The Dual Role of PDCD10 in Cancers: A Promising Therapeutic Target
Abstract
Simple Summary
Abstract
1. Introduction
2. Structure, Interactors, and Regulators of PDCD10
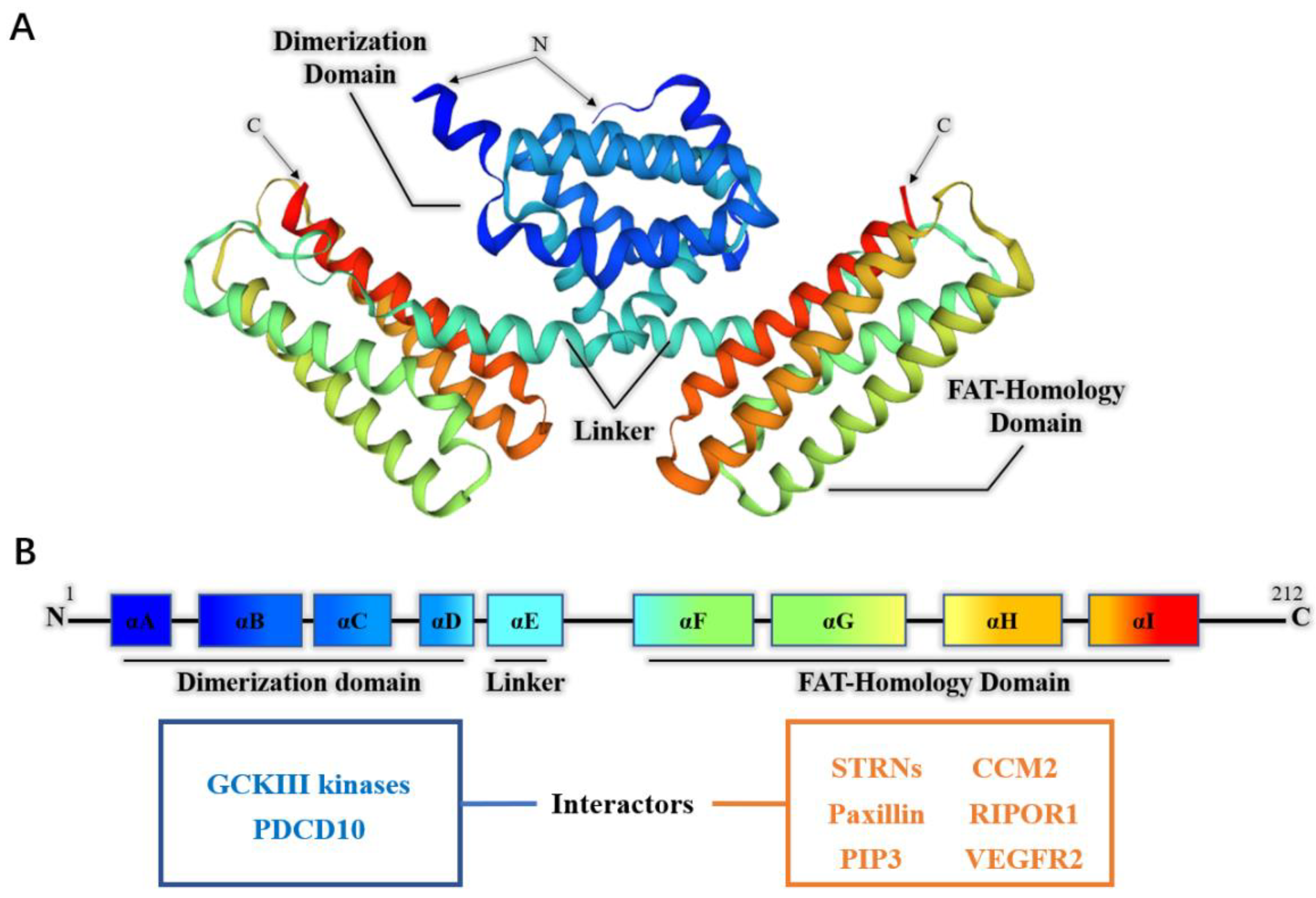
2.1. The Structure of PDCD10
2.2. The Interactors of PDCD10
2.2.1. The Interactors of PDCD10 in Physiological Processes
2.2.2. The Interactors of PDCD10 in Pathological Processes
2.3. The Regulators of PDCD10
3. The Dual Role of PDCD10 in Cancer
3.1. PDCD10 Exerts Pro-Tumorigenic Effects
3.1.1. PDCD10 Promotes Survival and Self-Renewal of Tumor Cells
3.1.2. PDCD10 Promotes Tumor Migration, Invasion, and Metastasis
3.2. PDCD10 Exerts Anti-Tumorigenic Effects
3.2.1. PDCD10 Depletion Is Associated with Meningioma
3.2.2. Loss of PDCD10 Promotes Tumor Metastasis by Activating CAFs
3.2.3. Loss of PDCD10 Promotes Chemoresistance of Tumors
3.2.4. Loss of PDCD10 Promotes Angiogenesis of Tumors
3.3. The Controversy Regarding the Role of PDCD10 in Gliomas
4. Perspective
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Wang, Y.; Liu, H.; Zhang, Y.; Ma, D. cDNA cloning and expression of an apoptosis-related gene, humanTFAR15 gene. Sci. China Ser. C Life Sci. 1999, 42, 323–329. [Google Scholar] [CrossRef]
- Guan, X.; Lu, J.; Sun, F.; Li, Q.; Pang, Y. The Molecular Evolution and Functional Divergence of Lamprey Programmed Cell Death Genes. Front. Immunol. 2019, 10, 1382. [Google Scholar] [CrossRef]
- Togashi, Y.; Shitara, K.; Nishikawa, H. Regulatory T cells in cancer immunosuppression—Implications for anticancer therapy. Nat. Rev. Clin. Oncol. 2019, 16, 356–371. [Google Scholar] [CrossRef]
- Dermani, F.K.; Samadi, P.; Rahmani, G.; Kohlan, A.K.; Najafi, R. PD-1/PD-L1 immune checkpoint: Potential target for cancer therapy. J. Cell. Physiol. 2018, 234, 1313–1325. [Google Scholar] [CrossRef]
- Wang, S.; Englund, E.; Kjellman, P.; Li, Z.; Ahnlide, J.K.; Rodriguez-Cupello, C.; Saggioro, M.; Kanzaki, R.; Pietras, K.; Lindgren, D.; et al. CCM3 is a gatekeeper in focal adhesions regulating mechanotransduction and YAP/TAZ signalling. Nat. Cell Biol. 2021, 23, 758–770. [Google Scholar] [CrossRef]
- Malinverno, M.; Maderna, C.; Taha, A.A.; Corada, M.; Orsenigo, F.; Valentino, M.; Pisati, F.; Fusco, C.; Graziano, P.; Giannotta, M.; et al. Endothelial cell clonal expansion in the development of cerebral cavernous malformations. Nat. Commun. 2019, 10, 2761. [Google Scholar] [CrossRef]
- Louvi, A.; Nishimura, S.; Günel, M. Ccm3, a gene associated with cerebral cavernous malformations, is required for neuronal migration. Development 2014, 141, 1404–1415. [Google Scholar] [CrossRef]
- Guerrero, A.; Iglesias, C.; Raguz, S.; Floridia, E.; Gil, J.; Pombo, C.M.; Zalvide, J. The cerebral cavernous malformation 3 gene is necessary for senescence induction. Aging Cell 2015, 14, 274–283. [Google Scholar] [CrossRef]
- Yaba, A.; Ordueri, N.E.G.; Tanriover, G.; Sahin, P.; Demir, N.; Celik-Ozenci, C. Expression of CCM2 and CCM3 during mouse gonadogenesis. J. Assist. Reprod. Genet. 2015, 32, 1497–1507. [Google Scholar] [CrossRef]
- Wang, R.; Wu, S.-T.; Yang, X.; Qian, Y.; Choi, J.P.; Gao, R.; Song, S.; Wang, Y.; Zhuang, T.; Wong, J.J.; et al. Pdcd10-Stk24/25 complex controls kidney water reabsorption by regulating Aqp2 membrane targeting. JCI Insight 2021, 6, e142838. [Google Scholar] [CrossRef]
- Craig, H.D.; Günel, M.; Cepeda, O.; Johnson, E.W.; Ptacek, L.; Steinberg, G.K.; Ogilvy, C.S.; Berg, M.J.; Crawford, S.C.; Scott, R.M.; et al. Multilocus linkage identifies two new loci for a mendelian form of stroke, cerebral cavernous malformation, at 7p15-13 and 3q25.2-27. Hum. Mol. Genet. 1998, 7, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Bergametti, F.; Denier, C.; Labauge, P.; Arnoult, M.; Boetto, S.; Clanet, M.; Coubes, P.; Echenne, B.; Ibrahim, R.; Irthum, B.; et al. Mutations within the Programmed Cell Death 10 Gene Cause Cerebral Cavernous Malformations. Am. J. Hum. Genet. 2005, 76, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Riolo, G.; Ricci, C.; Battistini, S. Molecular Genetic Features of Cerebral Cavernous Malformations (CCM) Patients: An Overall View from Genes to Endothelial Cells. Cells 2021, 10, 704. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Qi, Y.; Guo, Z.; Li, P.; Zhou, D. miR-613 suppresses ischemia-reperfusion-induced cardiomyocyte apoptosis by targeting the programmed cell death 10 gene. Biosci. Trends 2016, 10, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, T.; Xiu, W.; Cao, W.; He, M.; Sun, W.; Zhao, W. MiR-107 overexpression attenuates neurotoxicity induced by 6-hydroxydopamine both in vitro and in vivo. Chem. Interactions 2019, 315, 108908. [Google Scholar] [CrossRef]
- Ni, W.-J.; Leng, X.-M. Down-regulated miR-495 can target programmed cell death 10 in ankylosing spondylitis. Mol. Med. 2020, 26, 50. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Hussen, B.M.; Mohaqiq, M.; Shoorei, H.; Baniahmad, A.; Taheri, M.; Jamali, E. Interplay Between Non-Coding RNAs and Programmed Cell Death Proteins. Front. Oncol. 2022, 12, 808475. [Google Scholar] [CrossRef]
- Li, X.; Zhang, R.; Zhang, H.; He, Y.; Ji, W.; Min, W.; Boggon, T.J. Crystal Structure of CCM3, a Cerebral Cavernous Malformation Protein Critical for Vascular Integrity. J. Biol. Chem. 2010, 285, 24099–24107. [Google Scholar] [CrossRef]
- Fidalgo, M.; Fraile, M.; Pires, A.; Force, T.; Pombo, C.; Zalvide, J. CCM3/PDCD10 stabilizes GCKIII proteins to promote Golgi assembly and cell orientation. J. Cell Sci. 2010, 123, 1274–1284. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, H.; Shan, J.; Long, F.; Chen, Y.; Chen, Y.; Zhang, Y.; Han, X.; Ma, D. PDCD10 Interacts with Ste20-related Kinase MST4 to Promote Cell Growth and Transformation via Modulation of the ERK Pathway. Mol. Biol. Cell 2007, 18, 1965–1978. [Google Scholar] [CrossRef]
- Mardakheh, F.K.; Self, A.; Marshall, C.J. RHO binding to FAM65A regulates Golgi reorientation during cell migration. J. Cell Sci. 2016, 129, 4466–4479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Dong, L.; Shi, Z.; Jiao, S.; Zhang, Z.; Zhang, W.; Liu, G.; Chen, C.; Feng, M.; Hao, Q.; et al. Structural Mechanism of CCM3 Heterodimerization with GCKIII Kinases. Structure 2013, 21, 680–688. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ceccarelli, D.; Laister, R.C.; Mulligan, V.; Kean, M.J.; Goudreault, M.; Scott, I.; Derry, B.; Chakrabartty, A.; Gingras, A.-C.; Sicheri, F. CCM3/PDCD10 Heterodimerizes with Germinal Center Kinase III (GCKIII) Proteins Using a Mechanism Analogous to CCM3 Homodimerization. J. Biol. Chem. 2011, 286, 25056–25064. [Google Scholar] [CrossRef] [PubMed]
- Draheim, K.M.; Li, X.; Zhang, R.; Fisher, O.S.; Villari, G.; Boggon, T.J.; Calderwood, D.A. CCM2–CCM3 interaction stabilizes their protein expression and permits endothelial network formation. J. Cell Biol. 2015, 208, 987–1001. [Google Scholar] [CrossRef] [PubMed]
- Kean, M.J.; Ceccarelli, D.F.; Goudreault, M.; Sanches, M.; Tate, S.; Larsen, B.; Gibson, L.C.; Derry, W.B.; Scott, I.C.; Pelletier, L.; et al. Structure-function analysis of core STRIPAK Proteins: A signaling complex implicated in Golgi polarization. J. Biol. Chem. 2011, 286, 25065–25075. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.F.; Horst, J.A.; Malone, M.H.; Park, K.; Temple, B.; Cheeseman, H.; Barbaro, J.R.; Johnson, G.L.; Bencharit, S. Defining the Functional Domain of Programmed Cell Death 10 through Its Interactions with Phosphatidylinositol-3,4,5-Trisphosphate. PLoS ONE 2010, 5, e11740. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, H.; Yu, L.; Gunel, M.; Boggon, T.J.; Chen, H.; Min, W. Stabilization of VEGFR2 Signaling by Cerebral Cavernous Malformation 3 Is Critical for Vascular Development. Sci. Signal. 2010, 3, ra26. [Google Scholar] [CrossRef]
- Qin, L.; Zhang, H.; Li, B.; Jiang, Q.; Lopez, F.; Min, W.; Zhou, J.H. CCM3 Loss-Induced Lymphatic Defect Is Mediated by the Augmented VEGFR3-ERK1/2 Signaling. Arter. Thromb. Vasc. Biol. 2021, 41, 2943–2960. [Google Scholar] [CrossRef]
- Tan, P.; Ye, Y.; He, L.; Xie, J.; Jing, J.; Ma, G.; Pan, H.; Han, L.; Han, W.; Zhou, Y. TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PLoS Biol. 2018, 16, e3000051. [Google Scholar] [CrossRef]
- Glatter, T.; Wepf, A.; Aebersold, R.; Gstaiger, M. An integrated workflow for charting the human interaction proteome: Insights into the PP2A system. Mol. Syst. Biol. 2009, 5, 237. [Google Scholar] [CrossRef]
- Goudreault, M.; D’Ambrosio, L.M.; Kean, M.J.; Mullin, M.J.; Larsen, B.G.; Sanchez, A.; Chaudhry, S.; Chen, G.I.; Sicheri, F.; Nesvizhskii, A.I.; et al. A PP2A Phosphatase High Density Interaction Network Identifies a Novel Striatin-interacting Phosphatase and Kinase Complex Linked to the Cerebral Cavernous Malformation 3 (CCM3) Protein. Mol. Cell. Proteom. 2009, 8, 157–171. [Google Scholar] [CrossRef]
- Ribeiro, P.S.; Josué, F.; Wepf, A.; Wehr, M.C.; Rinner, O.; Kelly, G.; Tapon, N.; Gstaiger, M. Combined Functional Genomic and Proteomic Approaches Identify a PP2A Complex as a Negative Regulator of Hippo Signaling. Mol. Cell 2010, 39, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Sents, W.; Ivanova, E.; Lambrecht, C.; Haesen, D.; Janssens, V. The biogenesis of active protein phosphatase 2A holoenzymes: A tightly regulated process creating phosphatase specificity. FEBS J. 2012, 280, 644–661. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.; Hwang, J.; Carrier, K.J.; Jones, C.A.; Kern, Q.L.; Moreno, C.S.; Karas, R.H.; Pallas, D.C. Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-Like kinase Mst3. BMC Biochem. 2011, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Pallas, D.C. STRIPAK complexes: Structure, biological function, and involvement in human diseases. Int. J. Biochem. Cell Biol. 2013, 47, 118–148. [Google Scholar] [CrossRef] [PubMed]
- Kück, U.; Beier, A.M.; Teichert, I. The composition and function of the striatin-interacting phosphatases and kinases (STRIPAK) complex in fungi. Fungal Genet. Biol. 2016, 90, 31–38. [Google Scholar] [CrossRef]
- Kück, U.; Radchenko, D.; Teichert, I. STRIPAK, a highly conserved signaling complex, controls multiple eukaryotic cellular and developmental processes and is linked with human diseases. Biol. Chem. 2019, 400, 1005–1022. [Google Scholar] [CrossRef]
- Frey, S.; Lahmann, Y.; Hartmann, T.; Seiler, S.; Pöggeler, S. Deletion of Smgpi1 encoding a GPI-anchored protein suppresses sterility of the STRIPAK mutant ΔSmmob3 in the filamentous ascomycete Sordaria macrospora. Mol. Microbiol. 2015, 97, 676–697. [Google Scholar] [CrossRef]
- Peng, W.; Wu, X.; Feng, D.; Zhang, Y.; Chen, X.; Ma, C.; Shen, H.; Li, X.; Li, H.; Zhang, J.; et al. Cerebral cavernous malformation 3 relieves subarachnoid hemorrhage-induced neuroinflammation in rats through inhibiting NF-kB signaling pathway. Brain Res. Bull. 2020, 160, 74–84. [Google Scholar] [CrossRef]
- Gong, L.; Zhu, H.; Li, T.; Ming, G.; Duan, X.; Wang, J.; Jiang, Y. Molecular signatures of cytotoxic effects in human embryonic kidney 293 cells treated with single and mixture of ochratoxin A and citrinin. Food Chem. Toxicol. 2018, 123, 374–384. [Google Scholar] [CrossRef]
- Kim, H.W.; Mallick, F.; Durrani, S.; Ashraf, M.; Jiang, S.; Haider, K.H. Concomitant Activation of miR-107/PDCD10 and Hypoxamir-210/Casp8ap2 and Their Role in Cytoprotection During Ischemic Preconditioning of Stem Cells. Antioxid. Redox Signal. 2012, 17, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Meng, S.; Zhu, T.; Wang, X. PDCD10/CCM3 Acts Downstream of γ-Protocadherins to Regulate Neuronal Survival. J. Biol. Chem. 2010, 285, 41675–41685. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wu, Q.; Xu, J.-F.; Miller, D.; Sandalcioglu, I.E.; Zhang, J.-M.; Sure, U. Differential angiogenesis function of CCM2 and CCM3 in cerebral cavernous malformations. Neurosurg. Focus 2010, 29, E1. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tanriover, G.; Yano, H.; Friedlander, R.; Louvi, A.; Gunel, M. Apoptotic Functions of PDCD10/CCM3, the Gene Mutated in Cerebral Cavernous Malformation 3. Stroke 2009, 40, 1474–1481. [Google Scholar] [CrossRef] [PubMed]
- Fidalgo, M.; Guerrero, A.; Fraile, M.; Iglesias, C.; Pombo, C.M.; Zalvide, J. Adaptor Protein Cerebral Cavernous Malformation 3 (CCM3) Mediates Phosphorylation of the Cytoskeletal Proteins Ezrin/Radixin/Moesin by Mammalian Ste20-4 to Protect Cells from Oxidative Stress. J. Biol. Chem. 2012, 287, 11556–11565. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, X.; Deng, X.; Chen, Y.; Mo, X.; Zhang, Y.; Zhao, H.; Ma, D. PDCD10 interacts with STK25 to accelerate cell apoptosis under oxidative stress. Front. Biosci. 2012, 17, 2295–3305. [Google Scholar] [CrossRef]
- Schleider, E.; Stahl, S.; Wüstehube, J.; Walter, U.; Fischer, A.; Felbor, U. Evidence for anti-angiogenic and pro-survival functions of the cerebral cavernous malformation protein 3. Neurogenetics 2010, 12, 83–86. [Google Scholar] [CrossRef][Green Version]
- Sugden, P.H.; McGuffin, L.J.; Clerk, A. SOcK, MiSTs, MASK and STicKs: The GCKIII (germinal centre kinase III) kinases and their heterologous protein–protein interactions. Biochem. J. 2013, 454, 13–30. [Google Scholar] [CrossRef]
- Zhou, H.J.; Qin, L.; Zhang, H.; Tang, W.; Ji, W.; He, Y.; Liang, X.; Wang, Z.; Yuan, Q.; Vortmeyer, A.; et al. Endothelial exocytosis of angiopoietin-2 resulting from CCM3 deficiency contributes to cerebral cavernous malformation. Nat. Med. 2016, 22, 1033–1042. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, W.; Zhang, H.; Niu, X.; Xu, Y.; Zhang, J.; Gao, K.; Pan, W.; Boggon, T.J.; Toomre, D.; et al. A Network of Interactions Enables CCM3 and STK24 to Coordinate UNC13D-Driven Vesicle Exocytosis in Neutrophils. Dev. Cell 2013, 27, 215–226. [Google Scholar] [CrossRef]
- Han, Y.; Wang, X. The emerging roles of KPNA2 in cancer. Life Sci. 2019, 241, 117140. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, T.; Johnson, E.W.; Thomas, J.W.; Kuehl, P.M.; Jones, T.L.; Dokken, C.G.; Touchman, J.W.; Gallione, C.J.; Lee-Lin, S.-Q.; Kosofsky, B.; et al. Mutations in the Gene Encoding KRIT1, a Krev-1/rap1a Binding Protein, Cause Cerebral Cavernous Malformations (CCM1). Hum. Mol. Genet. 1999, 8, 2325–2333. [Google Scholar] [CrossRef] [PubMed]
- Liquori, C.L.; Berg, M.J.; Siegel, A.M.; Huang, E.; Zawistowski, J.S.; Stoffer, T.; Verlaan, D.; Balogun, F.; Hughes, L.; Leedom, T.P.; et al. Mutations in a Gene Encoding a Novel Protein Containing a Phosphotyrosine-Binding Domain Cause Type 2 Cerebral Cavernous Malformations. Am. J. Hum. Genet. 2003, 73, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Dashti, S.; Hoffer, A.; Hu, Y.C.; Selman, W.R. Molecular genetics of familial cerebral cavernous malformations. Neurosurg. Focus 2006, 21, 1–5. [Google Scholar] [CrossRef]
- Stockton, R.A.; Shenkar, R.; Awad, I.A.; Ginsberg, M.H. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J. Exp. Med. 2010, 207, 881–896. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Y.; Whitehead, K.J. Evaluating Strategies for the Treatment of Cerebral Cavernous Malformations. Stroke 2010, 41, S92–S94. [Google Scholar] [CrossRef]
- Whitehead, K.J.; Chan, A.C.; Navankasattusas, S.; Koh, W.; London, N.R.; Ling, J.; Mayo, A.H.; Drakos, S.G.; Jones, C.A.; Zhu, W.; et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat. Med. 2009, 15, 177–184. [Google Scholar] [CrossRef]
- Cullere, X.; Plovie, E.; Bennett, P.M.; MacRae, C.A.; Mayadas, T.N. The cerebral cavernous malformation proteins CCM2L and CCM2 prevent the activation of the MAP kinase MEKK3. Proc. Natl. Acad. Sci. USA 2015, 112, 14284–14289. [Google Scholar] [CrossRef]
- Fisher, O.S.; Deng, H.; Liu, D.; Zhang, Y.; Wei, R.; Deng, Y.; Zhang, F.; Louvi, A.; Turk, B.E.; Boggon, T.J.; et al. Structure and vascular function of MEKK3–cerebral cavernous malformations 2 complex. Nat. Commun. 2015, 6, 7937. [Google Scholar] [CrossRef]
- Zhou, Z.; Tang, A.T.; Wong, W.-Y.; Bamezai, S.; Goddard, L.M.; Shenkar, R.; Zhou, S.; Yang, J.; Wright, A.C.; Foley, M.; et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3–KLF2/4 signalling. Nature 2016, 532, 122–126. [Google Scholar] [CrossRef]
- Cuttano, R.; Rudini, N.; Bravi, L.; Corada, M.; Giampietro, C.; Papa, E.; Morini, M.F.; Maddaluno, L.; Baeyens, N.; Adams, R.H.; et al. KLF 4 is a key determinant in the development and progression of cerebral cavernous malformations. EMBO Mol. Med. 2015, 8, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Maddaluno, L.; Rudini, N.; Cuttano, R.; Bravi, L.; Giampietro, C.; Corada, M.; Ferrarini, L.; Orsenigo, F.; Papa, E.; Boulday, G.; et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature 2013, 498, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Wüstehube, J.; Bartol, A.; Liebler, S.S.; Brütsch, R.; Zhu, Y.; Felbor, U.; Sure, U.; Augustin, H.G.; Fischer, A. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 12640–12645. [Google Scholar] [CrossRef]
- Goitre, L.; Balzac, F.; Degani, S.; Degan, P.; Marchi, S.; Pinton, P.; Retta, S.F. KRIT1 Regulates the Homeostasis of Intracellular Reactive Oxygen Species. PLoS ONE 2010, 5, e11786. [Google Scholar] [CrossRef]
- Marchi, S.; Corricelli, M.; Trapani, E.; Bravi, L.; Pittaro, A.; Monache, S.D.; Ferroni, L.; Patergnani, S.; Missiroli, S.; Goitre, L.; et al. Defective autophagy is a key feature of cerebral cavernous malformations. EMBO Mol. Med. 2015, 7, 1403–1417. [Google Scholar] [CrossRef]
- You, C.; Sandalcioglu, I.E.; Dammann, P.; Felbor, U.; Sure, U.; Zhu, Y. Loss of CCM3 impairs DLL4-Notch signalling: Implication in endothelial angiogenesis and in inherited cerebral cavernous malformations. J. Cell. Mol. Med. 2013, 17, 407–418. [Google Scholar] [CrossRef]
- Wang, K.; Zhou, H.J.; Wang, M. CCM3 and cerebral cavernous malformation disease. Stroke Vasc. Neurol. 2019, 4, 67–70. [Google Scholar] [CrossRef]
- Shenkar, R.; Shi, C.; Rebeiz, T.; Stockton, R.A.; McDonald, D.A.; Mikati, A.G.; Zhang, L.; Austin, C.; Akers, A.L.; Gallione, C.J.; et al. Exceptional aggressiveness of cerebral cavernous malformation disease associated with PDCD10 mutations. Genet. Med. 2015, 17, 188–196. [Google Scholar] [CrossRef]
- Chan, A.C.; Drakos, S.G.; Ruiz, O.E.; Smith, A.C.; Gibson, C.C.; Ling, J.; Passi, S.F.; Stratman, A.N.; Sacharidou, A.; Revelo, M.P.; et al. Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J. Clin. Investig. 2011, 121, 1871–1881. [Google Scholar] [CrossRef]
- Peach, C.J.; Mignone, V.W.; Arruda, M.A.; Alcobia, D.C.; Hill, S.J.; Kilpatrick, L.E.; Woolard, J. Molecular Pharmacology of VEGF-A Isoforms: Binding and Signalling at VEGFR2. Int. J. Mol. Sci. 2018, 19, 1264. [Google Scholar] [CrossRef]
- Ripamonti, M.; Wehrle-Haller, B.; de Curtis, I. Paxillin: A Hub for Mechano-Transduction from the β3 Integrin-Talin-Kindlin Axis. Front. Cell Dev. Biol. 2022, 10, 852016. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ji, W.; Zhang, R.; Folta-Stogniew, E.; Min, W.; Boggon, T.J. Molecular Recognition of Leucine-Aspartate Repeat (LD) Motifs by the Focal Adhesion Targeting Homology Domain of Cerebral Cavernous Malformation 3 (CCM3). J. Biol. Chem. 2011, 286, 26138–26147. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, H.; He, Y.; Jiang, Q.; Tanaka, Y.; Park, I.-H.; Pober, J.S.; Min, W.; Zhou, H.J. Mural Cell-Specific Deletion of Cerebral Cavernous Malformation 3 in the Brain Induces Cerebral Cavernous Malformations. Arter. Thromb. Vasc. Biol. 2020, 40, 2171–2186. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.J.; Qin, L.; Jiang, Q.; Murray, K.N.; Zhang, H.; Li, B.; Lin, Q.; Graham, M.; Liu, X.; Grutzendler, J.; et al. Caveolae-mediated Tie2 signaling contributes to CCM pathogenesis in a brain endothelial cell-specific Pdcd10-deficient mouse model. Nat. Commun. 2021, 12, 1–22. [Google Scholar] [CrossRef] [PubMed]
- You, C.; Zhao, K.; Dammann, P.; Keyvani, K.; Kreitschmann-Andermahr, I.; Sure, U.; Zhu, Y. EphB4 forward signalling mediates angiogenesis caused by CCM3/PDCD10 -ablation. J. Cell. Mol. Med. 2017, 21, 1848–1858. [Google Scholar] [CrossRef] [PubMed]
- Tang, A.T.; Sullivan, K.R.; Hong, C.C.; Goddard, L.M.; Mahadevan, A.; Ren, A.; Pardo, H.; Peiper, A.; Griffin, E.; Tanes, C.; et al. Distinct cellular roles for PDCD10 define a gut-brain axis in cerebral cavernous malformation. Sci. Transl. Med. 2019, 11, eaaw3521. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhong, F.-J.; Xu, C.; Li, Y.-M.; Zhao, Y.-R.; Cao, M.-M.; Yang, L.-Y. Programmed cell death 10 promotes metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma via PP2Ac-mediated YAP activation. Cell Death Dis. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Madsen, C.D.; Hooper, S.; Tozluoglu, M.; Bruckbauer, A.; Fletcher, G.; Erler, J.; Bates, P.; Thompson, B.; Sahai, E. STRIPAK components determine mode of cancer cell migration and metastasis. Nature 2014, 17, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Moqrich, A.; Mattei, M.; Bartoli, M.; Rakitina, T.; Baillat, G.; Monneron, A.; Castets, F. Cloning of Human Striatin cDNA (STRN), Gene Mapping to 2p22–p21, and Preferential Expression in Brain. Genomics 1998, 51, 136–139. [Google Scholar] [CrossRef]
- Castets, F.; Rakitina, T.; Gaillard, S.; Moqrich, A.; Mattei, M.-G.; Monneron, A. Zinedin, SG2NA, and Striatin Are Calmodulin-binding, WD Repeat Proteins Principally Expressed in the Brain. J. Biol. Chem. 2000, 275, 19970–19977. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, X.; Peng, S.; Nan, X.; Zhao, H. Differential expression of MST4, STK25 and PDCD10 between benign prostatic hyperplasia and prostate cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 8105–8111. [Google Scholar] [PubMed]
- Mandell, M.A.; Jain, A.; Arko-Mensah, J.; Chauhan, S.; Kimura, T.; Dinkins, C.; Silvestri, G.; Münch, J.; Kirchhoff, F.; Simonsen, A.; et al. TRIM Proteins Regulate Autophagy and Can Target Autophagic Substrates by Direct Recognition. Dev. Cell 2014, 30, 394–409. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; He, L.; Zhou, Y. TRIM59 deficiency curtails breast cancer metastasis through SQSTM1-selective autophagic degradation of PDCD10. Autophagy 2019, 15, 747–749. [Google Scholar] [CrossRef]
- Wu, K.; Mu, X.; Jiang, J.; Tan, M.; Wang, R.; Zhou, W.; Wang, X.; He, Y.; Li, M.; Liu, Z. miRNA-26a-5p and miR-26b-5p inhibit the proliferation of bladder cancer cells by regulating PDCD10. Oncol. Rep. 2018, 40, 3523–3532. [Google Scholar] [CrossRef]
- Yang, D.; Wang, J.-J.; Li, J.-S.; Xu, Q.-Y. miR-103 Functions as a Tumor Suppressor by Directly Targeting Programmed Cell Death 10 in NSCLC. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 26, 519–528. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, W.; Su, Y.; Lu, L.; Wang, D.; Wang, H. MicroRNA-103 suppresses tumor cell proliferation by targeting PDCD10 in prostate cancer. Prostate 2016, 76, 543–551. [Google Scholar] [CrossRef]
- Dong, C.; Fan, B.; Ren, Z.; Liu, B.; Wang, Y. CircSMARCA5 Facilitates the Progression of Prostate Cancer Through miR-432/PDCD10 Axis. Cancer Biother. Radiopharm. 2021, 36, 70–83. [Google Scholar] [CrossRef]
- Feng, Z.-M.; Qiu, J.; Chen, X.-W.; Liao, R.-X.; Liao, X.-Y.; Zhang, L.-P.; Chen, X.; Li, Y.; Chen, Z.-T.; Sun, J.-G. Essential role of miR-200c in regulating self-renewal of breast cancer stem cells and their counterparts of mammary epithelium. BMC Cancer 2015, 15, 1–12. [Google Scholar] [CrossRef][Green Version]
- Xu, X.; Ge, S.; Jia, R.; Zhou, Y.; Song, X.; Zhang, H.; Fan, X. Hypoxia-induced miR-181b enhances angiogenesis of retinoblastoma cells by targeting PDCD10 and GATA6. Oncol. Rep. 2015, 33, 2789–2796. [Google Scholar] [CrossRef]
- Fan, L.; Lei, H.; Zhang, S.; Peng, Y.; Fu, C.; Shu, G.; Yin, G. Non-canonical signaling pathway of SNAI2 induces EMT in ovarian cancer cells by suppressing miR-222-3p transcription and upregulating PDCD10. Theranostics 2020, 10, 5895–5913. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, X.; Miao, X.; Zhu, K.; Cui, S.; Meng, Q.; Sun, J.; Wang, T. Micro RNA -425-5p regulates chemoresistance in colorectal cancer cells via regulation of Programmed Cell Death 10. J. Cell. Mol. Med. 2016, 20, 360–369. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, M. Novel exosomal miR-46146 transfer oxaliplatin chemoresistance in colorectal cancer. Clin. Transl. Oncol. 2019, 22, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Lauenborg, B.; Kopp, K.L.; Krejsgaard, T.; Eriksen, K.W.; Geisler, C.; Dabelsteen, S.; Gniadecki, R.; Zhang, Q.; Wasik, M.A.; Woetmann, A.; et al. Programmed cell death-10 enhances proliferation and protects malignant T cells from apoptosis. APMIS 2010, 118, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, J.; Yao, X.; Wu, S.; Tian, W.; Gan, C.; Wan, X.; You, C.; Hu, F.; Zhang, S.; et al. Programmed Cell Death 10 Mediated CXCL2-CXCR2 Signaling in Regulating Tumor-Associated Microglia/Macrophages Recruitment in Glioblastoma. Front. Immunol. 2021, 12, 637053. [Google Scholar] [CrossRef] [PubMed]
- De Marco, C.; Zoppoli, P.; Rinaldo, N.; Morganella, S.; Morello, M.; Zuccalà, V.; Carriero, M.V.; Malanga, D.; Chirillo, R.; Bruni, P.; et al. Genome-wide analysis of copy number alterations led to the characterisation of PDCD10 as oncogene in ovarian cancer. Transl. Oncol. 2021, 14, 101013. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, J.; Tian, W.; Xu, Y.; Li, R.; Zhao, K.; You, C.; Zhu, Y.; Bartsch, J.W.; Niu, H.; et al. PDCD10 promotes the aggressive behaviors of pituitary adenomas by up-regulating CXCR2 and activating downstream AKT/ERK signaling. Aging 2022, 14, 6066–6080. [Google Scholar] [CrossRef]
- Urfali-Mamatoglu, C.; Kazan, H.H.; Gündüz, U. Dual function of programmed cell death 10 (PDCD10) in drug resistance. Biomed. Pharmacother. 2018, 101, 129–136. [Google Scholar] [CrossRef]
- Riant, F.; Bergametti, F.; Fournier, H.-D.; Chapon, F.; Michalak-Provost, S.; Cecillon, M.; Lejeune, P.; Hosseini, H.; Choe, C.; Orth, M.; et al. CCM3 Mutations Are Associated with Early-Onset Cerebral Hemorrhage and Multiple Meningiomas. Mol. Syndr. 2013, 4, 165–172. [Google Scholar] [CrossRef]
- Garaci, F.; Marsili, L.; Riant, F.; Marziali, S.; Cécillon, M.; Pasquarelli, R.; Sangiuolo, F.; Floris, R.; Novelli, G.; Tournier-Lasserve, E.; et al. Cerebral cavernous malformations associated to meningioma: High penetrance in a novel family mutated in the PDCD10 gene. Neuroradiol. J. 2015, 28, 289–293. [Google Scholar] [CrossRef]
- Fauth, C.; Rostasy, K.; Rath, M.; Gizewski, E.R.; Lederer, A.; Sure, U.; Zschocke, J.; Felbor, U. Highly variable intrafamilial manifestations of a CCM3 mutation ranging from acute childhood cerebral haemorrhage to late-onset meningiomas. Clin. Neurol. Neurosurg. 2015, 128, 41–43. [Google Scholar] [CrossRef]
- Nickel, A.-C.; Wan, X.-Y.; Saban, D.-V.; Weng, Y.-L.; Zhang, S.; Keyvani, K.; Sure, U.; Zhu, Y. Loss of programmed cell death 10 activates tumor cells and leads to temozolomide-resistance in glioblastoma. J. Neuro-Oncol. 2018, 141, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Saban, D.V.; Na Kim, S.; Weng, Y.; Dammann, P.; Keyvani, K.; Sure, U.; Zhu, Y. PDCD10-Deficiency Promotes Malignant Behaviors and Tumor Growth via Triggering EphB4 Kinase Activity in Glioblastoma. Front. Oncol. 2020, 10, 1377. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhao, K.; Prinz, A.; Keyvani, K.; Lambertz, N.; Kreitschmann-Andermahr, I.; Lei, T.; Sure, U. Loss of endothelial programmed cell death 10 activates glioblastoma cells and promotes tumor growth. Neuro-Oncol. 2015, 18, 538–548. [Google Scholar] [CrossRef]
- Lawal, B.; Kuo, Y.-C.; Tang, S.-L.; Liu, F.-C.; Wu, A.T.H.; Lin, H.-Y.; Huang, H.-S. Transcriptomic-Based Identification of the Immuno-Oncogenic Signature of Cholangiocarcinoma for HLC-018 Multi-Target Therapy Exploration. Cells 2021, 10, 2873. [Google Scholar] [CrossRef]
- Nanjundan, M.; Nakayama, Y.; Cheng, K.W.; Lahad, J.; Liu, J.; Lu, K.; Kuo, W.-L.; Smith-McCune, K.; Fishman, D.; Gray, J.W.; et al. Amplification of MDS1/EVI1 and EVI1, Located in the 3q26.2 Amplicon, Is Associated with Favorable Patient Prognosis in Ovarian Cancer. Cancer Res. 2007, 67, 3074–3084. [Google Scholar] [CrossRef]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef]
- Yeung, K.T.; Yang, J. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2016, 11, 28–39. [Google Scholar] [CrossRef]
- Mittal, V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu. Rev. Pathol. 2018, 13, 395–412. [Google Scholar] [CrossRef]
- Singh, M.; Yelle, N.; Venugopal, C.; Singh, S.K. EMT: Mechanisms and therapeutic implications. Pharmacol. Ther. 2018, 182, 80–94. [Google Scholar] [CrossRef]
- Biffi, G.; Tuveson, D.A. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol. Rev. 2021, 101, 147–176. [Google Scholar] [CrossRef]
- Miller, K.D.; Ostrom, Q.T.; Kruchko, C.; Patil, N.; Tihan, T.; Cioffi, G.; Fuchs, H.E.; Waite, K.A.; Jemal, A.; Siegel, R.L.; et al. Brain and other central nervous system tumor statistics, 2021. CA A Cancer J. Clin. 2021, 71, 381–406. [Google Scholar] [CrossRef] [PubMed]
- Louvi, A.; Chen, L.; Two, A.M.; Zhang, H.; Min, W.; Günel, M. Loss of cerebral cavernous malformation 3 ( Ccm3 ) in neuroglia leads to CCM and vascular pathology. Proc. Natl. Acad. Sci. USA 2011, 108, 3737–3742. [Google Scholar] [CrossRef] [PubMed]
- Lambertz, N.; El Hindy, N.; Kreitschmann-Andermahr, I.; Stein, K.P.; Dammann, P.; Oezkan, N.; Mueller, O.; Sure, U.; Zhu, Y. Downregulation of programmed cell death 10 is associated with tumor cell proliferation, hyperangiogenesis and peritumoral edema in human glioblastoma. BMC Cancer 2015, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2020, 221, 107753. [Google Scholar] [CrossRef]
- Boutilier, A.; Elsawa, S. Macrophage Polarization States in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef]
- Peyre, M.; Miyagishima, D.; Bielle, F.; Chapon, F.; Sierant, M.; Venot, Q.; Lerond, J.; Marijon, P.; Abi-Jaoude, S.; Le Van, T.; et al. Somatic PIK3CA Mutations in Sporadic Cerebral Cavernous Malformations. N. Engl. J. Med. 2021, 385, 996–1004. [Google Scholar] [CrossRef]
- Ren, A.A.; Snellings, D.A.; Su, Y.S.; Hong, C.C.; Castro, M.; Tang, A.T.; Detter, M.R.; Hobson, N.; Girard, R.; Romanos, S.; et al. PIK3CA and CCM mutations fuel cavernomas through a cancer-like mechanism. Nature 2021, 594, 271–276. [Google Scholar] [CrossRef]
- Ercoli, J.; Finetti, F.; Woodby, B.; Belmonte, G.; Miracco, C.; Valacchi, G.; Trabalzini, L. KRIT1 as a possible new player in melanoma aggressiveness. Arch. Biochem. Biophys. 2020, 691, 108483. [Google Scholar] [CrossRef]
- Park, S.; Lee, H.-Y.; Kim, J.; Park, H.; Ju, Y.; Kim, E.-G.; Kim, J. Cerebral Cavernous Malformation 1 Determines YAP/TAZ Signaling-Dependent Metastatic Hallmarks of Prostate Cancer Cells. Cancers 2021, 13, 1125. [Google Scholar] [CrossRef]
- Harel, L.; Costa, B.; Tcherpakov, M.; Zapatka, M.; Oberthuer, A.; Hansford, L.M.; Vojvodic, M.; Levy, Z.; Chen, Z.-Y.; Lee, F.S.; et al. CCM2 Mediates Death Signaling by the TrkA Receptor Tyrosine Kinase. Neuron 2009, 63, 585–591. [Google Scholar] [CrossRef]
- Abou-Fadel, J.; Qu, Y.; Gonzalez, E.M.; Smith, M.; Zhang, J. Emerging roles of CCM genes during tumorigenesis with potential application as novel biomarkers across major types of cancers. Oncol. Rep. 2020, 43, 1945–1963. [Google Scholar] [CrossRef] [PubMed]

| MicroRNA | Disease/Cell | Function by Targeting PDCD10 | Upstream Regulators | Ref |
|---|---|---|---|---|
| miR-1-3p | HEK293 cell | associated with cell death induced by ochratoxin A | / | [40] |
| miR-107 | Mesenchymal stem cell | promotes cell survival during ischemic preconditioning | HIF-1α | [41] |
| miR-107 | Alzheimer’s disease | attenuates neurotoxicity | / | [15] |
| miR-495 | Ankylosing spondylitis | involved in pathogenesis of ankylosing spondylitis | / | [16] |
| miR-613 | Myocardial ischemia/reperfusion injury | suppresses ischemia-reperfusion-induced cardiomyocyte apoptosis | / | [14] |
| miR-26-5p | Bladder cancer | inhibits the tumor | / | [84] |
| miR-103 | Non-small cell lung cancer | inhibits cell proliferation, migration, and invasion | / | [85] |
| miR-103 | Prostate cancer | inhibits cell proliferation and invasion | / | [86] |
| miR-432 | Prostate cancer | inhibits proliferation, metastasis, and glycolysis | CircSMARCA5 | [87] |
| miR-200c | Breast cancer stem cell | inhibits self-renewal of stem cell | / | [88] |
| miR-181b | Retinoblastoma | promotes angiogenesis induced by hypoxia | / | [89] |
| miR-222-3p | Ovarian cancer | inhibits EMT | SNAI2 | [90] |
| miR-425-5p | Colorectal cancer | enhances chemoresistance | / | [91] |
| miR-46146 | Colorectal cancer | enhances chemoresistance | / | [92] |
| Pro-Tumorigenic Effects of PDCD10 | |||
| Cancer/Cell Types | Interactions/Pathways | Effects | Ref |
| Lymphomas | - | promoted proliferation and decreased apoptosis | [93] |
| Glioma | CXCL2-CXCR2 | promoted recruitment and M2 polarization of glioma-associated microglia/macrophages | [94] |
| Non-small cell lung cancer | - | promoted cell proliferation and migration | [85] |
| Breast cancer | TRIM59; STK24&26; Rho/ROCK signaling | promoted survival, stemness, EMT, and metastasis of tumor cells | [29,78,88] |
| Liver cancer | PP2A-YAP signaling | promoted cell proliferation, migration, invasion/metastasis, and EMT | [77] |
| Ovarian cancer | ERK signaling; RhoA signaling; β-catenin signaling | promoted cell proliferation, migration, and EMT | [90,95] |
| Prostate cancer | - | Promoted cell proliferation, migration, EMT, and glycolysis | [86,87] |
| Bladder cancer | - | promoted cell proliferation | [84] |
| Pituitary adenomas | CXCR2-Akt/Erk signaling | promoted cell proliferation, migration, and invasion | [96] |
| Cervical cancer | - | promoted chemoresistance of tumor cells | [97] |
| Anti-Tumorigenic Effects of PDCD10 | |||
| Cancer/Cell Types | Interactions/Pathways | Effects | Ref |
| Meningioma | - | PDCD10 deletion was specifically associated with multiple meningiomas | [98,99,100] |
| Glioma | Soluble factors; EphB4- Erk1/2 | Knockdown of PDCD10 facilitated tumor cell proliferation, migration, invasion, and stimulated angiogenesis. | [101,102] |
| Retinoblastoma | - | Downregulation of PDCD10 in retinoblastoma cells promoted angiogenesis | [89] |
| Colorectal cancer | - | Loss of PDCD10 enhanced chemoresistance of tumor cells | [91,92] |
| Breast cancer | - | Loss of PDCD10 enhanced chemoresistance of tumor cells | [97] |
| Endothelial cells | Growth factors signaling | Knockdown of endothelial PDCD10 stimulated angiogenesis and tumor growth of glioma | [103] |
| Cancer-associated fibroblasts | Paxillin | Loss of PDCD10 in CAFs drove remodeling of extracellular matrix network and increased metastatic spread of tumor | [5] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Zhao, K.; Wu, S.; Li, C.; You, C.; Wang, J.; Shu, K.; Lei, T. The Dual Role of PDCD10 in Cancers: A Promising Therapeutic Target. Cancers 2022, 14, 5986. https://doi.org/10.3390/cancers14235986
Liu J, Zhao K, Wu S, Li C, You C, Wang J, Shu K, Lei T. The Dual Role of PDCD10 in Cancers: A Promising Therapeutic Target. Cancers. 2022; 14(23):5986. https://doi.org/10.3390/cancers14235986
Chicago/Turabian StyleLiu, Jingdian, Kai Zhao, Sisi Wu, Chaoxi Li, Chao You, Junwen Wang, Kai Shu, and Ting Lei. 2022. "The Dual Role of PDCD10 in Cancers: A Promising Therapeutic Target" Cancers 14, no. 23: 5986. https://doi.org/10.3390/cancers14235986
APA StyleLiu, J., Zhao, K., Wu, S., Li, C., You, C., Wang, J., Shu, K., & Lei, T. (2022). The Dual Role of PDCD10 in Cancers: A Promising Therapeutic Target. Cancers, 14(23), 5986. https://doi.org/10.3390/cancers14235986






