Interobserver Variability Prediction of Primary Gross Tumor in a Patient with Non-Small Cell Lung Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods and Materials
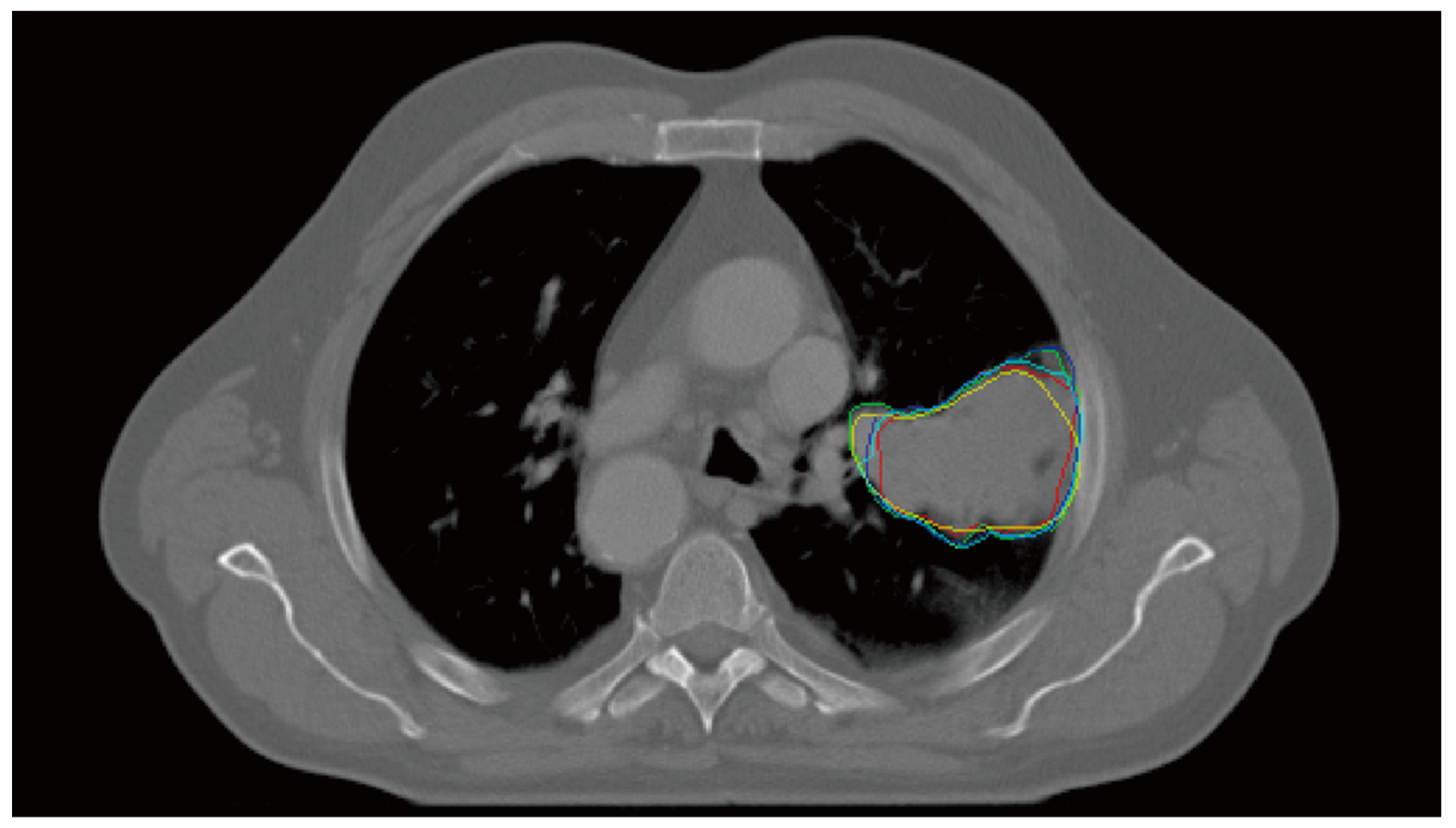
2.1. Multiple Delineation Database
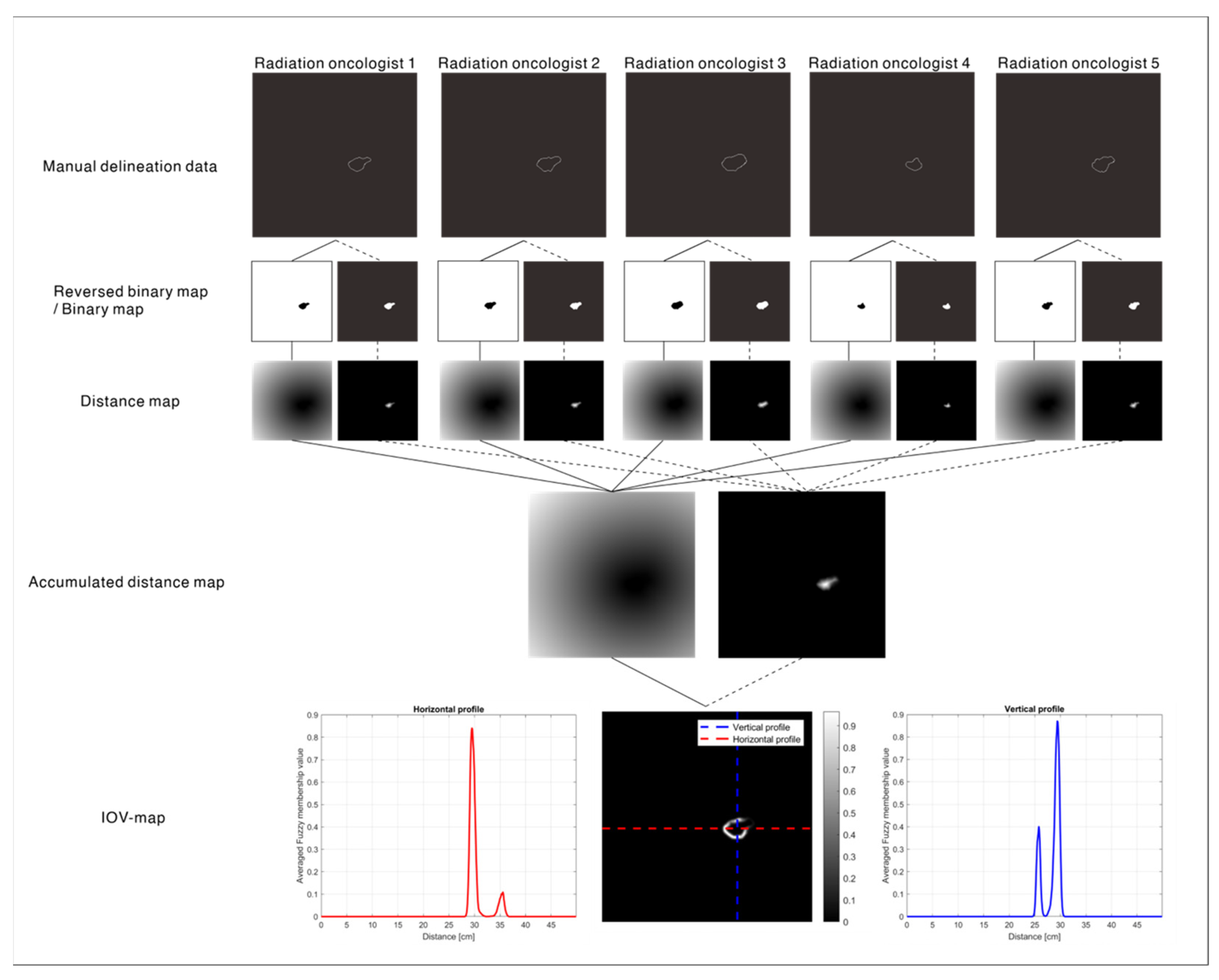
2.2. IOV Map: Ground Truth
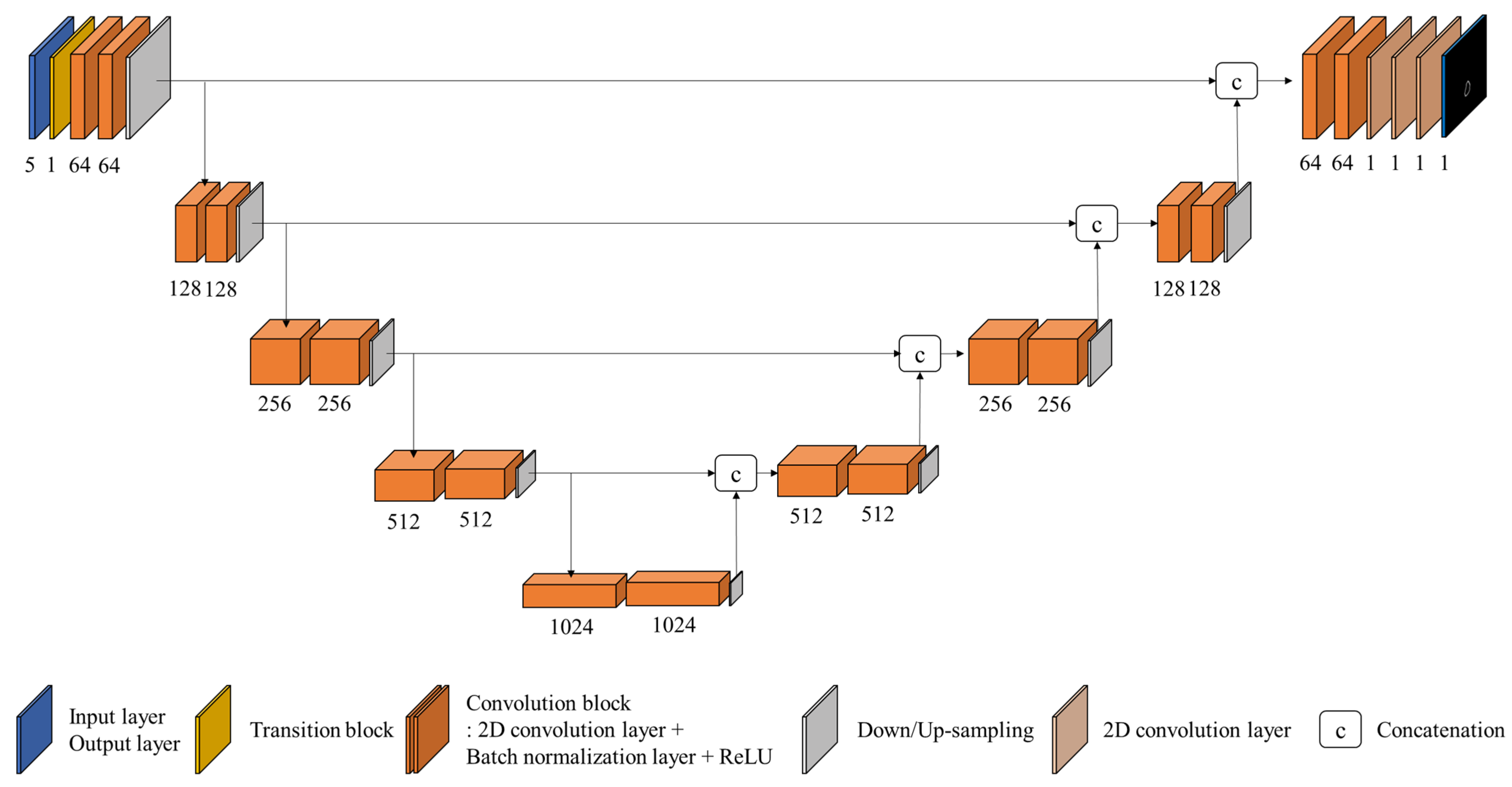
2.3. IOV Prediction Model: IOV-Net
2.4. Data Preparation for Training and Clinical Feasibility Validation
2.5. Training and Validation of the IOV-Net
2.6. Evaluation Metrics for the IOV-Net and Clinical Validation
3. Results
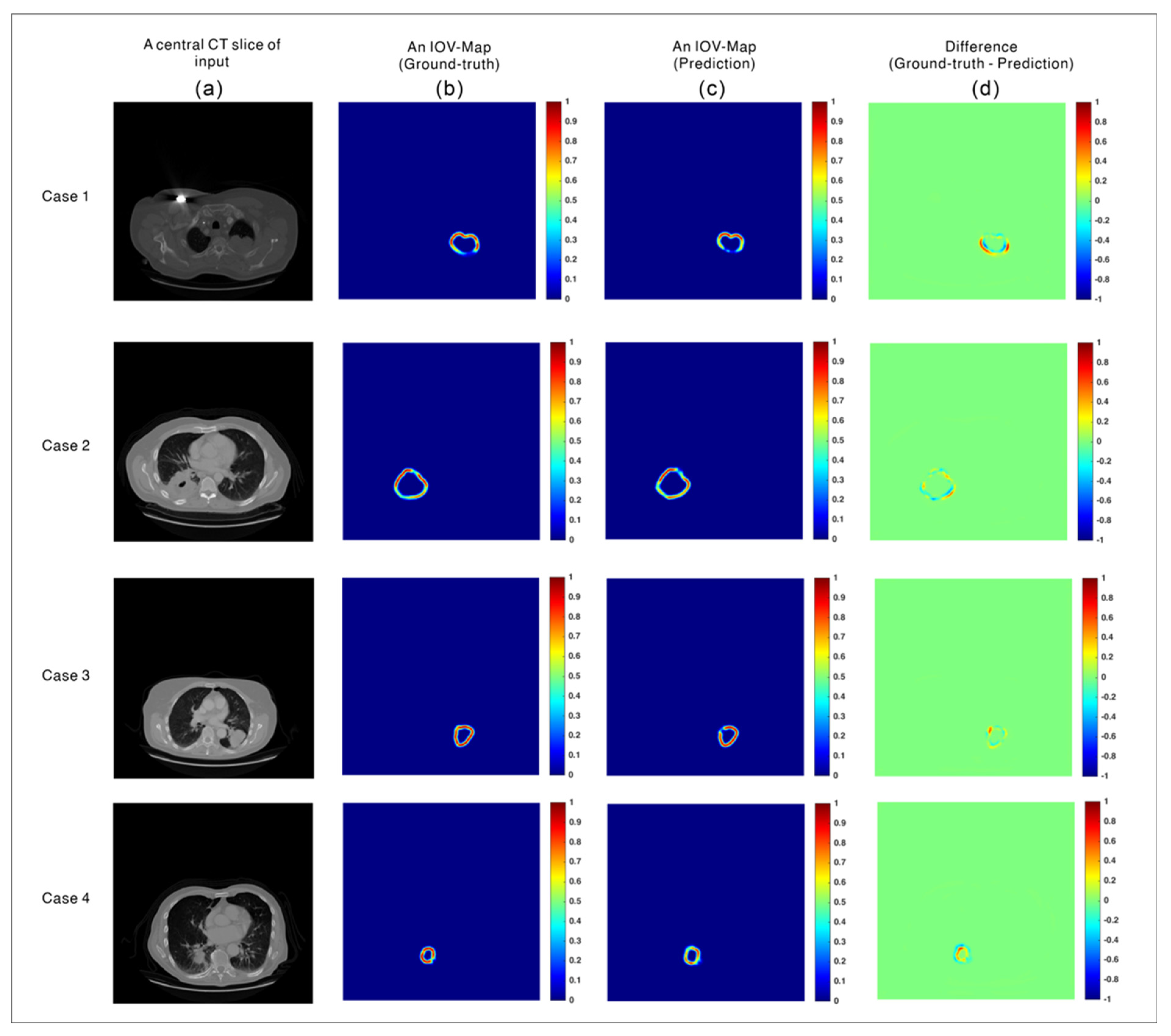
3.1. Evaluation of the Prediction Accuracy of the IOV-Net
3.2. Analysis of the Effect of the IOV Map on the Reduction of IOV in Clinics
4. Discussion
- IOV-Net, which applies an innovative implementation of the signed Euclidean distance transform and fuzzy membership function to predict the IOV map from input CT images without the need for additional human resources, image data, and time;
- A clinically effective method of reducing the IOV of clinician pGTV contour annotations, as validated for a set of NSCLC patients.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hong, S.; Won, Y.J.; Lee, J.J.; Jung, K.W.; Kong, H.J.; Im, J.S.; Seo, H.G. Community of population-based regional cancer registries. Cancer statistics in Korea: Incidence, mortality, survival, and prevalence in 2018. Cancer Res. Treat. 2021, 53, 301–315. [Google Scholar] [CrossRef]
- Liu, X.; Huang, E.; Wang, Y.; He, Y.; Luo, H.; Zhong, M.; Qiu, D.; Li, C.; Yang, H.; He, G.; et al. Dosimetric comparison of helical tomotherapy, VMAT, fixed-field IMRT, and 3D-conformal radiotherapy for stage I-II nasal natural killer T-cell lymphoma. Radiat. Oncol. 2017, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, S.; Sung, J.; Yoon, M. Radiotherapy-induced secondary cancer risk for breast cancer: 3D conformal therapy versus IMRT versus VMAT. J. Radiol. Protect. 2014, 34, 325. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xie, C.; Hu, M.; Han, C.; Yi, J.; Zhou, Y.; Yan, H.; Jin, X. Dosimetric benefits of IMRT and VMAT in the treatment of middle thoracic esophageal cancer: Is the conformal radiotherapy still an alternative option? J. Appl. Clin. Med. Phys. 2014, 15, 93–101. [Google Scholar] [CrossRef]
- Beltran, C.; Roca, M.; Merchant, T.E. On the benefits and risks of proton therapy in pediatric craniopharyngioma. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, e281–e287. [Google Scholar] [CrossRef]
- Widesott, L.; Pierelli, A.; Fiorino, C.; Dell’Oca, I.; Broggi, S.; Cattaneo, G.M.; Di Muzio, N.; Fazio, F.; Calandrino, R.; Schwarz, M. Intensity-modulated proton therapy versus helical tomotherapy in nasopharynx cancer: Planning comparison and NTCP evaluation. Int. J. Radiat. Oncol. Biol. Phys. 2008, 72, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Lim, G.J.; Lee, A.; Li, Y.; Liu, W.; Zhu, R.X.; Zhang, X. Uncertainty incorporated beam angle optimization for IMPT treatment planning. Med. Phys. 2012, 39, 5248–5256. [Google Scholar] [CrossRef] [PubMed]
- Lomax, A.J.; Boehringer, T.; Coray, A.; Egger, E.; Goitein, G.; Grossmann, M.; Juelke, P.; Lin, S.; Pedroni, E.; Rohrer, B.; et al. Intensity modulated proton therapy: A clinical example. Med. Phys. 2001, 28, 317–324. [Google Scholar] [CrossRef]
- Liu, C.; Schild, S.E.; Chang, J.Y.; Liao, Z.; Korte, S.; Shen, J.; Ding, X.; Hu, Y.; Kang, Y.; Keole, S.R.; et al. Impact of spot size and spacing on the quality of robustly optimized intensity modulated proton therapy plans for lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 2018, 101, 479–489. [Google Scholar] [CrossRef]
- Gorgisyan, J.; Rosenschold, P.M.A.; Perrin, R.; Persson, G.F.; Josipovic, M.; Belosi, M.F.; Engelholm, S.A.; Weber, D.C.; Lomax, A.J. Feasibility of pencil beam scanned intensity modulated proton therapy in breath-hold for locally advanced non-small cell lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, 1121–1128. [Google Scholar] [CrossRef]
- Van Marlen, P.; Dahele, M.; Folkerts, M.; Abel, E.; Slotman, B.J.; Verbakel, W.F. Bringing FLASH to the clinic: Treatment planning considerations for ultrahigh dose-rate proton beams. Int. J. Radiat. Oncol. Biol. Phys. 2020, 106, 621–629. [Google Scholar] [CrossRef]
- Tinganelli, W.; Sokol, O.; Quartieri, M.; Puspitasari, A.; Dokic, I.; Abdollahi, A.; Durante, M.; Haberer, T.; Debus, J.; Boscolo, D.; et al. Ultra-high dose rate (FLASH) carbon ion irradiation: Dosimetry and first cell experiments. Int. J. Radiat. Oncol. Biol. Phys. 2022, 112, 1012–1022. [Google Scholar] [CrossRef] [PubMed]
- De Kruijff, R.M. FLASH radiotherapy: Ultra-high dose rates to spare healthy tissue. Int. J. Radiat. Biol. 2020, 96, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Hofmaier, J.; Walter, F.; Hadi, I.; Rottler, M.; von Bestenbostel, R.; Dedes, G.; Parodi, K.; Niyazi, M.; Belka, C.; Kamp, F. Combining inter-observer variability, range, and setup uncertainty in a variance-based sensitivity analysis for proton therapy. Phys. Imaging Radiat. Oncol. 2021, 20, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.C.; Liu, M.; Chan, E.; Lund, C.; Truong, P.T.; Loewen, S.; Cao, J.; Schellenberg, D.; Carolan, H.; Berrang, T.; et al. The impact of peer review of volume delineation in stereotactic body radiation therapy planning for primary lung cancer: A multicenter quality assurance study. J. Thorac. Oncol. 2014, 9, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Jameson, M.G.; Kumar, S.; Vinod, S.K.; Metcalfe, P.E.; Holloway, L.C. Correlation of contouring variation with modeled outcome for conformal non-small cell lung cancer radiotherapy. Radiother. Oncol. 2014, 112, 332–336. [Google Scholar] [CrossRef]
- Vinod, S.K.; Min, M.; Jameson, M.G.; Holloway, L.C. A review of interventions to reduce inter-observer variability in volume delineation in radiation oncology. J. Med. Imaging Radiat. Oncol. 2016, 60, 393–406. [Google Scholar] [CrossRef]
- Dadone, P. Design Optimization of Fuzzy Logic Systems. Ph.D. Thesis, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 2001. [Google Scholar]
- Chen, L.; Liang, X.; Shen, C.; Jiang, S.; Wang, J. Synthetic CT generation from CBCT images via deep learning. Med. Phys. 2020, 47, 1115–1125. [Google Scholar] [CrossRef]
- Zhang, Y.; Yue, N.; Su, M.Y.; Liu, B.; Ding, Y.; Zhou, Y.; Wang, H.; Kuang, Y.; Nie, K. Improving CBCT quality to CT level using deep learning with generative adversarial network. Med. Phys. 2021, 48, 2816–2826. [Google Scholar] [CrossRef]
- Liu, Y.; Lei, Y.; Wang, Y.; Shafai-Erfani, G.; Wang, T.; Tian, S.; Patel, P.; Jani, A.B.; McDonald, M.; Curran, W.J.; et al. Evaluation of a deep learning-based pelvic synthetic CT generation technique for MRI-based prostate proton treatment planning. Phys. Med. Biol. 2019, 64, 205022. [Google Scholar] [CrossRef]
- Liu, Y.; Lei, Y.; Wang, T.; Kayode, O.; Tian, S.; Liu, T.; Patel, P.; Curran, W.J.; Ren, L.; Yang, X. MRI-based treatment planning for liver stereotactic body radiotherapy: Validation of a deep learning-based synthetic CT generation method. Brit. J. Radiol. 2019, 92, 20190067. [Google Scholar] [CrossRef]
- Cheon, W.; Kim, S.J.; Hwang, U.J.; Min, B.J.; Han, Y. Feasibility study of the fluence-to-dose network (FDNet) for patient-specific IMRT quality assurance. J. Korean Phys. Soc. 2019, 75, 724–734. [Google Scholar] [CrossRef]
- Zhu, W.; Huang, Y.; Zeng, L.; Chen, X.; Liu, Y.; Qian, Z.; Du, N.; Fan, W.; Xie, X. AnatomyNet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med. Phys. 2019, 46, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, J.; Qin, G.; Shen, L.; Sun, Y.; Ying, H.; Zhang, Z.; Hu, W. A deep learning-based auto-segmentation of rectal tumors in MR images. Med. Phys. 2018, 45, 2560–2564. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chang, Y.; Peng, Z.; Lv, Y.; Shi, W.; Wang, F.; Pei, X.; Xu, X.G. Evaluation of deep learning-based auto-segmentation algorithms for delineating clinical target volume and organs at risk involving data for 125 cervical cancer patients. J. Appl. Clin. Med. Phys. 2020, 21, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, P.; Gupta, S.; Koundal, D.; Zaguia, A.; Enbeyle, W. Deep neural networks for medical image segmentation. J. Healthc. Eng. 2022, 2022, 9580991. [Google Scholar] [CrossRef] [PubMed]
- Vu, M.H.; Grimbergen, G.; Nyholm, T.; Löfstedt, T. Evaluation of multislice inputs to convolutional neural networks for medical image segmentation. Med. Phys. 2020, 47, 6216–6231. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17 October 2016; Springer: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional Networks for Biomedical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- El-Sappagh, S.; Ali, F.; Abuhmed, T.; Singh, J.; Alonso, J.M. Automatic detection of Alzheimer’s disease progression: An efficient information fusion approach with heterogeneous ensemble classifiers. Neurocomputing 2022, 512, 203–224. [Google Scholar] [CrossRef]
- Vulli, A.; Srinivasu, P.N.; Sashank, M.S.; Shafi, J.; Choi, J.; Ijaz, M.F. Fine-Tuned DenseNet-169 for Breast Cancer Metastasis Prediction Using FastAI and 1-Cycle Policy. Sensors 2022, 22, 2988. [Google Scholar] [CrossRef]
- Van Baardwijk, A.; Bosmans, G.; Boersma, L.; Buijsen, J.; Wanders, S.; Hochstenbag, M.; Van Suylen, R.J.; Dekker, A.; Dehing-Oberije, C.; Houben, R.; et al. PET-CT–based auto-contouring in non-small-cell lung cancer correlates with pathology and reduces interobserver variability in the delineation of the primary tumor and involved nodal volumes. Int. J. Radiat. Oncol. Biol. Phys. 2007, 68, 771–778. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumor phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.Z. Signed Euclidean distance transform applied to shape analysis. In Issues on Machine Vision (Austria, Vienna); Springer: Vienna, Austria, 1989; pp. 249–262. [Google Scholar]
- Pei, L.; Liu, Y. Multimodal Brain Tumor Segmentation Using a 3D ResUNet in BraTS 2021. In International MICCAI Brain Lesion Workshop; Springer: Cham, Switzerland, 2022; pp. 315–323. [Google Scholar]
- Orr, G.B.; Müller, K.R. (Eds.) Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 1998. [Google Scholar]
- Sutanto, A.R.; Kang, D.K. A Novel Diminish Smooth L1 Loss Model with Generative Adversarial Network. In International Conference on Intelligent Human Computer Interaction; Springer: Cham, Switzerland, 2020; pp. 361–368. [Google Scholar]
- Cheon, W.; Ahn, S.H.; Jeong, S.; Lee, S.B.; Shin, D.; Lim, Y.K.; Jeong, J.H.; Youn, S.H.; Lee, S.U.; Moon, S.H.; et al. Beam angle optimization for double-scattering proton delivery technique using an eclipse application programming interface and convolutional neural network. Front. Oncol. 2021, 11, 707464. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An imperative style, high-performance deep learning library. In Proceedings of the Advances in Neural Information Processing Systems 32 (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019; Volume 32. [Google Scholar]
- Warfield, S.K.; Zou, K.H.; Wells, W.M. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans. Med. Imaging 2004, 23, 903–921. [Google Scholar] [CrossRef]
- Allozi, R.; Li, X.A.; White, J.; Apte, A.; Tai, A.; Michalski, J.M.; Bosch, W.R.; El Naqa, I. Tools for consensus analysis of experts’ contours for radiotherapy structure definitions. Radiother. Oncol. 2010, 97, 572–578. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Ebbinghaus, H. Memory: A contribution to experimental psychology. Ann. Neurosci. 2013, 20, 155. [Google Scholar] [CrossRef]
| Initial Study | CT with the Predicted IOV Map * | CT Only | ||||
| Radiation oncologists | DSC | JI | HD | DSC | JI | HD |
| A | 0.96 | 0.93 | 3.34 | 0.91 | 0.87 | 4.68 |
| B | 0.91 | 0.84 | 8.26 | 0.82 | 0.76 | 7.51 |
| C | 0.94 | 0.91 | 4.72 | 0.78 | 0.73 | 7.32 |
| D | 0.76 | 0.68 | 10.60 | 0.86 | 0.79 | 7.12 |
| Repeated Study (Six Months Later) | CT Only | CT with Predicted IOV Map | ||||
| Radiation oncologists | DSC | JI | HD | DSC | JI | HD |
| A | 0.95 | 0.92 | 4.60 | 0.97 | 0.95 | 3.74 |
| B | 0.82 | 0.72 | 13.54 | 0.85 | 0.77 | 7.24 |
| C | 0.91 | 0.86 | 5.86 | 0.94 | 0.89 | 5.95 |
| D | 0.77 | 0.69 | 9.21 | 0.85 | 0.76 | 7.74 |
| DSC | JI | HD | ||||
|---|---|---|---|---|---|---|
| Difference | p-Value | Difference | p-Value | Difference | p-Value | |
| Radiation oncologist A | 0.058 ± 0.217 | 0.0220 | 0.063 ± 0.229 | 0.0287 | −1.631 ± 4.231 | 0.0369 |
| Radiation oncologist B | 0.051 ± 0.241 | <0.0001 | 0.057 ± 0.237 | 0.0002 | −2.819 ± 9.896 | 0.0433 |
| Radiation oncologist C | 0.094 ± 0.261 | <0.0001 | 0.108 ± 0.248 | <0.0001 | −1.375 ± 5.542 | 0.0217 |
| Radiation oncologist D | 0.0 ± 0.196 | 0.5173 | −0.004 ± 0.197 | 0.4687 | 0.649 ± 6.377 | 0.3260 |
| Total | 0.051 ± 0.232 | <0.0001 | 0.056 ± 0.231 | <0.0001 | −1.042 ± 6.908 | 0.0356 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheon, W.; Jeong, S.; Jeong, J.H.; Lim, Y.K.; Shin, D.; Lee, S.B.; Lee, D.Y.; Lee, S.U.; Suh, Y.G.; Moon, S.H.; et al. Interobserver Variability Prediction of Primary Gross Tumor in a Patient with Non-Small Cell Lung Cancer. Cancers 2022, 14, 5893. https://doi.org/10.3390/cancers14235893
Cheon W, Jeong S, Jeong JH, Lim YK, Shin D, Lee SB, Lee DY, Lee SU, Suh YG, Moon SH, et al. Interobserver Variability Prediction of Primary Gross Tumor in a Patient with Non-Small Cell Lung Cancer. Cancers. 2022; 14(23):5893. https://doi.org/10.3390/cancers14235893
Chicago/Turabian StyleCheon, Wonjoong, Seonghoon Jeong, Jong Hwi Jeong, Young Kyung Lim, Dongho Shin, Se Byeong Lee, Doo Yeul Lee, Sung Uk Lee, Yang Gun Suh, Sung Ho Moon, and et al. 2022. "Interobserver Variability Prediction of Primary Gross Tumor in a Patient with Non-Small Cell Lung Cancer" Cancers 14, no. 23: 5893. https://doi.org/10.3390/cancers14235893
APA StyleCheon, W., Jeong, S., Jeong, J. H., Lim, Y. K., Shin, D., Lee, S. B., Lee, D. Y., Lee, S. U., Suh, Y. G., Moon, S. H., Kim, T. H., & Kim, H. (2022). Interobserver Variability Prediction of Primary Gross Tumor in a Patient with Non-Small Cell Lung Cancer. Cancers, 14(23), 5893. https://doi.org/10.3390/cancers14235893





