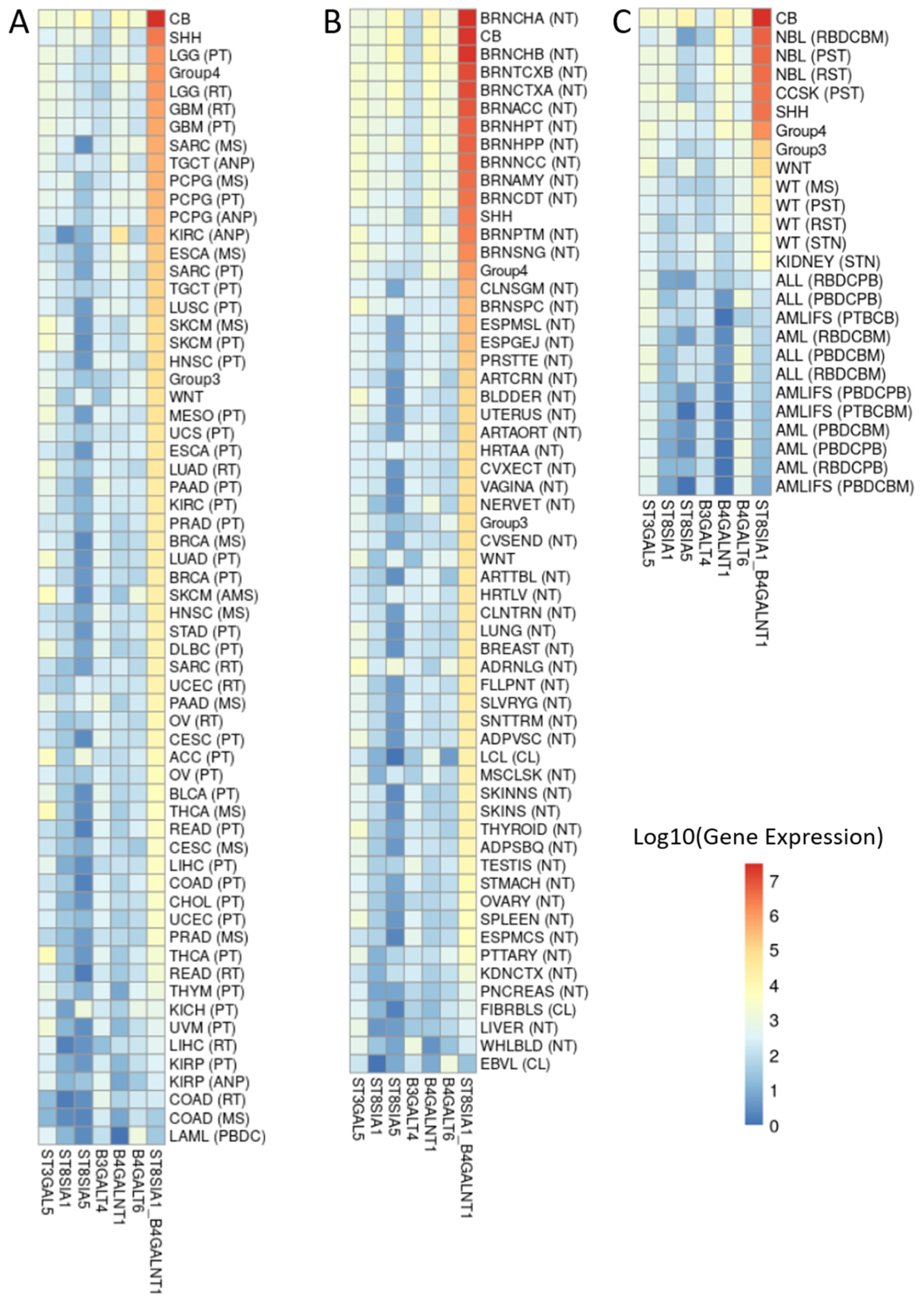
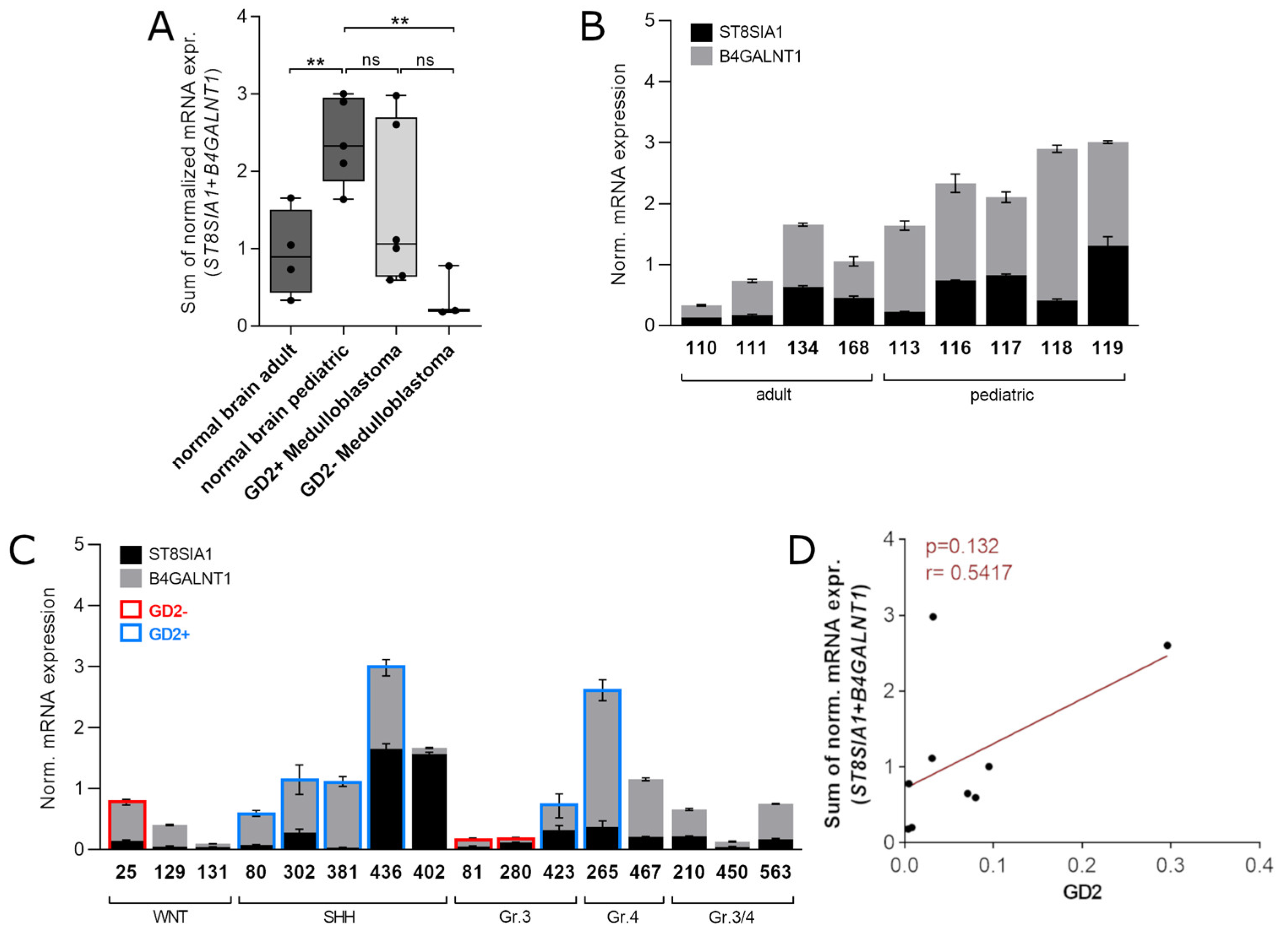
3.4. ST8SIA1 + B4GALNT1 Expression Is a Surrogate Marker for the Detection of GD2-Positive MB in RNAseq Data
3.5. Validation of the Two-Gene Signature Confirms a MB Subtype-Dependent GD2 Expression
4. Discussion
4.1. TLC and LC-MS2 Based Analysis of Gangliosides
4.2. Gangliosides Patterns in NBL and MB
4.3. Relevance of Gangliosides Expression for Therapy of MB and NBL
4.4. Biomarkers for the Detection of GD2-Positive MB
4.5. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fredman, P.; Hedberg, K.; Brezicka, T. Gangliosides as Therapeutic Targets for Cancer. BioDrugs 2003, 17, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Malykh, Y.N.; Schauer, R.; Shaw, L. N-Glycolylneuraminic acid in human tumours. Biochimie 2001, 83, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Nazha, B.; Inal, C.; Owonikoko, T.K. Disialoganglioside GD2 Expression in Solid Tumors and Role as a Target for Cancer Therapy. Front. Oncol. 2020, 10, 1000. [Google Scholar] [CrossRef] [PubMed]
- Slatnick, L.; Jimeno, A.; Gore, L.; Macy, M. Naxitamab: A humanized anti-glycolipid disialoganglioside (anti-GD2) monoclonal antibody for treatment of neuroblastoma. Drugs Today 2021, 57, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Hoy, S.M. Dinutuximab: A Review in High-Risk Neuroblastoma. Target. Oncol. 2016, 11, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Cáceres-Lavernia, H.H.; Nenínger-Vinageras, E.; Varona-Rodríguez, L.M.; Olivares-Romero, Y.A.; Sánchez-Rojas, I.; Mazorra-Herrera, Z.; Basanta-Bergolla, D.; Duvergel-Calderín, D.; Torres-Cuevas, B.L.; del Castillo-Carrillo, C. Racotumomab in Non-Small Cell Lung Cancer as Maintenanceand Second-Line Treatment. MEDICC Rev. 2021, 23, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, M.; Kholodenko, I.; Kalinovsky, D.; Shamanskaya, T.; Doronin, I.; Konovalov, D.; Mironov, A.; Kuzmin, D.; Nikitin, D.; Deyev, S.; et al. RNA Sequencing-Based Identification of Ganglioside GD2-Positive Cancer Phenotype. Biomedicines 2020, 8, 142. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Bagatell, R.; London, W.B.; Maris, J.M.; Cohn, S.L.; Mattay, K.M.; Hogarty, M.; on behalf of the COG Neuroblastoma Committee. Children’s Oncology Group’s 2013 blueprint for research: Neuroblastoma. Pediatr. Blood Cancer 2013, 60, 985–993. [Google Scholar] [CrossRef]
- Northcott, P.A.; Korshunov, A.; Pfister, S.; Taylor, M. The clinical implications of medulloblastoma subgroups. Nat. Rev. Neurol. 2012, 8, 340–351. [Google Scholar] [CrossRef]
- Lazow, M.A.; Palmer, J.D.; Fouladi, M.; Salloum, R. Medulloblastoma in the Modern Era: Review of Contemporary Trials, Molecular Advances, and Updates in Management. Neurotherapeutics 2022, 19, 1733–1751. [Google Scholar] [CrossRef]
- Van Tilburg, C.M.; Pfaff, E.; Pajtler, K.W.; Langenberg, K.P.; Fiesel, P.; Jones, B.C.; Balasubramanian, G.P.; Stark, S.; Johann, P.D.; Blattner-Johnson, M.; et al. The Pediatric Precision Oncology INFORM Registry: Clinical Outcome and Benefit for Patients with Very High-Evidence Targets. Cancer Discov. 2021, 11, 2764–2779. [Google Scholar] [CrossRef] [PubMed]
- Cinatl, J.; Kotchetkov, R.; Pouckova, P.; Vogel, J.U.; Rabenau, H.; Michaelis, M.; Kornhuber, B. Cytotoxicity of L-cycloserine against human neuroblastoma and medulloblastoma cells is associated with the suppression of ganglioside expression. Anticancer Res. 1999, 19, 5349–5354. [Google Scholar] [PubMed]
- Longee, D.C.; Wikstrand, C.J.; Månsson, J.E.; He, X.; Fuller, G.N.; Bigner, S.H.; Fredman, P.; Svennerholm, L.; Bigner, D.D. Disialoganglioside GD2 in human neuroectodermal tumor cell lines and gliomas. Acta Neuropathol. 1991, 82, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K.; Humm, J.L.; Souweidane, M.M.; Zanzonico, P.B.; Dunkel, I.J.; Gerald, W.L.; Khakoo, Y.; Yeh, S.D.; Yeung, H.W.; Finn, R.D.; et al. Phase I Study of Targeted Radioimmunotherapy for Leptomeningeal Cancers Using Intra-Ommaya 131-I-3F8. J. Clin. Oncol. 2007, 25, 5465–5470. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K.; Pandit-Taskar, N.; Humm, J.L.; Zanzonico, P.B.; Haque, S.; Dunkel, I.J.; Wolden, S.L.; Donzelli, M.; Goldman, D.A.; Lewis, J.S.; et al. A phase II study of radioimmunotherapy with intraventricular 131 I-3F8 for medulloblastoma. Pediatr. Blood Cancer 2018, 65, e26754. [Google Scholar] [CrossRef] [PubMed]
- Bayerle, A.; Marsching, C.; Rabionet, M.; Dworski, S.; Kamani, M.A.; Chitraju, C.; Gluchowski, N.L.; Gabriel, K.R.; Herzer, S.; Jennemann, R.; et al. Endogenous levels of 1-O-acylceramides increase upon acidic ceramidase deficiency and decrease due to loss of Dgat1 in a tissue-dependent manner. Biochim. et Biophys. Acta BBA Mol. Cell Biol. Lipids 2020, 1865, 158741. [Google Scholar] [CrossRef]
- Jennemann, R.; Rodden, A.; Bauer, B.L.; Mennel, H.D.; Wiegandt, H. Glycosphingolipids of human gliomas. Cancer Res. 1990, 50, 7444–7449. [Google Scholar]
- Azatyan, A.; Zaphiropoulos, P.G. Circular and Fusion RNAs in Medulloblastoma Development. Cancers 2022, 14, 3134. [Google Scholar] [CrossRef]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ahlmann-Eltze, C.; Patil, I. ggsignif: R Package for Displaying Significance Brackets for ‘ggplot2’. psyArxiv 2021. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef]
- Levitt, G.; Platt, K.; De Byrne, R.; Sebire, N.; Owens, C. 4S neuroblastoma: The long-term outcome. Pediatr. Blood Cancer 2004, 43, 120–125. [Google Scholar] [CrossRef]
- Simon, T.; Hero, B.; Schulte, J.H.; Deubzer, H.; Hundsdoerfer, P.; von Schweinitz, D.; Fuchs, J.; Schmidt, M.; Prasad, V.; Krug, B.; et al. 2017 GPOH Guidelines for Diagnosis and Treatment of Patients with Neuroblastic Tumors. Klin. Padiatr. 2017, 229, 147–167. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Majzner, R.G.; Rietberg, S.P.; Sotillo, E.; Dong, R.; Vachharajani, V.T.; Labanieh, L.; Myklebust, J.H.; Kadapakkam, M.; Weber, E.W.; Tousley, A.M.; et al. Tuning the Antigen Density Requirement for CAR T-cell Activity. Cancer Discov. 2020, 10, 702–723. [Google Scholar] [CrossRef]
- Sung, C.-C.; Pearl, D.K.; Coons, S.W.; Scheithauer, B.W.; Johnson, P.C.; Yates, A.J. Gangliosides as diagnostic markers of human astrocytomas and primitive neuroectodermal tumors. Cancer 1994, 74, 3010–3022. [Google Scholar] [CrossRef]
- Lin, Y.-J.; Mashouf, L.A.; Lim, M. CAR T Cell Therapy in Primary Brain Tumors: Current Investigations and the Future. Front. Immunol. 2022, 13, 817296. [Google Scholar] [CrossRef]
- Blanco, R.; Rengifo, C.E.; Cedeño, M.; Frómeta, M.; Rengifo, E.; Carr, A. Immunoreactivity of the 14F7 Mab (Raised against N-Glycolyl GM3 Ganglioside) as a Positive Prognostic Factor in Non-Small-Cell Lung Cancer. Pathol. Res. Int. 2012, 2012, 235418. [Google Scholar] [CrossRef] [PubMed]
- Blanco, R.; Rengifo, E.; Rengifo, C.E.; Cedeño, M.; Frómeta, M.; Carr, A. Immunohistochemical Reactivity of the 14F7 Monoclonal Antibody Raised against N-Glycolyl GM3 Ganglioside in Some Benign and Malignant Skin Neoplasms. ISRN Dermatol. 2011, 2011, 848909. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carr, A.; Mullet, A.; Mazorra, Z.; Vázquez, A.M.; Alfonso, M.; Mesa, C.; Rengifo, E.; Pérez, R.; Fernández, L.E. A Mouse IgG1Monoclonal Antibody Specific for N-Glycolyl GM3 Ganglioside Recognized Breast and Melanoma Tumors. Hybridoma 2000, 19, 241–247. [Google Scholar] [CrossRef]
- Scursoni, A.M.; Galluzzo, L.; Camarero, S.; Lopez, J.; Lubieniecki, F.; Sampor, C.; Segatori, V.I.; Gabri, M.R.; Alonso, D.F.; Chantada, G.; et al. Detection of N-Glycolyl GM3 Ganglioside in Neuroectodermal Tumors by Immunohistochemistry: An Attractive Vaccine Target for Aggressive Pediatric Cancer. J. Immunol. Res. 2011, 2011, 245181. [Google Scholar] [CrossRef][Green Version]
- Sariola, M.H.; Harri, M.T.; Rapola, M.J.; Saarinen, M.U.M. Cell-Surface Ganglioside GD2 in the Immunohistochemical Detection and Differential Diagnosis of Neuroblastoma. Am. J. Clin. Pathol. 1991, 96, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Wingerter, A.; El Malki, K.; Sandhoff, R.; Seidmann, L.; Wagner, D.-C.; Lehmann, N.; Vewinger, N.; Frauenknecht, K.; Sommer, C.; Traub, F.; et al. Exploiting Gangliosides for the Therapy of Ewing’s Sarcoma and H3K27M-Mutant Diffuse Midline Glioma. Cancers 2021, 13, 520. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Saxton, R.E.; Morton, D.L.; Irie, R.F. Gangliosides of Human Melanoma2. Gynecol. Oncol. 1987, 78, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.D.; Larson, S.M.; Burch, L.; Kushner, B.H.; Laquaglia, M.; Finn, R.; Cheung, N.K. Radioimmunodetection of neuroblastoma with iodine-131-3F8: Correlation with biopsy, iodine-131-metaiodobenzylguanidine and standard diagnostic modalities. J. Nucl. Med. 1991, 32, 769–776. [Google Scholar]
- Jennemann, R.; Rothermel, U.; Wang, S.; Sandhoff, R.; Kaden, S.; Out, R.; van Berkel, T.J.; Aerts, J.M.; Ghauharali, K.; Sticht, C.; et al. Hepatic glycosphingolipid deficiency and liver function in mice. Hepatology 2010, 51, 1799–1809. [Google Scholar] [CrossRef]
- Ikeda, K.; Shimizu, T.; Taguchi, R. Targeted analysis of ganglioside and sulfatide molecular species by LC/ESI-MS/MS with theoretically expanded multiple reaction monitoring. J. Lipid Res. 2008, 49, 2678–2689. [Google Scholar] [CrossRef]
- Lee, J.; Hwang, H.; Kim, S.; Hwang, J.; Yoon, J.; Yin, D.; Choi, S.I.; Kim, Y.-H.; Kim, Y.-S.; An, H.J. Comprehensive Profiling of Surface Gangliosides Extracted from Various Cell Lines by LC-MS/MS. Cells 2019, 8, 1323. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, M.; Yu, R.K. The expression and functions of glycoconjugates in neural stem cells. Glycobiology 2007, 17, 57R–74R. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, M.; Patel, A.G.; Dyer, M.A. Neuroblastoma: When differentiation goes awry. Neuron 2022, 110, 2916–2928. [Google Scholar] [CrossRef] [PubMed]
- Iwamori, M.; Nagai, Y. A new chromatographic approach to the resolution of individual gangliosides. Ganglioside mapping. Biochim. Biophys. Acta 1978, 528, 257–267. [Google Scholar]
- Schengrund, C.L.; Repman, M.A.; Shochat, S.J. Ganglioside composition of human neuroblastomas. Correlation with prognosis. A Pediatric Oncology Group Study. Cancer 1985, 56, 2640–2646. [Google Scholar] [CrossRef]
- Cavalli, F.M.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e6. [Google Scholar] [CrossRef]
- Schwalbe, E.C.; Lindsey, J.C.; Nakjang, S.; Crosier, S.; Smith, A.J.; Hicks, D.; Rafiee, G.; Hill, R.M.; Iliasova, A.; Stone, T.; et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: A cohort study. Lancet Oncol. 2017, 18, 958–971. [Google Scholar] [CrossRef]
- Hovestadt, V.; Smith, K.S.; Bihannic, L.; Filbin, M.G.; Shaw, M.L.; Baumgartner, A.; DeWitt, J.C.; Groves, A.; Mayr, L.; Weisman, H.R.; et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 2019, 572, 74–79. [Google Scholar] [CrossRef]
- Terzic, T.; Cordeau, M.; Herblot, S.; Teira, P.; Cournoyer, S.; Beaunoyer, M.; Peuchmaur, M.; Duval, M.; Sartelet, H. Expression of Disialoganglioside (GD2) in Neuroblastic Tumors: A Prognostic Value for Patients Treated with Anti-GD2 Immunotherapy. Pediatr. Dev. Pathol. 2018, 21, 355–362. [Google Scholar] [CrossRef]
- Edelman, M.J.; Dvorkin, M.; Laktionov, K.; Navarro, A.; Juan-Vidal, O.; Kozlov, V.; Golden, G.; Jordan, O.; Deng, C.; Bentsion, D.; et al. Randomized phase 3 study of the anti-disialoganglioside antibody dinutuximab and irinotecan vs irinotecan or topotecan for second-line treatment of small cell lung cancer. Lung Cancer 2022, 166, 135–142. [Google Scholar] [CrossRef]
- Saleh, M.N.; Khazaeli, M.B.; Wheeler, R.H.; Dropcho, E.; Liu, T.; Urist, M.; Miller, D.M.; Lawson, S.; Dixon, P.; Russell, C.H. Phase I trial of the murine monoclonal anti-GD2 antibody 14G2a in metastatic melanoma. Cancer Res. 1992, 52, 4342–4347. [Google Scholar] [PubMed]
- Heitzeneder, S.; Bosse, K.R.; Zhu, Z.; Zhelev, D.; Majzner, R.G.; Radosevich, M.T.; Dhingra, S.; Sotillo, E.; Buongervino, S.; Pascual-Pasto, G.; et al. GPC2-CAR T cells tuned for low antigen density mediate potent activity against neuroblastoma without toxicity. Cancer Cell 2022, 40, 53–69.e59. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.-J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2011, 123, 465–472. [Google Scholar] [CrossRef]
- Donovan, L.K.; Delaidelli, A.; Joseph, S.K.; Bielamowicz, K.; Fousek, K.; Holgado, B.L.; Manno, A.; Srikanthan, D.; Gad, A.Z.; Van Ommeren, R.; et al. Locoregional delivery of CAR T cells to the cerebrospinal fluid for treatment of metastatic medulloblastoma and ependymoma. Nat. Med. 2020, 26, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Lammie, G.; Cheung, N.; Gerald, W.; Rosenblum, M.; Cordoncardo, C. Ganglioside Gd(2) Expression In The Human Nervous-System And In Neuroblastomas—An Immunohistochemical Study. Int. J. Oncol. 1993, 3, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Richman, S.A.; Nunez-Cruz, S.; Moghimi, B.; Li, L.Z.; Gershenson, Z.T.; Mourelatos, Z.; Barrett, D.M.; Grupp, S.A.; Milone, M.C. High-Affinity GD2-Specific CAR T Cells Induce Fatal Encephalitis in a Preclinical Neuroblastoma Model. Cancer Immunol. Res. 2018, 6, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Majzner, R.G.; Weber, E.W.; Lynn, R.C.; Xu, P.; Mackall, C.L. Neurotoxicity Associated with a High-Affinity GD2 CAR—Letter. Cancer Immunol. Res. 2018, 6, 494–495. [Google Scholar] [CrossRef]
- Gabri, M.R.; Cacciavillano, W.; Chantada, G.L.; Alonso, D.F. Racotumomab for treating lung cancer and pediatric refractory malignancies. Expert Opin. Biol. Ther. 2016, 16, 573–578. [Google Scholar] [CrossRef]
- Itokazu, Y.; Wang, J.; Yu, R.K. Gangliosides in Nerve Cell Specification. Prog. Mol. Biol. Transl. Sci. 2018, 156, 241–263. [Google Scholar] [CrossRef]
- Grayson, G.; Ladisch, S. Immunosuppression by human gangliosides: II. Carbohydrate structure and inhibition of human NK activity. Cell. Immunol. 1992, 139, 18–29. [Google Scholar] [CrossRef]
- Shurin, G.V.; Shurin, M.R.; Bykovskaia, S.; Shogan, J.; Lotze, M.T.; Barksdale, E.M., Jr. Neuroblastoma-derived gangliosides inhibit dendritic cell generation and function. Cancer Res. 2001, 61, 363–369. [Google Scholar] [PubMed]
- Mount, C.; Majzner, R.G.; Sundaresh, S.; Arnold, E.P.; Kadapakkam, M.; Haile, S.; Labanieh, L.; Hulleman, E.; Woo, P.J.; Rietberg, S.P.; et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M+ diffuse midline gliomas. Nat. Med. 2018, 24, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Lo Piccolo, M.S.; Cheung, N.K.; Cheung, I.Y. GD2 synthase: A new molecular marker for detecting neuroblastoma. Cancer 2001, 92, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Han, L.; Sun, B.; Zhao, Q. Identification of a Glycosyltransferase Signature for Predicting Prognosis and Immune Microenvironment in Neuroblastoma. Front. Cell Dev. Biol. 2021, 9, 769580. [Google Scholar] [CrossRef]

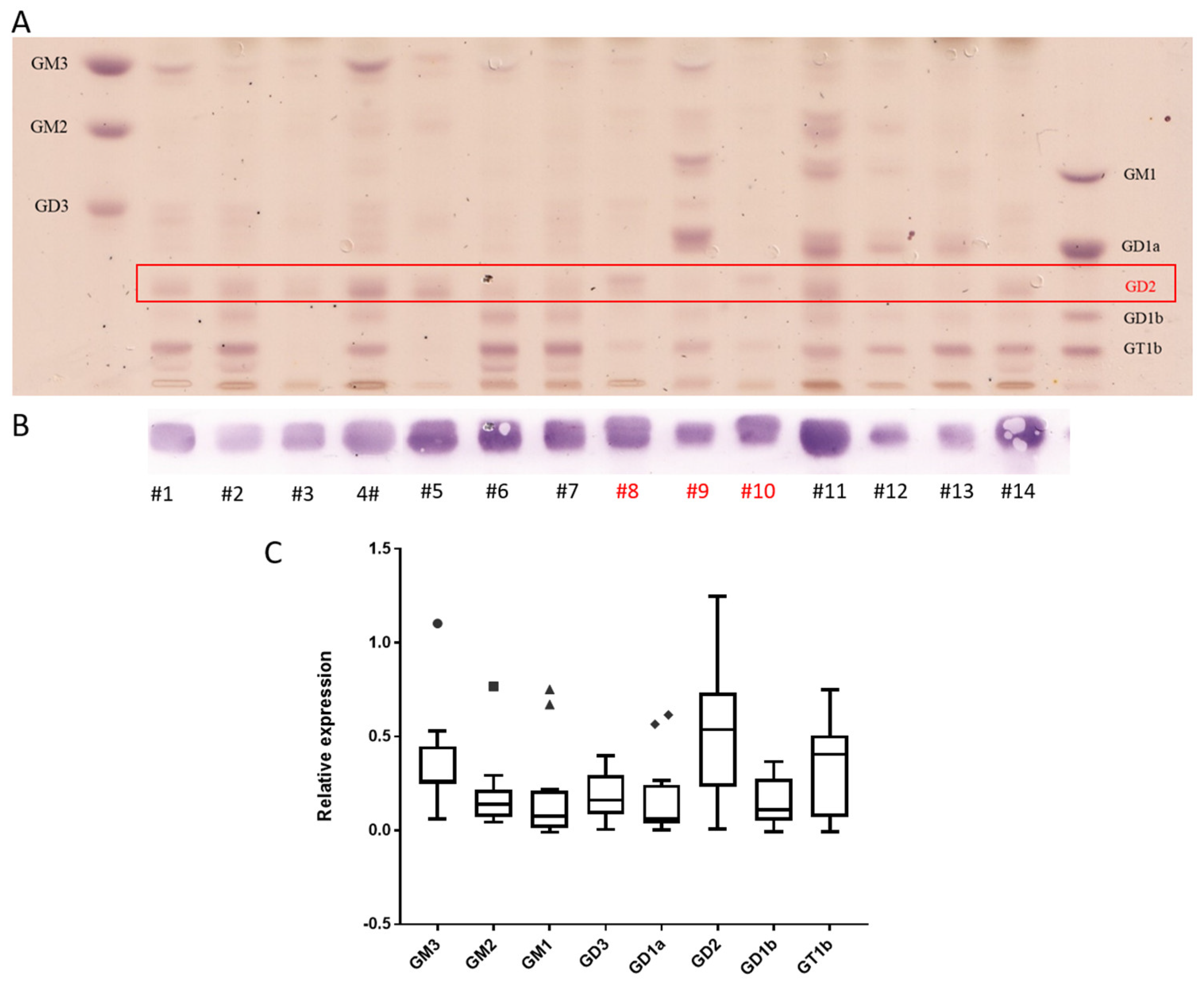


| Nr | Age | Sex | Histology | Localization | Sample Type | INSS | MYCN | GD2 |
|---|---|---|---|---|---|---|---|---|
| #1 | 5 m | m | NBL, pd | adrenal gland | PT | nd | NA | 0.64 |
| #2 | 1 m | f | NBL, pd | adrenal gland | PT | 1 | NA | 0.95 |
| #3 | 1 y | m | NBL, pd | retroperitoneal | PT | 3/4 | A | 0.59 |
| #4 | 5 m | m | NBL, pd | adrenal gland | PT | nd | nd | 1.93 |
| #5 | 1 y | m | NBL, pd | retroperitoneal | PT | 3/4 | A | 1.06 |
| #6 | 8 m | m | NBL, pd | lumbal soft tissue | PM | 4S | NA | 0.49 |
| #7 | 2 m | m | NBL, pd | adrenal gland | PT | 4S | NA | 0.43 |
| #8 | 8 y | m | NBL, pd | lymphnode | PM | 4 | NA | 1.1 |
| #9 | 8 y | m | GNB, int | adrenal gland | PT | 4 | NA | 0.16 |
| #10 | 10 y | m | NBL, pd | intracerebral | R | 4 | nd | 0.31 |
| #11 | 8 y | m | NBL, dif | retroperitoneal | PT | 4 | nd | 1.58 |
| #12 | 2 y | f | NBL, dif | para/intraspinal | PT | nd | NA | 0.15 |
| #13 | 4 y | f | NBL, dif/mature GN | adrenal gland | PT | nd | nd | 0.04 |
| #14 | 5 m | f | NBL, pd | adrenal gland | PT | nd | NA | 0.49 |
| Nr | Age | Sex | Histological Subtype | Molecular Subtype | Sample Type | MYCC | MYCN | GD2 |
|---|---|---|---|---|---|---|---|---|
| 25 | 15 y | f | CBM | WNT | PT | nd | nd | 0.005 |
| 80 | 6 y | f | AMB | SHH | PT | nd | nd | 0.080 |
| 81 | 5 y | m | CBM | Gr.3/III | PT | nd | NA | 0.004 |
| 129 | 8 y | f | CBM | WNT | PT | nd | nd | nd |
| 131 | 8 y | f | CBM | WNT | PT | nd | nd | nd |
| 210 | 5 y | m | CBM | Gr.3/4 | PT | NA | NA | nd |
| 265 | 11 y | m | AMB | Gr.4/V | PT | NA | Gain | 0.296 |
| 280 | 6 y | f | CBM | Gr.3/IV | PT | NA | NA | 0.008 |
| 302 | 1 y | f | DMB | SHH | PT | NA | NA | 0.095 |
| 381 | 1 y | f | DMB | SHH | PT | nd | nd | 0.031 |
| 402 | 1 y | f | DMB | SHH | R | nd | nd | nd |
| 423 | 6 y | m | CBM | Gr.3/II | PT | A | NA | 0.071 |
| 436 | 3 y | f | DMB | SHH | PT | NA | NA | 0.032 |
| 450 | 2 y | f | CBM | Gr.3/4 | PT | NA | NA | nd |
| 467 | 15 y | f | CBM | Gr.4 | PT | NA | NA | nd |
| 563 | 5 y | m | CBM | Gr.3/4 | PT | NA | NA | nd |
| Sample | Age | Sex | Localization | GD2 |
|---|---|---|---|---|
| 110 | 22, 26, 27, 28, 29 y | m | Frontal lobe | nd |
| 111 | Adult (age unknown) | unknown | Temporal lobe | nd |
| 113 | 2 y | m | Temporal lobe | nd |
| 116 | 2 y | m | Hippocampus | nd |
| 117 | 8 m | m | Hippocampus | nd |
| 118 | 8 m | m | Temporal lobe | nd |
| 119 | 2 y | m | Brain stem | nd |
| 134 | 24 y | m | Cerebellum | nd |
| 168 | 60 y | f | Pons | nd |
| 560 | 1 d | m | Pons | 0.395 |
| 561 | 29 y | m | Pons | 0.065 |
| 562 | 61 y | m | Pons | 0.159 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paret, C.; Ustjanzew, A.; Ersali, S.; Seidmann, L.; Jennemann, R.; Ziegler, N.; Malki, K.E.; Russo, A.; Wingerter, A.; Ortmüller, F.; et al. GD2 Expression in Medulloblastoma and Neuroblastoma for Personalized Immunotherapy: A Matter of Subtype. Cancers 2022, 14, 6051. https://doi.org/10.3390/cancers14246051
Paret C, Ustjanzew A, Ersali S, Seidmann L, Jennemann R, Ziegler N, Malki KE, Russo A, Wingerter A, Ortmüller F, et al. GD2 Expression in Medulloblastoma and Neuroblastoma for Personalized Immunotherapy: A Matter of Subtype. Cancers. 2022; 14(24):6051. https://doi.org/10.3390/cancers14246051
Chicago/Turabian StyleParet, Claudia, Arsenij Ustjanzew, Sara Ersali, Larissa Seidmann, Richard Jennemann, Nicole Ziegler, Khalifa El Malki, Alexandra Russo, Arthur Wingerter, Franziska Ortmüller, and et al. 2022. "GD2 Expression in Medulloblastoma and Neuroblastoma for Personalized Immunotherapy: A Matter of Subtype" Cancers 14, no. 24: 6051. https://doi.org/10.3390/cancers14246051
APA StyleParet, C., Ustjanzew, A., Ersali, S., Seidmann, L., Jennemann, R., Ziegler, N., Malki, K. E., Russo, A., Wingerter, A., Ortmüller, F., Bornas, A., Wehling, P. C., Lepădatu, A., Ottenhausen, M., Roth, W., Sommer, C., Fliss, B., Frauenknecht, K. B. M., Sandhoff, R., & Faber, J. (2022). GD2 Expression in Medulloblastoma and Neuroblastoma for Personalized Immunotherapy: A Matter of Subtype. Cancers, 14(24), 6051. https://doi.org/10.3390/cancers14246051