Suppression of Calcium Entry Modulates the Expression of TRβ1 and Runx2 in Thyroid Cancer Cells, Two Transcription Factors That Regulate Invasion, Proliferation and Thyroid-Specific Protein Levels
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Lentiviral Transduction and Generation of Stable Cell Lines
2.4. Transient Transfections
2.5. Western Blot Analysis
2.6. Proliferation Assays
2.7. FACS Analysis
2.8. Migration and Invasion Assays
2.9. Measurement of Intracellular [Ca2+]i
2.10. TRβ1 and Runx2 Staining of Patient Samples
2.11. High Throughput Data Mining and Gene Expression Analyses
2.12. Fast Gene Enrichment Analysis
2.13. Survival Analysis
2.14. Statistical Analysis
3. Results
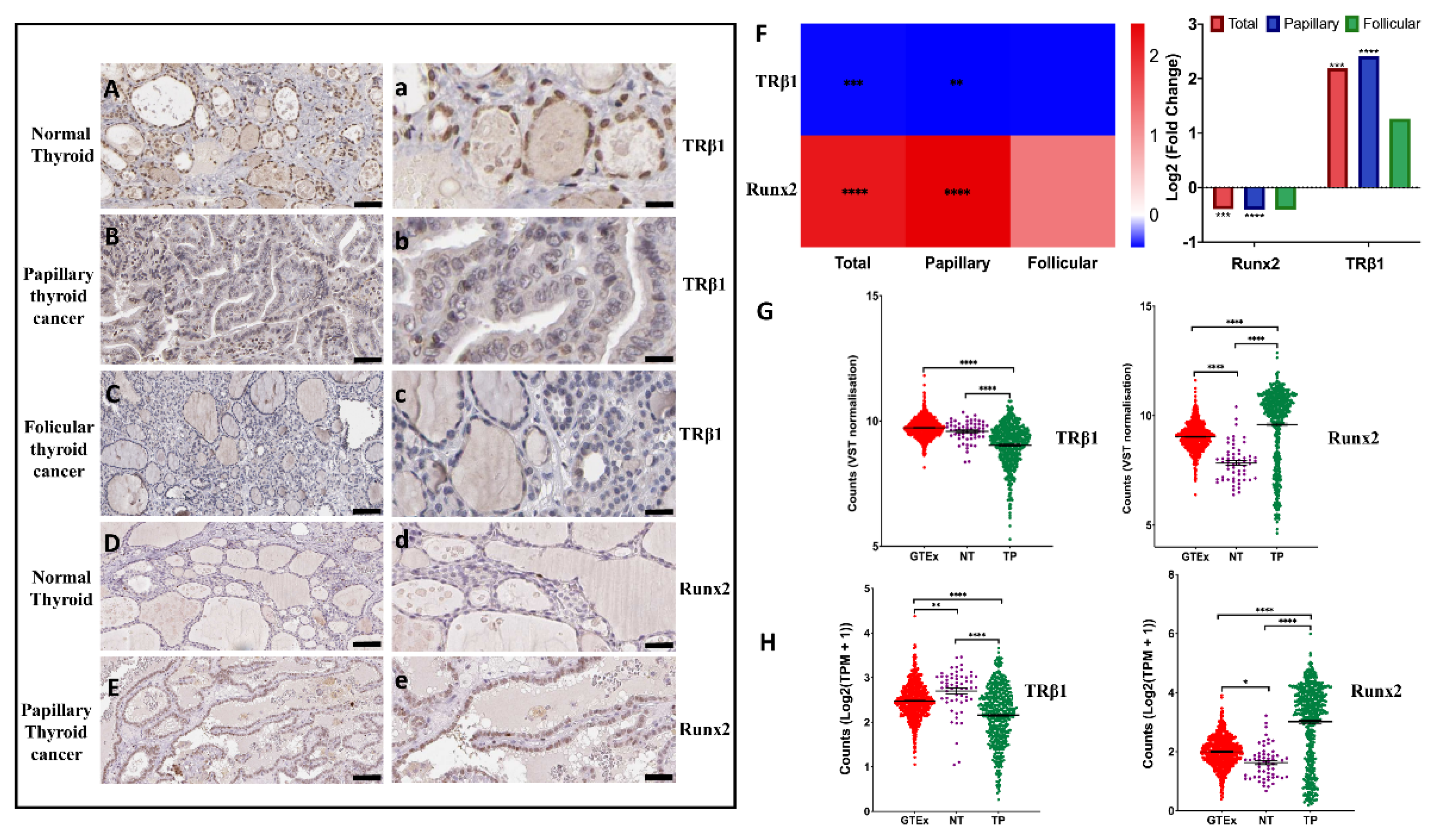
3.1. Expression of TRβ1 and Runx2 in Normal and Thyroid Cancer Tissues
3.2. Differential Expression of TRβ1 and Runx2 Genes in Normal Thyroid Tissues vs. Thyroid Cancer
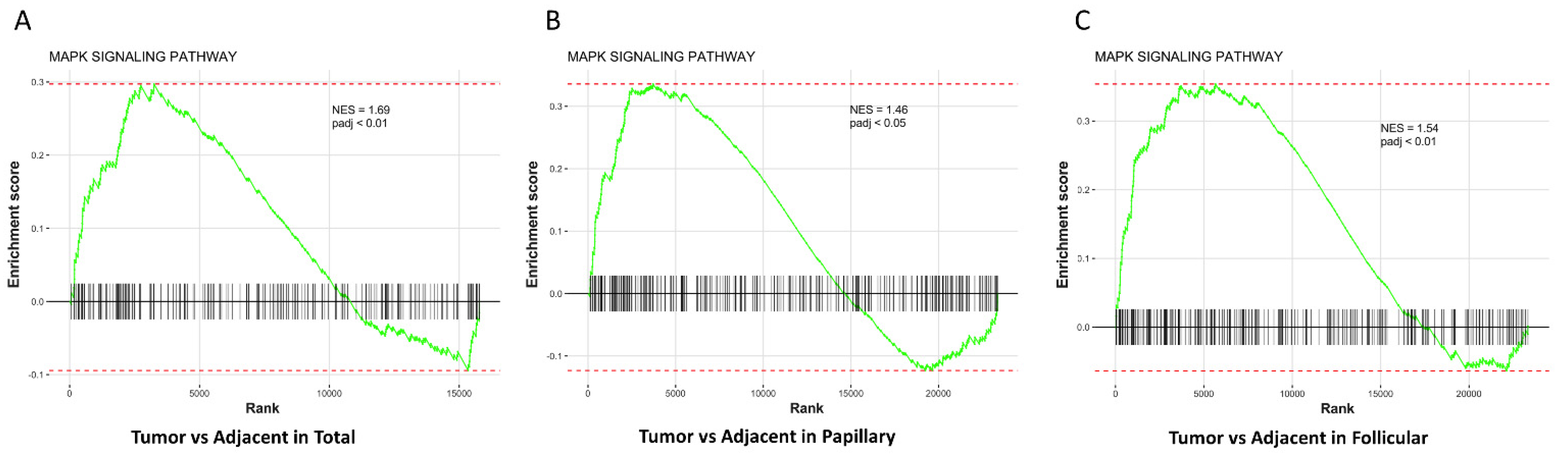
3.3. MAPK Pathway Is Up-Regulated in Thyroid Tumors Compared with Adjacent Normal Thyroid Tissues
3.4. High or Low Expression of TRβ1 and Runx2 Has no Effect on Survival of Thyroid Cancer
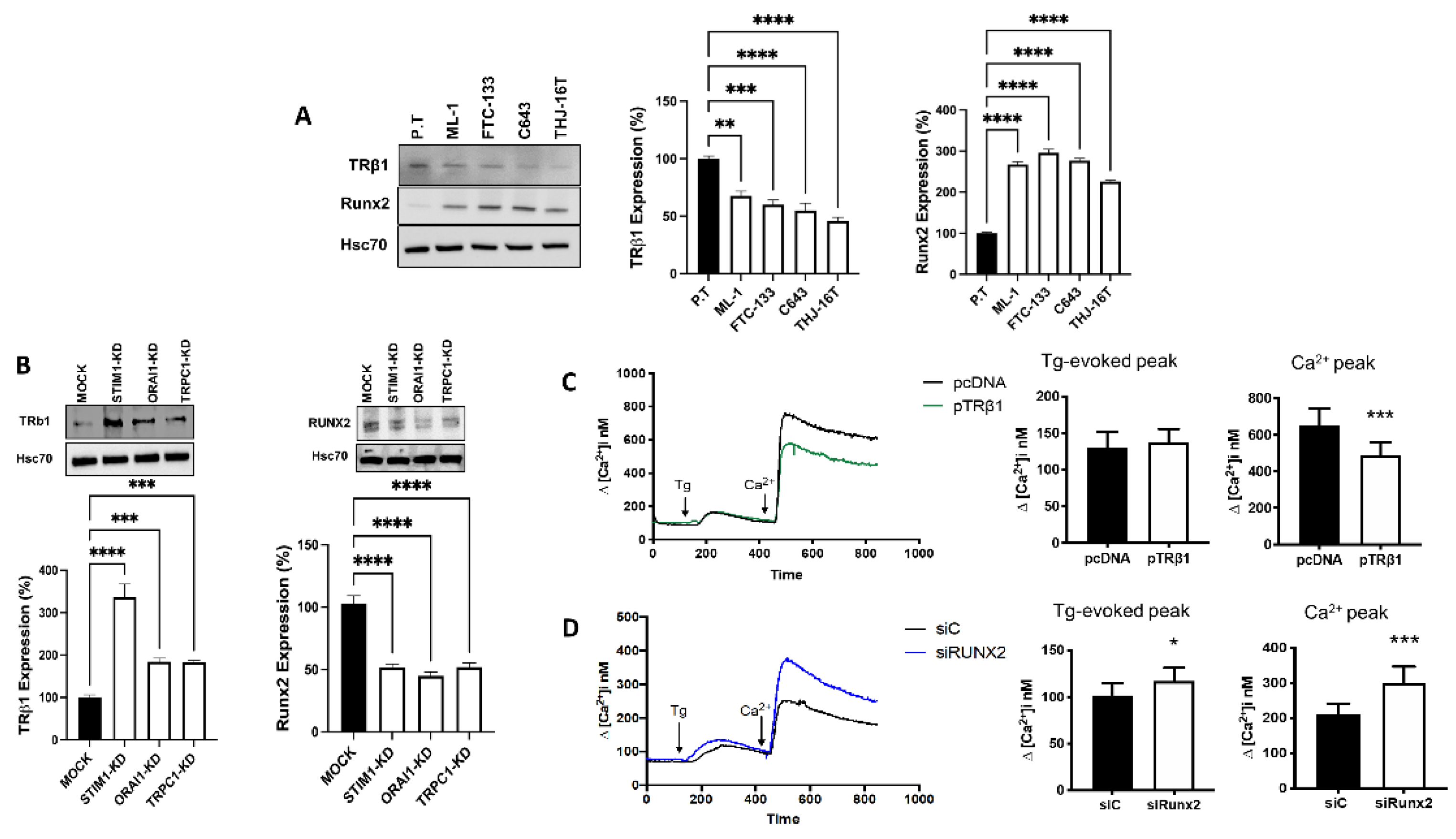
3.5. Store-Operated Calcium Entry Modulates the Calcium Dependent Expression of TRβ1 and Runx2
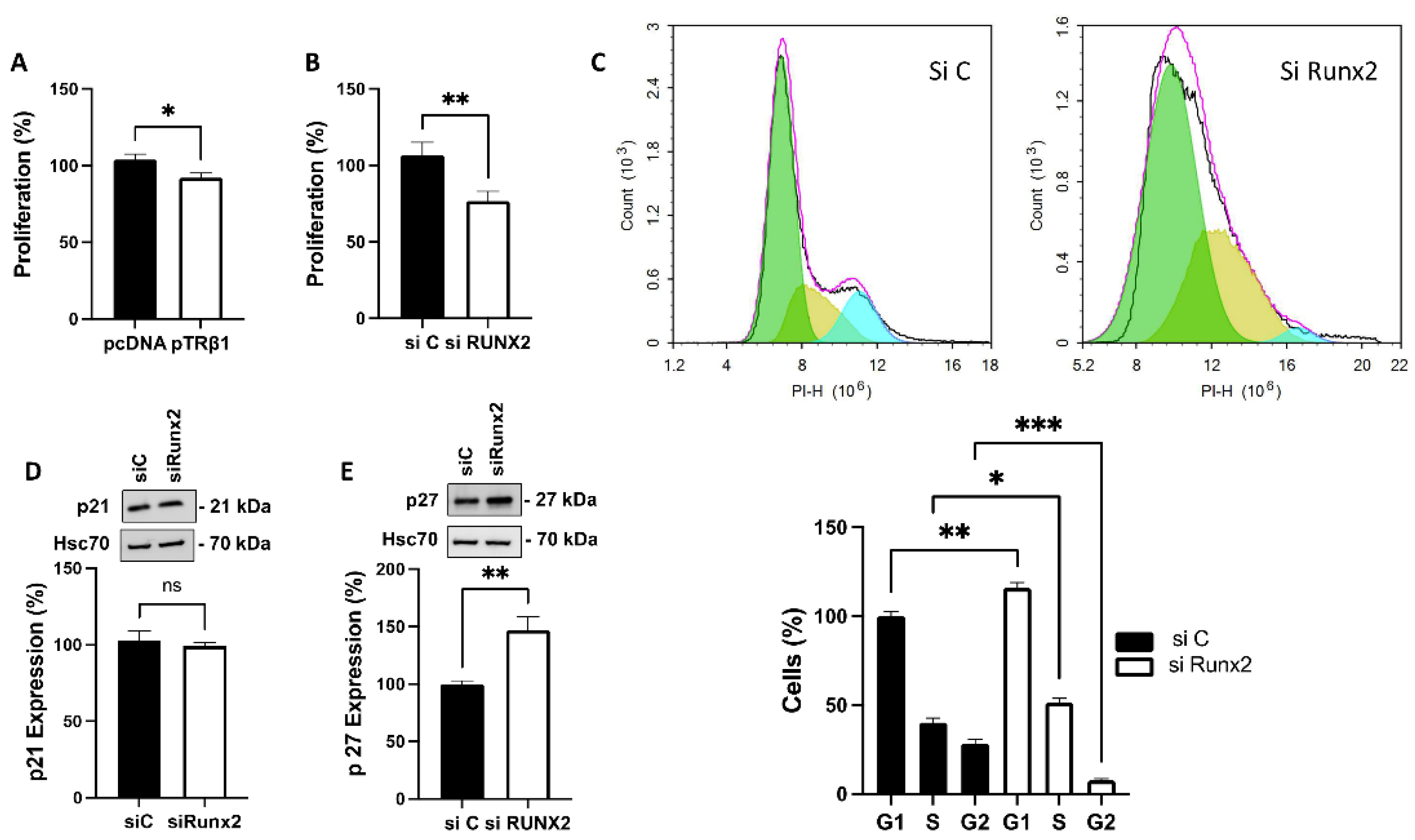
3.6. TRβ1 Overexpression and Knockdown of Runx2 Attenuate Proliferation of ML-1 Cells
3.7. TRβ1 Overexpression or Knockdown of Runx2 Abolish S1P Evoked Invasion of ML-1 Cells
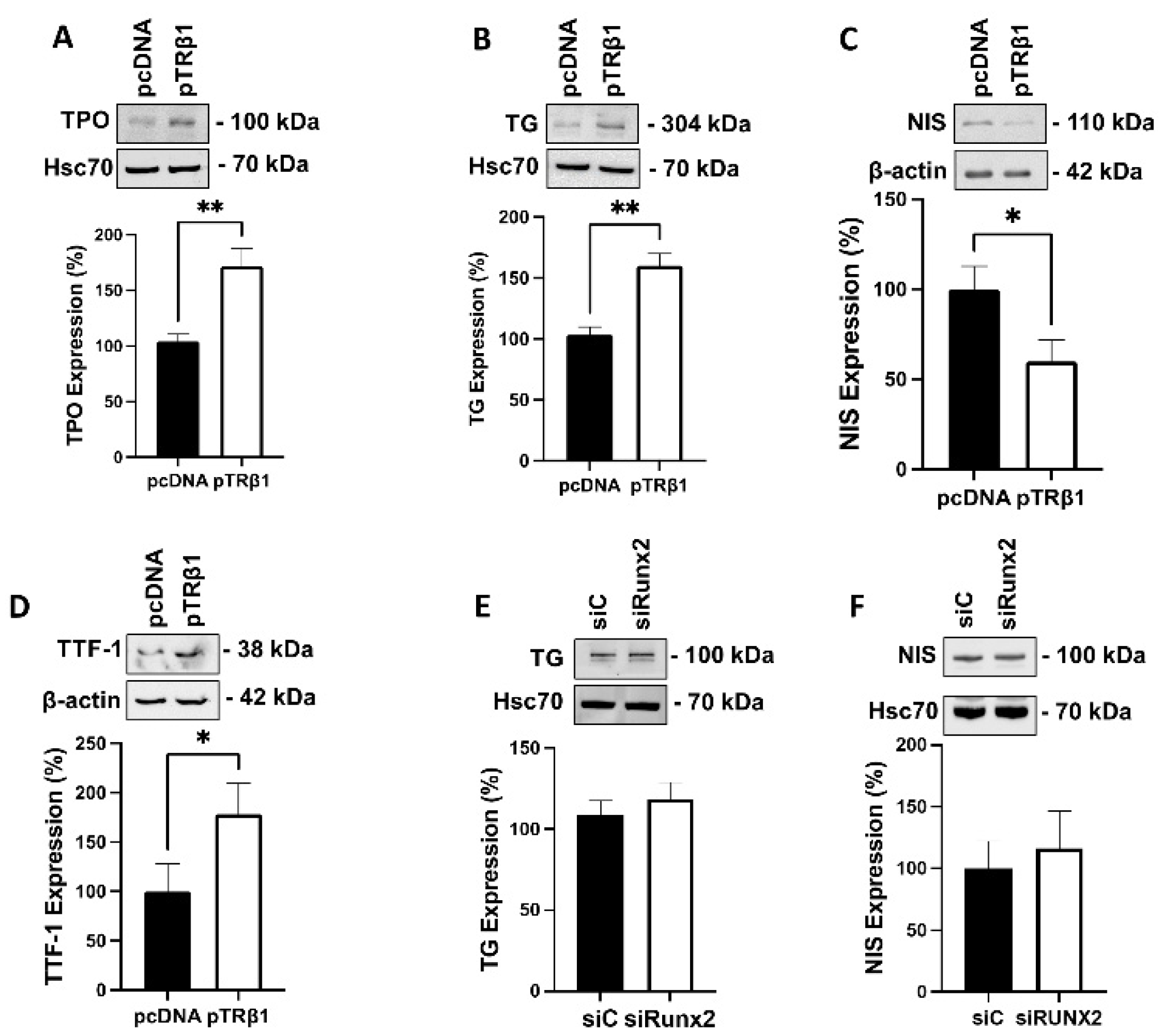
3.8. Overexpression of TRβ1, but Not Runx2, Restored Thyroid Specific Protein Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pitkäniemi, J.; Malila, N.; Tanskanen, T.; Degerlund, H.; Heikkinen, S.; Seppä, K. Cancer in Finland 2018; Cancer Society of Finland Publication: Helsinki, Finland, 2022. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Nagaiah, G.; Hossain, A.; Mooney, C.J.; Parmentier, J.; Remick, S.C. Anaplastic thyroid cancer: A review of epidemiology, pathogenesis, and treatment. J. Oncol. 2011, 2011, 542358. [Google Scholar] [CrossRef] [PubMed]
- Laha, D.; Nilubol, N.; Boufraqech, M. New Therapies for Advanced Thyroid Cancer. Front. Endocrinol. 2020, 11, 82. [Google Scholar] [CrossRef] [PubMed]
- Smallridge, R.C.; Marlow, L.A.; Copland, J.A. Anaplastic thyroid cancer: Molecular pathogenesis and emerging therapies. Endocr.-Relat. Cancer 2009, 16, 17–44. [Google Scholar] [CrossRef]
- Bochukova, E.; Schoenmakers, N.; Agostini, M.; Schoenmakers, E.; Rajanayagam, O.; Keogh, J.M.; Henning, E.; Reinemund, J.; Gevers, E.; Sarri, M.; et al. A Mutation in the Thyroid Hormone Receptor Alpha Gene. N. Engl. J. Med. 2012, 366, 243–249. [Google Scholar] [CrossRef]
- Fraichard, A.; Chassande, O.; Plateroti, M.; Roux, J.P.; Trouillas, J.; Dehay, C.; Legrand, C.; Gauthier, K.; Kedinger, M.; Malaval, L.; et al. The T3Rα gene encoding a thyroid hormone receptor is essential for post-natal development and thyroid hormone production. EMBO J. 1997, 16, 4412–4420. [Google Scholar] [CrossRef]
- Hall, L.C.; Salazar, E.P.; Kane, S.R.; Liu, N. Effects of thyroid hormones on human breast cancer cell proliferation. J. Steroid Biochem. Mol. Biol. 2008, 109, 57–66. [Google Scholar] [CrossRef]
- Kowalik, M.A.; Perra, A.; Pibiri, M.; Cocco, M.T.; Samarut, J.; Plateroti, M.; Ledda-Columbano, G.M.; Columbano, A. TRβ is the critical thyroid hormone receptor isoform in T3-induced proliferation of hepatocytes and pancreatic acinar cells. J. Hepatol. 2010, 53, 686–692. [Google Scholar] [CrossRef]
- Maruvada, P.; Dmitrieva, N.I.; East-Palmer, J.; Yen, P.M. Cell Cycle-dependent Expression of Thyroid Hormone Receptor-β Is a Mechanism for Variable Hormone Sensitivity. Mol. Biol. Cell 2004, 15, 1895–1903. [Google Scholar] [CrossRef][Green Version]
- Zambrano, A.; García-Carpizo, V.; Gallardo, M.E.; Villamuera, R.; Gómez-Ferrería, M.A.; Pascual, A.; Buisine, N.; Sachs, L.M.; Garesse, R.; Aranda, A. The thyroid hormone receptor β induces DNA damage and premature senescence. J. Cell Biol. 2014, 204, 129–146. [Google Scholar] [CrossRef]
- Moeller, L.C.; Führer, D. Thyroid hormone, thyroid hormone receptors, and cancer: A clinical perspective. Endocr.-Relat. Cancer 2013, 20, R19–R29. [Google Scholar] [CrossRef] [PubMed]
- Carr, F.E.; Tai, P.W.L.; Barnum, M.S.; Gillis, N.E.; Evans, K.G.; Taber, T.H.; White, J.H.; Tomczak, J.A.; Jaworski, D.M.; Zaidi, S.K.; et al. Thyroid hormone receptor-β (TRβ) mediates runt-related transcription factor 2 (Runx2) expression in thyroid cancer cells: A novel signaling pathway in thyroid cancer. Endocrinology 2016, 157, 3278–3292. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.G.; Cheng, S.Y. Thyroid hormone receptors and cancer. Biochim. Biophys. Acta-Gen. Subj. 2013, 1830, 3928–3936. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.I.; Shiga, T.; Ito, Y. Runx transcription factors in neuronal development. Neural Dev. 2008, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Stifani, S. Roles of Runx genes in nervous system development. Adv. Exp. Med. Biol. 2017, 962, 103–116. [Google Scholar]
- Cohen-Solal, K.A.; Boregowda, R.K.; Lasfar, A. RUNX2 and the PI3K/AKT axis reciprocal activation as a driving force for tumor progression. Mol. Cancer 2015, 14, 137. [Google Scholar] [CrossRef]
- Kim, J.M.; Yang, Y.S.; Park, K.H.; Ge, X.; Xu, R.; Li, N.; Song, M.; Chun, H.; Bok, S.; Charles, J.F.; et al. A RUNX2 stabilization pathway mediates physiologic and pathologic bone formation. Nat. Commun. 2020, 11, 2289. [Google Scholar] [CrossRef]
- Ozaki, T.; Wu, D.; Sugimoto, H.; Nagase, H.; Nakagawara, A. Runt-related transcription factor 2 (RUNX2) inhibits p53-dependent apoptosis through the collaboration with HDAC6 in response to DNA damage. Cell Death Dis. 2013, 4, e610. [Google Scholar] [CrossRef]
- Niu, D.F.; Kondo, T.; Nakazawa, T.; Oishi, N.; Kawasaki, T.; Mochizuki, K.; Yamane, T.; Katoh, R. Transcription factor Runx2 is a regulator of epithelial-mesenchymal transition and invasion in thyroid carcinomas. Lab. Investig. 2012, 92, 1181–1190. [Google Scholar] [CrossRef]
- Sase, T.; Suzuki, T.; Miura, K.; Shiiba, K.; Sato, I.; Nakamura, Y.; Takagi, K.; Onodera, Y.; Miki, Y.; Watanabe, M.; et al. Runt-related transcription factor 2 in human colon carcinoma: A potent prognostic factor associated with estrogen receptor. Int. J. Cancer 2012, 131, 2284–2293. [Google Scholar] [CrossRef]
- Asghar, M.Y.; Magnusson, M.; Kemppainen, K.; Sukumaran, P.; Löf, C.; Pulli, I.; Kalhori, V.; Törnquist, K. Transient receptor potential canonical 1 (TRPC1) channels as regulators of sphingolipid and VEGF receptor expression: Implications for thyroid cancer cell migration and proliferation. J. Biol. Chem. 2015, 290, 16116–16131. [Google Scholar] [CrossRef]
- Asghar, M.Y.; Lassila, T.; Paatero, I.; Nguyen, V.D.; Kronqvist, P.; Zhang, J.; Slita, A.; Löf, C.; Zhou, Y.; Rosenholm, J.; et al. Stromal interaction molecule 1 (STIM1) knock down attenuates invasion and proliferation and enhances the expression of thyroid-specific proteins in human follicular thyroid cancer cells. Cell. Mol. Life Sci. 2021, 78, 5827–5846. [Google Scholar] [CrossRef] [PubMed]
- Balthasar, S.; Samulin, J.; Ahlgren, H.; Bergelin, N.; Lundqvist, M.; Toescu, E.C.; Eggo, M.C.; Törnquist, K. Sphingosine 1-phosphate receptor expression profile and regulation of migration in human thyroid cancer cells. Biochem. J. 2006, 398, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Sivertsson, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2021, 060012. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. The Cox Model BT—Modeling Survival Data: Extending the Cox Model. In Statistics for Biology and Health; Springer: Berlin/Heidelberg, Germany, 2000. [Google Scholar]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Zaballos, M.A.; Santisteban, P. Key signaling pathways in thyroid cancer. J. Endocrinol. 2017, 235, R43–R61. [Google Scholar] [CrossRef]
- Braicu, C.; Buse, M.; Busuioc, C.; Drula, R.; Gulei, D.; Raduly, L.; Rusu, A.; Irimie, A.; Atanasov, A.G.; Slaby, O.; et al. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers 2019, 11, 1618. [Google Scholar] [CrossRef]
- Koveitypour, Z.; Panahi, F.; Vakilian, M.; Peymani, M.; Seyed Forootan, F.; Nasr Esfahani, M.H.; Ghaedi, K. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019, 9, 97. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, X.; Zhang, C.; Xue, L.; Yang, L. Expression and clinical significance of MAPK and EGFR in triple–negative breast cancer. Oncol. Lett. 2020, 19, 1842–1848. [Google Scholar] [CrossRef] [PubMed]
- Bergelin, N.; Blom, T.; Heikkilä, J.; Löf, C.; Alam, C.; Balthasar, S.; Slotte, J.P.; Hinkkanen, A.; Törnquist, K. Sphingosine kinase as an oncogene: Autocrine sphingosine 1-phoshate modulates ML-1 thyroid carcinoma cell migration by a mechanism dependent on protein kinase C-α and ERK1/2. Endocrinology 2009, 150, 2055–2063. [Google Scholar] [CrossRef] [PubMed]
- Lazar, V.; Bidart, J.M.; Caillou, B.; Mahé, C.; Lacroix, L.; Filetti, S.; Schlumberger, M. Expression of the Na+/I− symporter gene in human thyroid tumors: A comparison study with other thyroid-specific genes. J. Clin. Endocrinol. Metab. 1999, 84, 3228–3234. [Google Scholar] [CrossRef]
- Ros, P.; Rossi, D.L.; Acebrón, A.; Santisteban, P. Thyroid-specific gene expression in the multi-step process of thyroid carcinogenesis. Biochimie 1999, 81, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Schweppe, R.E.; Klopper, J.P.; Korch, C.; Pugazhenthi, U.; Benezra, M.; Knauf, J.A.; Fagin, J.A.; Marlow, L.A.; Copland, J.A.; Smallridge, R.C.; et al. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J. Clin. Endocrinol. Metab. 2008, 93, 4331–4341. [Google Scholar] [CrossRef]
- Marlow, L.A.; D’Innocenzi, J.; Zhang, Y.; Rohl, S.D.; Cooper, S.J.; Sebo, T.; Grant, C.; McIver, B.; Kasperbauer, J.L.; Wadsworth, J.T.; et al. Detailed molecular fingerprinting of four new anaplastic thyroid carcinoma cell lines and their use for verification of RhoB as a molecular therapeutic target. J. Clin. Endocrinol. Metab. 2010, 95, 5338–5347. [Google Scholar] [CrossRef] [PubMed]
- Bolf, E.L.; Gillis, N.E.; Barnum, M.S.; Beaudet, C.M.; Yu, G.Y.; Tomczak, J.A.; Stein, J.L.; Lian, J.B.; Stein, G.S.; Carr, F.E. The Thyroid Hormone Receptor-RUNX2 Axis: A Novel Tumor Suppressive Pathway in Breast Cancer. Horm. Cancer 2020, 11, 34–41. [Google Scholar] [CrossRef]
- Herreño, A.M.; Ramírez, A.C.; Chaparro, V.P.; Fernandez, M.J.; Cañas, A.; Morantes, C.F.; Moreno, O.M.; Brugés, R.E.; Mejía, J.A.; Bustos, F.J.; et al. Role of RUNX2 transcription factor in epithelial mesenchymal transition in non-small cell lung cancer lung cancer: Epigenetic control of the RUNX2 P1 promoter. Tumor Biol. 2019, 41, 1010428319851014. [Google Scholar] [CrossRef]
- Gillis, N.; Bolf, E.; Davidson, C.; Tomczak, J.; Frietze, S.; Carr, F. Restoration of Thyroid Hormone Receptor Beta Signaling Reprograms Anaplastic Thyroid Cancer Cells. J. Endocr. Soc. 2019, 3, SAT-571. [Google Scholar] [CrossRef]
- Davidson, C.D.; Gillis, N.E.; Carr, F.E. Thyroid hormone receptor beta as tumor suppressor: Untapped potential in treatment and diagnostics in solid tumors. Cancers 2021, 13, 4254. [Google Scholar] [CrossRef]
- Zhu, L.; Tian, G.; Yang, Q.; De, G.; Zhang, Z.; Wang, Y.; Nie, H.; Zhang, Y.; Yang, X.; Li, J. Thyroid hormone receptor β1 suppresses proliferation and migration by inhibiting PI3K/Akt signaling in human colorectal cancer cells. Oncol. Rep. 2016, 36, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Iglesias, O.; Garcia-Silva, S.; Tenbaum, S.P.; Regadera, J.; Larcher, F.; Paramio, J.M.; Vennström, B.; Aranda, A. Thyroid hormone receptor β1 acts as a potent suppressor of tumor invasiveness and metastasis. Cancer Res. 2009, 69, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Khoo, M.L.C.; Beasley, N.J.P.; Ezzat, S.; Freeman, J.L.; Asa, S.L. Overexpression of cyclin D1 and underexpression of p27 predict lymph node metastases in papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2002, 87, 1814–1818. [Google Scholar] [CrossRef] [PubMed]
- Pyne, N.J.; El Buri, A.; Adams, D.R.; Pyne, S. Sphingosine 1-phosphate and cancer. Adv. Biol. Regul. 2018, 68, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Fiorio Pla, A.; Kondratska, K.; Prevarskaya, N. STIM and ORAI proteins: Crucial roles in hallmarks of cancer. Am. J. Physiol.-Cell Physiol. 2016, 310, C509–C519. [Google Scholar] [CrossRef]
- Mo, P.; Yang, S. The store-operated calcium channels in cancer metastasis: From cell migration, invasion to metastatic colonization. Front. Biosci.-Landmark 2018, 23, 1241–1256. [Google Scholar] [CrossRef]
- Roos, J.; DiGregorio, P.J.; Yeromin, A.V.; Ohlsen, K.; Lioudyno, M.; Zhang, S.; Safrina, O.; Kozak, J.A.; Wagner, S.L.; Cahalan, M.D.; et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005, 169, 435–445. [Google Scholar] [CrossRef]
- Hewavitharana, T.; Deng, X.; Soboloff, J.; Gill, D.L. Role of STIM and Orai proteins in the store-operated calcium signaling pathway. Cell Calcium 2007, 42, 173–182. [Google Scholar] [CrossRef]
- Niemelä, E.; Desai, D.; Niemi, R.; Doroszko, M.; Özliseli, E.; Kemppainen, K.; Rahman, N.A.; Sahlgren, C.; Törnquist, K.; Eriksson, J.E.; et al. Nanoparticles carrying fingolimod and methotrexate enables targeted induction of apoptosis and immobilization of invasive thyroid cancer. Eur. J. Pharm. Biopharm. 2020, 148, 1–9. [Google Scholar] [CrossRef]

| Product | Gene ID | Nucleotide Sequence (5′-3′) |
|---|---|---|
| shRNA | Tcf25, TRCN0000190494, 66855 STIM1, TRCN0000149588, 6786 ORAI1, TRCN0000161221, 84876 TRPC1, TRCN0000043998, 7220 | CCGGGCCTCTGTCTCCCAAATGTTACTCGAGTAACATTTGGGAGACAGAGGCTTTTTTG CCGGCGATGAGATCAACCTTGCTAACTCGAGTTAGCAAGGTTGATCTCATCGTTTTTTG CCGGGAAACTGTCCTCTAAGAGAATCTCGAGATTCTCTTAGAGGACAGTTTCTTTTTTG CCGGGCCCACCTGTAAGAAGATAATCTCGAGATTATCTTCTTACAGGTGGGCTTTTTG |
| Primers | TRβ1 forward Tm (°C) 57.3 TRβ1 Reverse Tm (°C) 57.3 | TCTTCCCCCCTTTGTTCTTG ACTACTTCCCTTTTCCCTCC |
| siRNA | Non-targetting control siRNA siRNA TRβ1 siRNA Runx2 | CCU ACA UCC CGA UCG AUG AUG AUC AUC ACA CCA GCA AUU A GCU ACC UAU CAC AGA GCA A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asghar, M.Y.; Knuutinen, T.; Holm, E.; Nordström, T.; Nguyen, V.D.; Zhou, Y.; Törnquist, K. Suppression of Calcium Entry Modulates the Expression of TRβ1 and Runx2 in Thyroid Cancer Cells, Two Transcription Factors That Regulate Invasion, Proliferation and Thyroid-Specific Protein Levels. Cancers 2022, 14, 5838. https://doi.org/10.3390/cancers14235838
Asghar MY, Knuutinen T, Holm E, Nordström T, Nguyen VD, Zhou Y, Törnquist K. Suppression of Calcium Entry Modulates the Expression of TRβ1 and Runx2 in Thyroid Cancer Cells, Two Transcription Factors That Regulate Invasion, Proliferation and Thyroid-Specific Protein Levels. Cancers. 2022; 14(23):5838. https://doi.org/10.3390/cancers14235838
Chicago/Turabian StyleAsghar, Muhammad Yasir, Taru Knuutinen, Emilia Holm, Tommy Nordström, Van Dien Nguyen, You Zhou, and Kid Törnquist. 2022. "Suppression of Calcium Entry Modulates the Expression of TRβ1 and Runx2 in Thyroid Cancer Cells, Two Transcription Factors That Regulate Invasion, Proliferation and Thyroid-Specific Protein Levels" Cancers 14, no. 23: 5838. https://doi.org/10.3390/cancers14235838
APA StyleAsghar, M. Y., Knuutinen, T., Holm, E., Nordström, T., Nguyen, V. D., Zhou, Y., & Törnquist, K. (2022). Suppression of Calcium Entry Modulates the Expression of TRβ1 and Runx2 in Thyroid Cancer Cells, Two Transcription Factors That Regulate Invasion, Proliferation and Thyroid-Specific Protein Levels. Cancers, 14(23), 5838. https://doi.org/10.3390/cancers14235838







