Targeted Next-Generation Sequencing Identifies Additional Mutations Other than BCR∷ABL in Chronic Myeloid Leukemia Patients: A Chinese Monocentric Retrospective Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. NGS Detection and Response Assessment
2.3. Statistical Analysis
3. Results
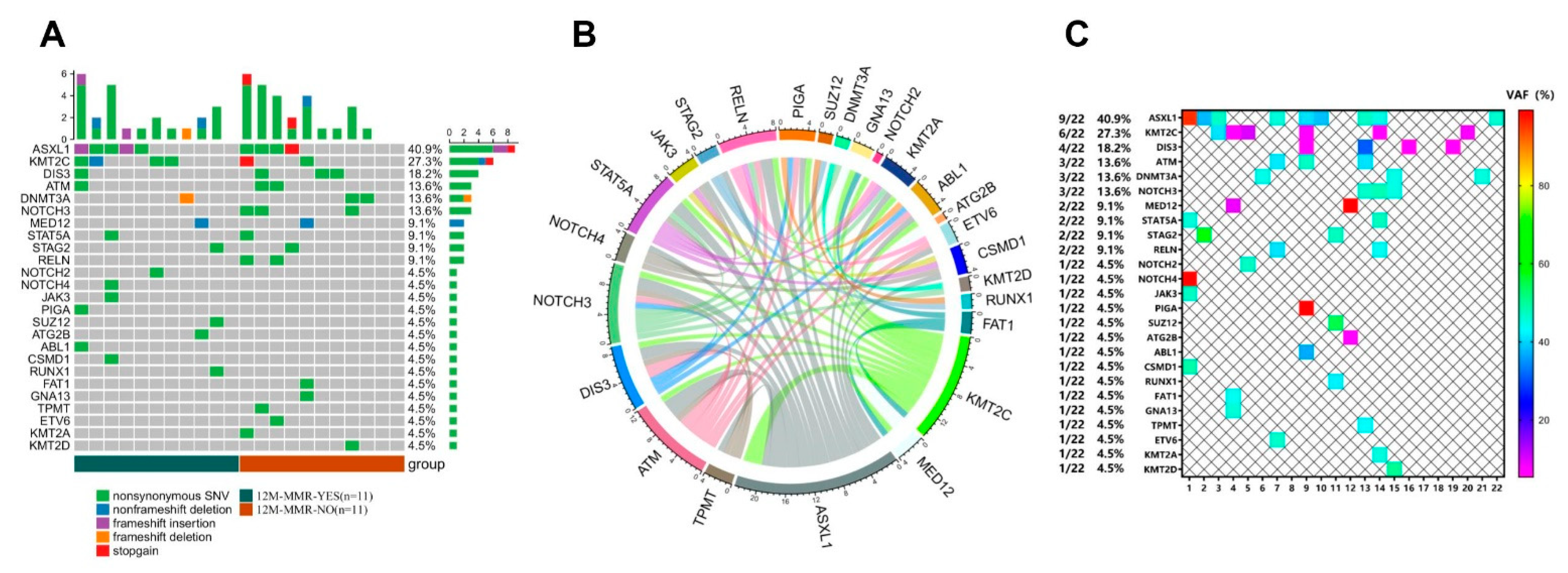
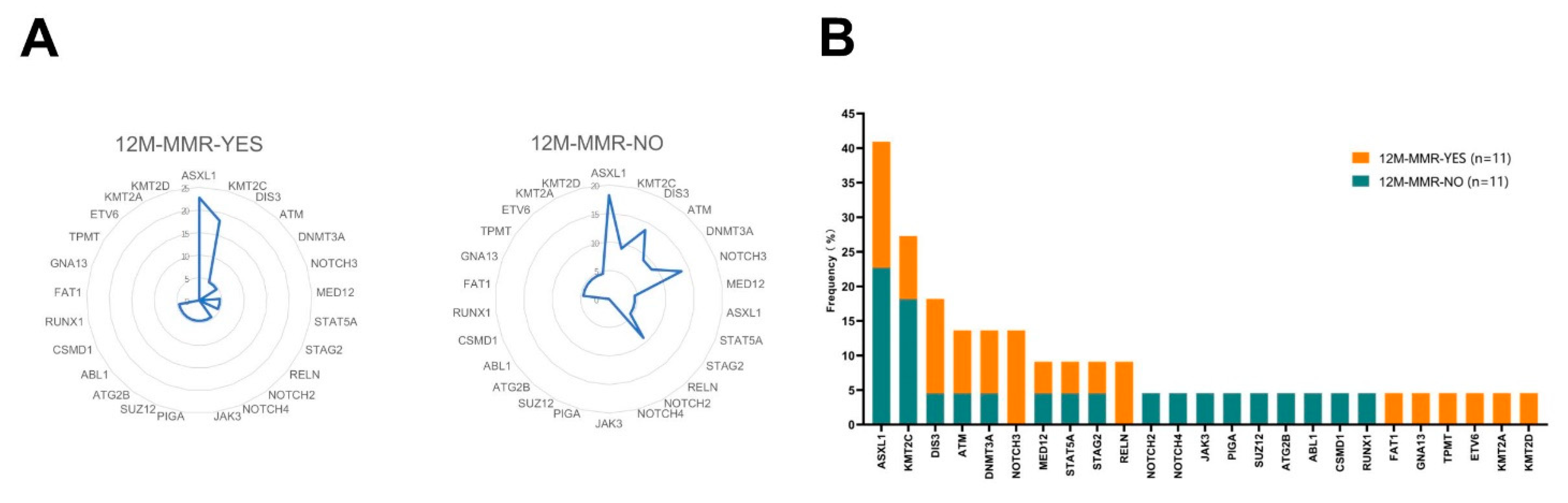
3.1. Mutation Analysis Based on NGS Detection
3.2. Mutation Analysis in the 12 M-MMR Group and Non12 M-MMR Group
3.3. Mutation Analysis in the 36 M-MR4.0 Group and Non-36 M-MR4.0 Group
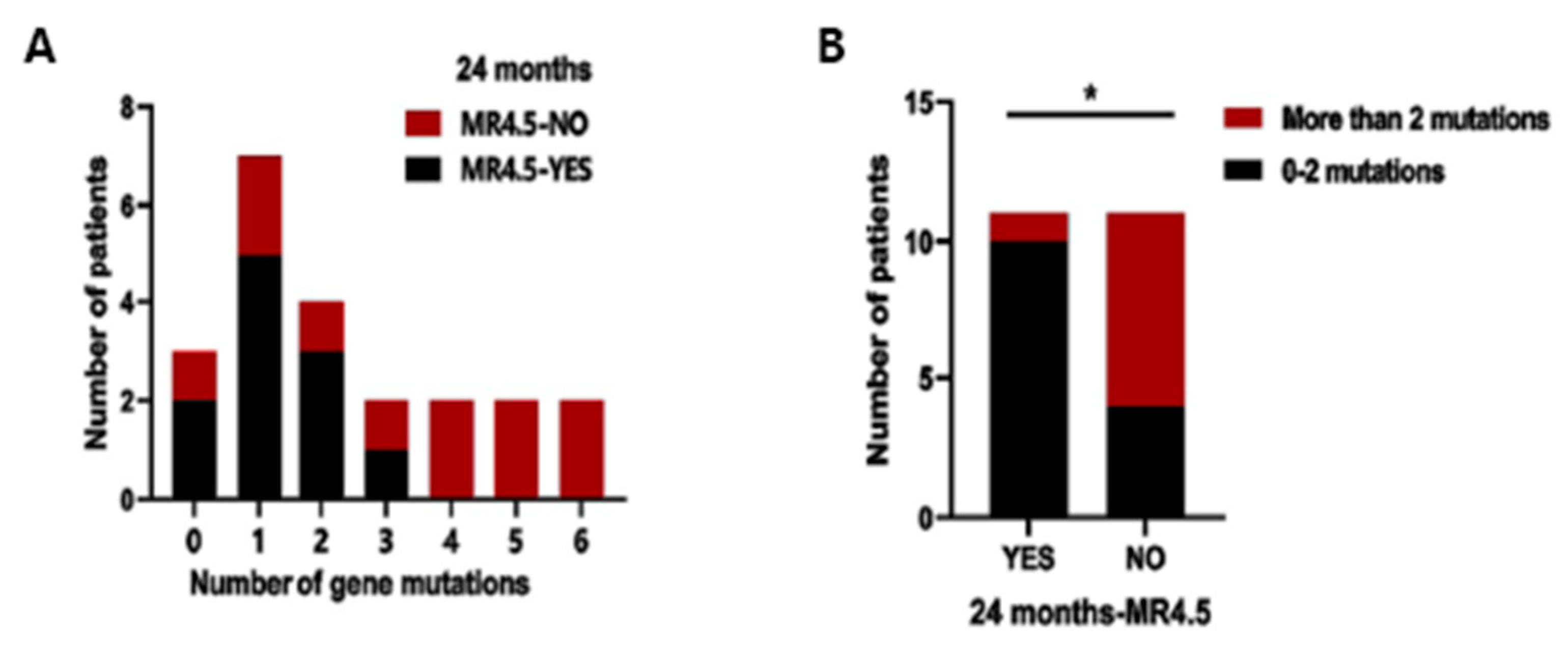
3.4. Analysis of the Number of Variants
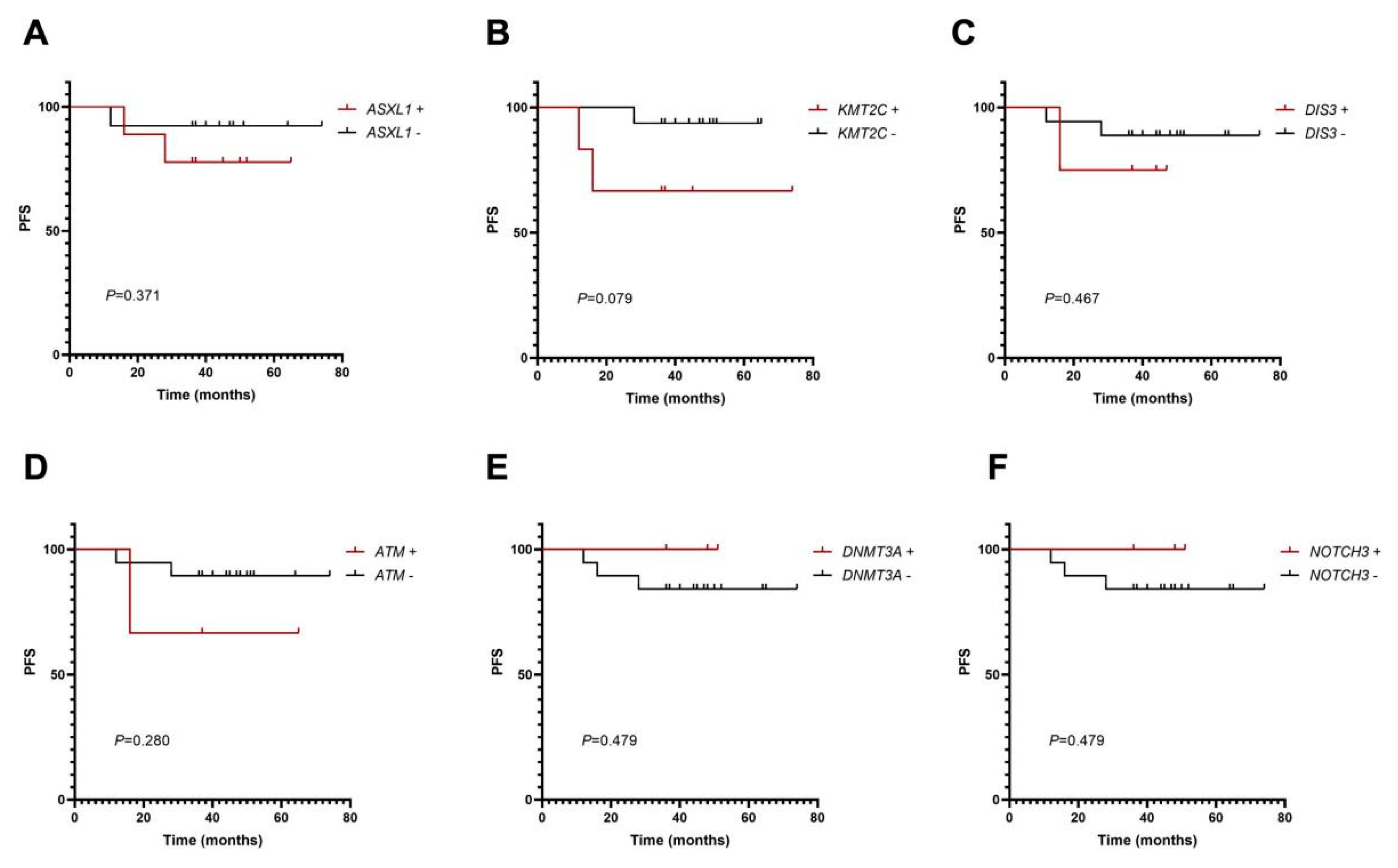
3.5. Mutation Analysis of PFS
3.6. Analysis of Clinical Characteristics on Molecular Response
3.7. Analysis of TKI Therapies on Molecular Response
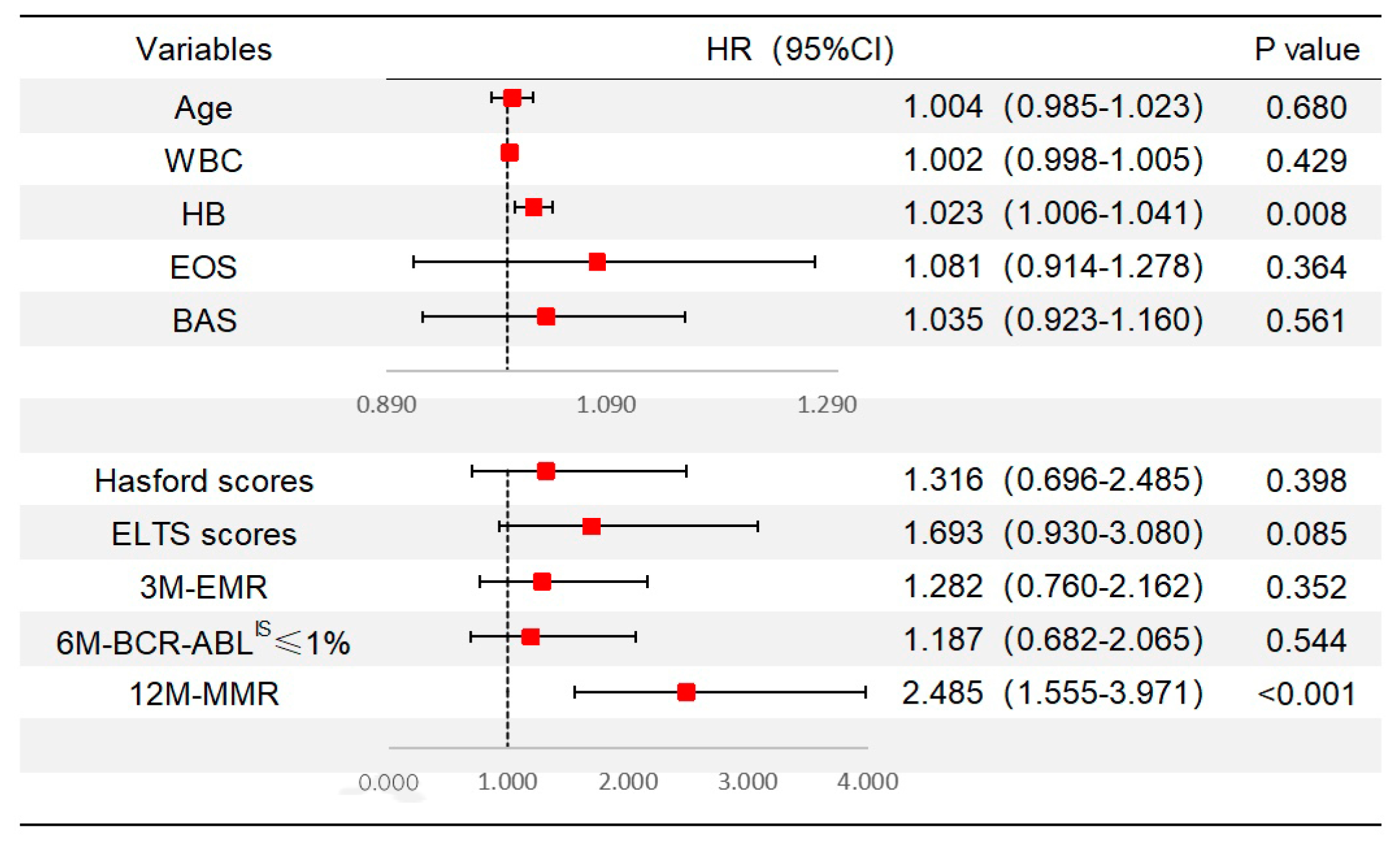
3.8. Analysis of Scoring Systems on Molecular Response and Mutation Status
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patnaik, M.M.; Tefferi, A. Chronic myelomonocytic leukemia: 2022 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2022, 97, 352–372. [Google Scholar] [CrossRef]
- Bower, H.; Bjorkholm, M.; Dickman, P.W.; Hoglund, M.; Lambert, P.C.; Andersson, T.M. Life Expectancy of Patients with Chronic Myeloid Leukemia Approaches the Life Expectancy of the General Population. J. Clin. Oncol. 2016, 34, 2851–2857. [Google Scholar] [CrossRef]
- Hughes, T.P.; Saglio, G.; Quintas-Cardama, A.; Mauro, M.J.; Kim, D.W.; Lipton, J.H.; Bradley-Garelik, M.B.; Ukropec, J.; Hochhaus, A. BCR-ABL1 mutation development during first-line treatment with dasatinib or imatinib for chronic myeloid leukemia in chronic phase. Leukemia 2015, 29, 1832–1838. [Google Scholar] [CrossRef]
- Branford, S.; Kim, D.D.H.; Apperley, J.F.; Eide, C.A.; Mustjoki, S.; Ong, S.T.; Nteliopoulos, G.; Ernst, T.; Chuah, C.; Gambacorti-Passerini, C.; et al. Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia. Leukemia 2019, 33, 1835–1850. [Google Scholar] [CrossRef]
- Nteliopoulos, G.; Bazeos, A.; Claudiani, S.; Gerrard, G.; Curry, E.; Szydlo, R.; Alikian, M.; Foong, H.E.; Nikolakopoulou, Z.; Loaiza, S.; et al. Somatic variants in epigenetic modifiers can predict failure of response to imatinib but not to second-generation tyrosine kinase inhibitors. Haematologica 2019, 104, 2400–2409. [Google Scholar] [CrossRef]
- Branford, S.; Wang, P.; Yeung, D.T.; Thomson, D.; Purins, A.; Wadham, C.; Shahrin, N.H.; Marum, J.E.; Nataren, N.; Parker, W.T.; et al. Integrative genomic analysis reveals cancer-associated mutations at diagnosis of CML in patients with high-risk disease. Blood 2018, 132, 948–961. [Google Scholar] [CrossRef]
- Jiang, H.; Zhi, L.T.; Hou, M.; Wang, J.X.; Wu, D.P.; Huang, X.J. Comparison of generic and original imatinib in the treatment of newly diagnosed patients with chronic myelogenous leukemia in chronic phase: A multicenter retrospective clinical study. Zhonghua Xue Ye Xue Za Zhi 2017, 38, 566–571. [Google Scholar] [CrossRef]
- Sokal, J.E.; Cox, E.B.; Baccarani, M.; Tura, S.; Gomez, G.A.; Robertson, J.E.; Tso, C.Y.; Braun, T.J.; Clarkson, B.D.; Cervantes, F.; et al. Prognostic discrimination in “good-risk” chronic granulocytic leukemia. Blood 1984, 63, 789–799. [Google Scholar] [CrossRef]
- Pfirrmann, M.; Baccarani, M.; Saussele, S.; Guilhot, J.; Cervantes, F.; Ossenkoppele, G.; Hoffmann, V.S.; Castagnetti, F.; Hasford, J.; Hehlmann, R.; et al. Prognosis of long-term survival considering disease-specific death in patients with chronic myeloid leukemia. Leukemia 2016, 30, 48–56. [Google Scholar] [CrossRef]
- Hasford, J.; Pfirrmann, M.; Hehlmann, R.; Allan, N.C.; Baccarani, M.; Kluin-Nelemans, J.C.; Alimena, G.; Steegmann, J.L.; Ansari, H. A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon alfa. Writing Committee for the Collaborative CML Prognostic Factors Project Group. J. Natl. Cancer Inst. 1998, 90, 850–858. [Google Scholar] [CrossRef]
- Hasford, J.; Baccarani, M.; Hoffmann, V.; Guilhot, J.; Saussele, S.; Rosti, G.; Guilhot, F.; Porkka, K.; Ossenkoppele, G.; Lindoerfer, D.; et al. Predicting complete cytogenetic response and subsequent progression-free survival in 2060 patients with CML on imatinib treatment: The EUTOS score. Blood 2011, 118, 686–692. [Google Scholar] [CrossRef]
- Gerds, A.T.; Gotlib, J.; Ali, H.; Bose, P.; Dunbar, A.; Elshoury, A.; George, T.I.; Gundabolu, K.; Hexner, E.; Hobbs, G.S.; et al. Myeloproliferative Neoplasms, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 1033–1062. [Google Scholar] [CrossRef]
- Menezes, J.; Salgado, R.N.; Acquadro, F.; Gomez-Lopez, G.; Carralero, M.C.; Barroso, A.; Mercadillo, F.; Espinosa-Hevia, L.; Talavera-Casanas, J.G.; Pisano, D.G.; et al. ASXL1, TP53 and IKZF3 mutations are present in the chronic phase and blast crisis of chronic myeloid leukemia. Blood Cancer J. 2013, 3, e157. [Google Scholar] [CrossRef]
- Xue, M.; Zeng, Z.; Wang, Q.; Wen, L.; Xu, Y.; Xie, J.; Wang, Q.; Ruan, C.; Wu, D.; Chen, S. Mutational Profiles during the Progression of Chronic Myeloid Leukemia. J. Blood 2021, 138, 3596. [Google Scholar] [CrossRef]
- Schonfeld, L.; Rinke, J.; Hinze, A.; Nagel, S.N.; Schafer, V.; Schenk, T.; Fabisch, C.; Brummendorf, T.H.; Burchert, A.; le Coutre, P.; et al. ASXL1 mutations predict inferior molecular response to nilotinib treatment in chronic myeloid leukemia. Leukemia 2022, 36, 2242–2249. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Z.; Qin, Y.; Gale, R.P.; Huang, X.; Jiang, Q. Correlations between Mutations in Cancer-Related Genes, Therapy Responses and Outcomes of the 3rd Generation Tyrosine Kinase-Inhibitor (TKI) in Persons with Chronic Myeloid Leukemia Failing Prior TKI-Therapy. Blood 2021, 138, 308. [Google Scholar] [CrossRef]
- Kim, T.; Tyndel, M.S.; Kim, H.J.; Ahn, J.S.; Choi, S.H.; Park, H.J.; Kim, Y.K.; Kim, S.Y.; Lipton, J.H.; Zhang, Z.; et al. Spectrum of somatic mutation dynamics in chronic myeloid leukemia following tyrosine kinase inhibitor therapy. Blood 2017, 129, 38–47. [Google Scholar] [CrossRef]
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; Miller, C.A.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478. [Google Scholar] [CrossRef]
- Bellavia, D.; Checquolo, S.; Campese, A.F.; Felli, M.P.; Gulino, A.; Screpanti, I. Notch3: From subtle structural differences to functional diversity. Oncogene 2008, 27, 5092–5098. [Google Scholar] [CrossRef]
- Ghosal, S.; Banerjee, S. In silico bioinformatics analysis for identification of differentially expressed genes and therapeutic drug molecules in Glucocorticoid-resistant Multiple myeloma. Med. Oncol. 2022, 39, 53. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, L.; Holmfeldt, L.; Wu, G.; Heatley, S.L.; Payne-Turner, D.; Easton, J.; Chen, X.; Wang, J.; Rusch, M.; et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012, 481, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, A.; Roeder, I.; Seifert, M. Comparative Gene Expression Analysis Reveals Similarities and Differences of Chronic Myeloid Leukemia Phases. Cancers 2022, 14, 256. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Cong, Y.; Li, C.; Zhang, C.; Lin, H. Predictive value of early molecular response for deep molecular response in chronic phase of chronic myeloid leukemia. Medicine 2019, 98, e15222. [Google Scholar] [CrossRef]
- Zhang, X.S.; Gale, R.P.; Zhang, M.J.; Huang, X.J.; Jiang, Q. A predictive scoring system for therapy-failure in persons with chronic myeloid leukemia receiving initial imatinib therapy. Leukemia 2022, 36, 1336–1342. [Google Scholar] [CrossRef] [PubMed]
- Hanfstein, B.; Muller, M.C.; Hehlmann, R.; Erben, P.; Lauseker, M.; Fabarius, A.; Schnittger, S.; Haferlach, C.; Gohring, G.; Proetel, U.; et al. Early molecular and cytogenetic response is predictive for long-term progression-free and overall survival in chronic myeloid leukemia (CML). Leukemia 2012, 26, 2096–2102. [Google Scholar] [CrossRef]
- Ko, P.S.; Yu, Y.B.; Liu, Y.C.; Wu, Y.T.; Hung, M.H.; Gau, J.P.; Liu, C.J.; Hsiao, L.T.; Chen, P.M.; Chiou, T.J.; et al. Moderate anemia at diagnosis is an independent prognostic marker of the EUTOS, Sokal, and Hasford scores for survival and treatment response in chronic-phase, chronic myeloid leukemia patients with frontline imatinib. Curr. Med. Res. Opin. 2017, 33, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shi, Y.; Yan, Z.; He, Z.; Ding, B.; Tao, S.; Li, Y.; Yu, L.; Wang, C. Impact of anemia on the outcomes of chronic phase chronic myeloid leukemia in TKI era. Hematology 2020, 25, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2018 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2018, 93, 442–459. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Saglio, G.; Kantarjian, H.M.; Baccarani, M.; Mayer, J.; Boque, C.; Shah, N.P.; Chuah, C.; Casanova, L.; Bradley-Garelik, B.; et al. Final 5-Year Study Results of DASISION: The Dasatinib Versus Imatinib Study in Treatment-Naive Chronic Myeloid Leukemia Patients Trial. J. Clin. Oncol. 2016, 34, 2333–2340. [Google Scholar] [CrossRef] [PubMed]
- Hochhaus, A.; Saglio, G.; Hughes, T.P.; Larson, R.A.; Kim, D.W.; Issaragrisil, S.; le Coutre, P.D.; Etienne, G.; Dorlhiac-Llacer, P.E.; Clark, R.E.; et al. Long-term benefits and risks of frontline nilotinib vs imatinib for chronic myeloid leukemia in chronic phase: 5-year update of the randomized ENESTnd trial. Leukemia 2016, 30, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Nakamae, H.; Yamamoto, M.; Sakaida, E.; Kanda, Y.; Ohmine, K.; Ono, T.; Matsumura, I.; Ishikawa, M.; Aoki, M.; Maki, A.; et al. Nilotinib vs. imatinib in Japanese patients with newly diagnosed chronic myeloid leukemia in chronic phase: 10-year followup of the Japanese subgroup of the randomized ENESTnd trial. Int. J. Hematol. 2022, 115, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Sato, E.; Iriyama, N.; Tokuhira, M.; Takaku, T.; Ishikawa, M.; Nakazato, T.; Sugimoto, K.J.; Fujita, H.; Kimura, Y.; Fujioka, I.; et al. The EUTOS long-term survival score predicts disease-specific mortality and molecular responses among patients with chronic myeloid leukemia in a practice-based cohort. Cancer Med. 2020, 9, 8931–8939. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.S.; Gale, R.P.; Huang, X.J.; Jiang, Q. Is the Sokal or EUTOS long-term survival (ELTS) score a better predictor of responses and outcomes in persons with chronic myeloid leukemia receiving tyrosine-kinase inhibitors? Leukemia 2022, 36, 482–491. [Google Scholar] [CrossRef] [PubMed]
| Variables | CML-CP | CML-CP with NGS |
|---|---|---|
| Age, years, median (range) | 45 (18–86) | 50 (25–84) |
| Sex | ||
| Male (n, %) | 83 (55.0%) | 11 (50.0%) |
| female (n, %) | 68 (45.0%) | 11 (50.0%) |
| WBC counts, ×109/L, median (range) | 94.8 (2.5–524.5) | 59 (11.4–367.0) |
| HB, g/L, median (range) | 112 (43–173) | 115 (62–162) |
| PLT counts, ×109/L, median (range) | 583 (14–3526) | 529 (110–1558) |
| Percentage of EOS, %, median (range) | 2.4 (0.0–14.0) | 2.5 (0–14) |
| Percentage of BAS, %, median (range) | 4.3 (0.0–15.3) | 5.4 (2–15.3) |
| Splenomegaly, cm, median (range) | 3.5 (0.0–20.0) | 4.7 (0–8.3) |
| Socal score | ||
| Low risk (n, %) | 71 (47.0%) | 7 (31.8%) |
| Medium risk (n, %) | 56 (37.1%) | 5 (22.7%) |
| High risk (n, %) | 24 (15.9%) | 10 (45.5%) |
| Hasford score | ||
| Low risk (n, %) | 82 (54.3%) | 8 (36.4%) |
| Medium risk (n, %) | 54 (35.8%) | 10 (45.5%) |
| High risk (n, %) | 15 (9.9%) | 4 (18.2%) |
| EUTOS score | ||
| Low risk (n, %) | 141 (93.4%) | 19 (86.4%) |
| High risk (n, %) | 10 (6.6%) | 3 (13.6%) |
| ELTS score | ||
| Low risk (n, %) | 101 (66.9%) | 11 (50.0%) |
| Medium risk (n, %) | 38 (25.2%) | 8 (36.4%) |
| High risk (n, %) | 12 (7.9%) | 3 (13.6%) |
| 3M-EMR | ||
| Yes (n, %) | 103 (68.2%) | 10 (45.5%) |
| No (n, %) | 48 (31.8%) | 12 (54.5%) |
| 6M-BCR∷ABLIS ≤ 1% | ||
| Yes (n, %) | 102 (67.5%) | 12 (54.5%) |
| No (n, %) | 49 (32.5%) | 10 (45.5%) |
| 12M-MMR | ||
| Yes (n, %) | 83 (55.0%) | 11 (50.0%) |
| No (n, %) | 68 (45.0%) | 11 (50.0%) |
| Gene | 12M-MMR | p Value | |
|---|---|---|---|
| YES (n = 11) | NO (n = 11) | ||
| NOTCH3 | 0 | 3 | 0.214 |
| RELN | 0 | 2 | 0.476 |
| DIS3 | 1 | 3 | 0.586 |
| KMT2C | 4 | 2 | 0.635 |
| ASXL1 | 5 | 4 | >0.999 |
| ATM | 1 | 2 | >0.999 |
| DNMT3A | 1 | 2 | >0.999 |
| MED12 | 1 | 1 | >0.999 |
| STAT5A | 1 | 1 | >0.999 |
| STAG2 | 1 | 1 | >0.999 |
| NOTCH2 | 1 | 0 | >0.999 |
| NOTCH4 | 1 | 0 | >0.999 |
| JAK3 | 1 | 0 | >0.999 |
| PIGA | 1 | 0 | >0.999 |
| SUZ12 | 1 | 0 | >0.999 |
| ATG2B | 1 | 0 | >0.999 |
| ABL1 | 1 | 0 | >0.999 |
| CSMD1 | 1 | 0 | >0.999 |
| RUNX1 | 1 | 0 | >0.999 |
| FAT1 | 0 | 1 | >0.999 |
| GNA13 | 0 | 1 | >0.999 |
| TPMT | 0 | 1 | >0.999 |
| ETV6 | 0 | 1 | >0.999 |
| KMT2A | 0 | 1 | >0.999 |
| KMT2D | 0 | 1 | >0.999 |
| Gene | 36 Months-MR4.0 | p Value | |
|---|---|---|---|
| YES (n = 16) | NO (n = 6) | ||
| ASXL1 | 4 | 5 | 0.023 |
| STAT5A | 0 | 2 | 0.065 |
| RELN | 0 | 2 | 0.065 |
| ATM | 1 | 2 | 0.169 |
| NOTCH3 | 1 | 2 | 0.169 |
| FAT1 | 0 | 1 | 0.273 |
| GNA13 | 0 | 1 | 0.273 |
| TPMT | 0 | 1 | 0.273 |
| NOTCH4 | 0 | 1 | 0.273 |
| JAK3 | 0 | 1 | 0.273 |
| ETV6 | 0 | 1 | 0.273 |
| CSMD1 | 0 | 1 | 0.273 |
| MED12 | 1 | 1 | 0.481 |
| STAG2 | 1 | 1 | 0.483 |
| DNMT3A | 3 | 0 | 0.532 |
| KMT2C | 4 | 2 | >0.999 |
| DIS3 | 3 | 1 | >0.999 |
| KMT2A | 0 | 1 | >0.999 |
| NOTCH2 | 1 | 0 | >0.999 |
| PIGA | 1 | 0 | >0.999 |
| SUZ12 | 1 | 0 | >0.999 |
| ATG2B | 1 | 0 | >0.999 |
| ABL1 | 1 | 0 | >0.999 |
| KMT2D | 1 | 0 | >0.999 |
| RUNX1 | 1 | 0 | >0.999 |
| Variables | MR4.5 | p Value | |
|---|---|---|---|
| No (n = 26) | Yes (n = 125) | ||
| Age, years, median (range) | 55 (25–86) | 44 (18–84) | 0.018 |
| Sex (male/female) | 16/10 | 67/58 | 0.459 |
| WBC counts, ×109/L, median (range) | 120.4 (2.5–366.9) | 93.8 (8.5–524.5) | 0.165 |
| HB, g/L, median (range) | 104 (43–146) | 114 (62–173) | 0.001 |
| PLT counts, ×109/L, median (range) | 602 (14–1409) | 579 (100–3526) | 0.783 |
| percentage of EOS, %, median (range) | 2.8 (0.4–10.0) | 2.3 (0.0–14.0) | 0.113 |
| percentage of BAS, %, median (range) | 3.7 (0.0–8.63) | 4.5 (0.0–15.3) | 0.056 |
| Splenomegaly, cm, median (range) | 5.1 (0.0–14.0) | 3.5 (0.0–20.0) | 0.432 |
| Socal score (Low/medium/high risk) | 9/13/4 | 62/43/20 | 0.294 |
| Hasford score (Low/medium/high risk) | 10/14/2 | 72/40/13 | 0.106 |
| EUTOS score (Low/high risk) | 25/1 | 116/9 | 0.848 |
| ELTS score (Low/medium/high risk) | 13/10/3 | 88/28/9 | 0.132 |
| 3M-EMR (Yes/No) | 11/15 | 92/33 | 0.002 |
| 6M-BCR∷ABLIS ≤ 1% (Yes/No) | 13/13 | 89/36 | 0.036 |
| 12M-MMR (Yes/No) | 1/25 | 82/43 | <0.001 |
| Treatment effect | First-Line First-Generation TKI n = 115 (A) | First-Line First-Generation Original TKI n = 68 (A1) | First-Line First-Generation Generic TKI n = 47 (A2) | Second-Line Second-Generation TKI n = 27 (B) | First-Line Second-Generation TKI n = 9 (C) | p Value | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| A vs. C | B vs. C | A1 vs. A2 | A1 vs. C | A2 vs. C | ||||||
| 3M-EMR (n, %) | 86 (74.8% ) | 56 (82.4% ) | 30 (63.8%r) | 9 (33.3%r) | 8 (88.9%) | 0.584 | 0.012 | 0.025 | 0.985 | 0.278 |
| 6M-BCR∷ABLIS ≤ 1% (n, %r) | 85 (73.9%r) | 48 (70.6%r) | 37 (78.7%r) | 9 (33.3%r) | 8 (88.9%) | 0.549 | 0.012 | 0.329 | 0.447 | 0.806 |
| 12M-MMR (n, %r) | 69 (60.0%r) | 45 (66.2%r) | 24 (48.0%r) | 7 (25.9%r) | 7 (77.8%) | 0.484 | 0.018 | 0.104 | 0.749 | 0.267 |
| MR4.5 (n, %r) | 97 (84.3%r) | 60 (88.2%r) | 37 (78.7%r) | 20 (74.1%r) | 8 (88.9%) | >0.999 | 0.643 | 0.168 | >0.999 | 0.806 |
| Scoring System | Risk Stratification | 3 Months ≤ 10% (Yes/Nor) | p Value | 6 Months ≤ 1% (Yes/Nor) | p Value | 12 Months ≤ 0.1% (Yes/Nor) | p Value |
|---|---|---|---|---|---|---|---|
| EUTOS score | Low risk | 99/42 | 0.103 | 97/44 | 0.220 | 81/60 | 0.049 |
| High risk | 4/6 | 5/5 | 2/8 | ||||
| Sokal score | Low risk | 54/17 | 0.140 | 49/22 | 0.795 | 41/30 | 0.777 |
| Medium risk | 35/21 | 36/20 | 30/26 | ||||
| High risk | 14/10 | 17/7 | 12/12 | ||||
| Hasford score | Low risk | 62/20 | 0.088 | 58/24 | 0.621 | 50/32 | 0.213 |
| Medium risk | 33/21 | 35/19 | 27/27 | ||||
| High risk | 8/7 | 9/6 | 6/9 | ||||
| ELTS score | Low risk | 78/23 | 0.001 | 74/27 | 0.090 | 65/36 | 0.004 |
| Medium risk | 21/17 | 22/16 | 13/25 | ||||
| High risk | 4/8 | 6/6 | 5/7 |
| Mutations (A1) | No Mutations (A2) | ASXL1 Mutations (A3) | Other Mutations (A4) | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| A1 vs. A2 | A2 vs. A3 | A2 vs. A4 | A3 vs. A4 | |||||
| EUTOS score (n, %) | ||||||||
| Low risk | 19 (86.4%) | 122 (94.6%) | 8 (88.9%) | 11 (84.6%) | 0.333 | >0.999 | 0.419 | >0.999 |
| High risk | 3 (13.6%) | 7 (5.4%) | 1 (11.1%) | 2 (15.4%) | ||||
| Socal score (n, %) | ||||||||
| Low risk | 7 (31.8%) | 64 (49.6%) | 3 (33.3%) | 4 (30.8%) | 0.001 | 0.051 | 0.012 | 0.992 |
| Medium risk | 5 (22.7%) | 51 (39.5%) | 2 (22.2%) | 3 (23.1%) | ||||
| High risk | 10 (45.5%) | 14 (10.9%) | 4 (44.4%) | 6 (46.2%) | ||||
| Hasford score (n, %) | ||||||||
| Low risk | 8 (36.4%) | 74 (57.4%) | 4 (44.4%) | 4 (30.8%) | 0.149 | 0.470 | 0.181 | 0.633 |
| Medium risk | 10 (45.5%) | 44 (34.1%) | 3 (33.3%) | 7 (53.8%) | ||||
| High risk | 4 (18.2%) | 11 (8.5%) | 2 (22.2%) | 2 (15.4%) | ||||
| ELTS score (n, %) | ||||||||
| Low risk | 11 (50.0%) | 90 (69.8%) | 6 (66.7%) | 5 (38.5%) | 0.198 | 0.836 | 0.062 | 0.247 |
| Medium risk | 8 (36.4%) | 30 (23.3%) | 3 (33.3%) | 5 (38.5%) | ||||
| High risk | 3 (13.6%) | 9 (7.0%) | 0 (0.0%) | 3 (23.1%) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Chen, D.; Xu, X.; Zhang, L.; Wang, S.; Jin, K.; Zheng, Y.; Zhu, X.; Jin, J.; Huang, J. Targeted Next-Generation Sequencing Identifies Additional Mutations Other than BCR∷ABL in Chronic Myeloid Leukemia Patients: A Chinese Monocentric Retrospective Study. Cancers 2022, 14, 5752. https://doi.org/10.3390/cancers14235752
Hu S, Chen D, Xu X, Zhang L, Wang S, Jin K, Zheng Y, Zhu X, Jin J, Huang J. Targeted Next-Generation Sequencing Identifies Additional Mutations Other than BCR∷ABL in Chronic Myeloid Leukemia Patients: A Chinese Monocentric Retrospective Study. Cancers. 2022; 14(23):5752. https://doi.org/10.3390/cancers14235752
Chicago/Turabian StyleHu, Shiwei, Dan Chen, Xiaofei Xu, Lan Zhang, Shengjie Wang, Keyi Jin, Yan Zheng, Xiaoqiong Zhu, Jie Jin, and Jian Huang. 2022. "Targeted Next-Generation Sequencing Identifies Additional Mutations Other than BCR∷ABL in Chronic Myeloid Leukemia Patients: A Chinese Monocentric Retrospective Study" Cancers 14, no. 23: 5752. https://doi.org/10.3390/cancers14235752
APA StyleHu, S., Chen, D., Xu, X., Zhang, L., Wang, S., Jin, K., Zheng, Y., Zhu, X., Jin, J., & Huang, J. (2022). Targeted Next-Generation Sequencing Identifies Additional Mutations Other than BCR∷ABL in Chronic Myeloid Leukemia Patients: A Chinese Monocentric Retrospective Study. Cancers, 14(23), 5752. https://doi.org/10.3390/cancers14235752






