The Somatic Mutation Landscape of UDP-Glycosyltransferase (UGT) Genes in Human Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Assessment of Somatic Mutations of UGT Genes in Human Cancers
2.1.1. The Multi-Center Mutation Calling in Multiple Cancers (MC3) Project
2.1.2. Extracting Individual Somatic Mutations from the MC3 MAF File and Assigning Them to Each of the 33 TCGA Cancer Types
2.1.3. Verifying and/or Correcting the Assignments of Individual Somatic Mutations from the MC3 MAF to Each of the 22 UGT Genes
2.2. Assessment of Somatic Mutations of UGT Genes in Human Cancer Cell Lines
2.3. Statistical Analysis
3. Results
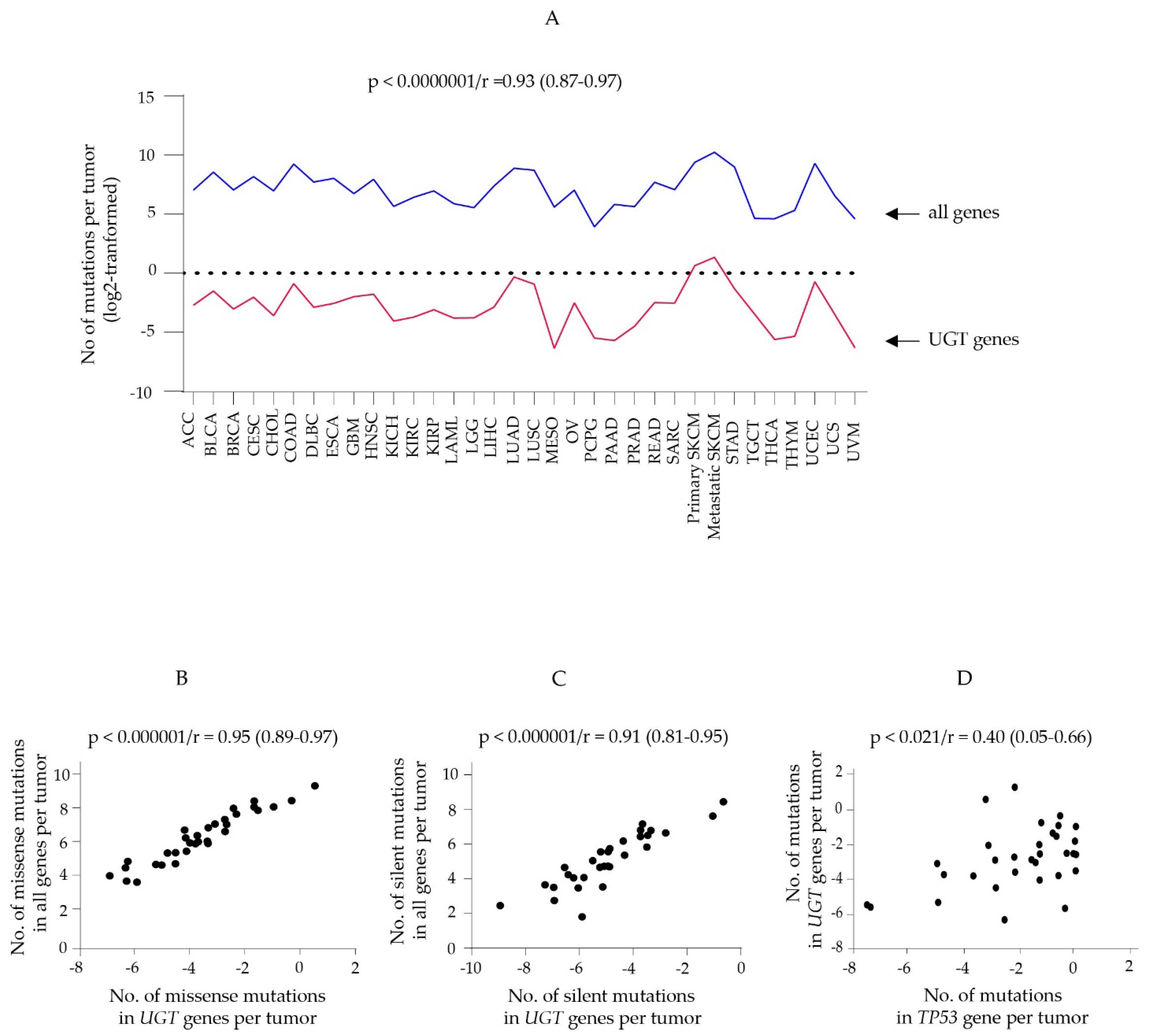
3.1. Somatic Mutations of Protein-Coding Genes in Human Cancers
3.2. Somatic Mutations in UGT Genes in Human Cancers
3.2.1. Summary
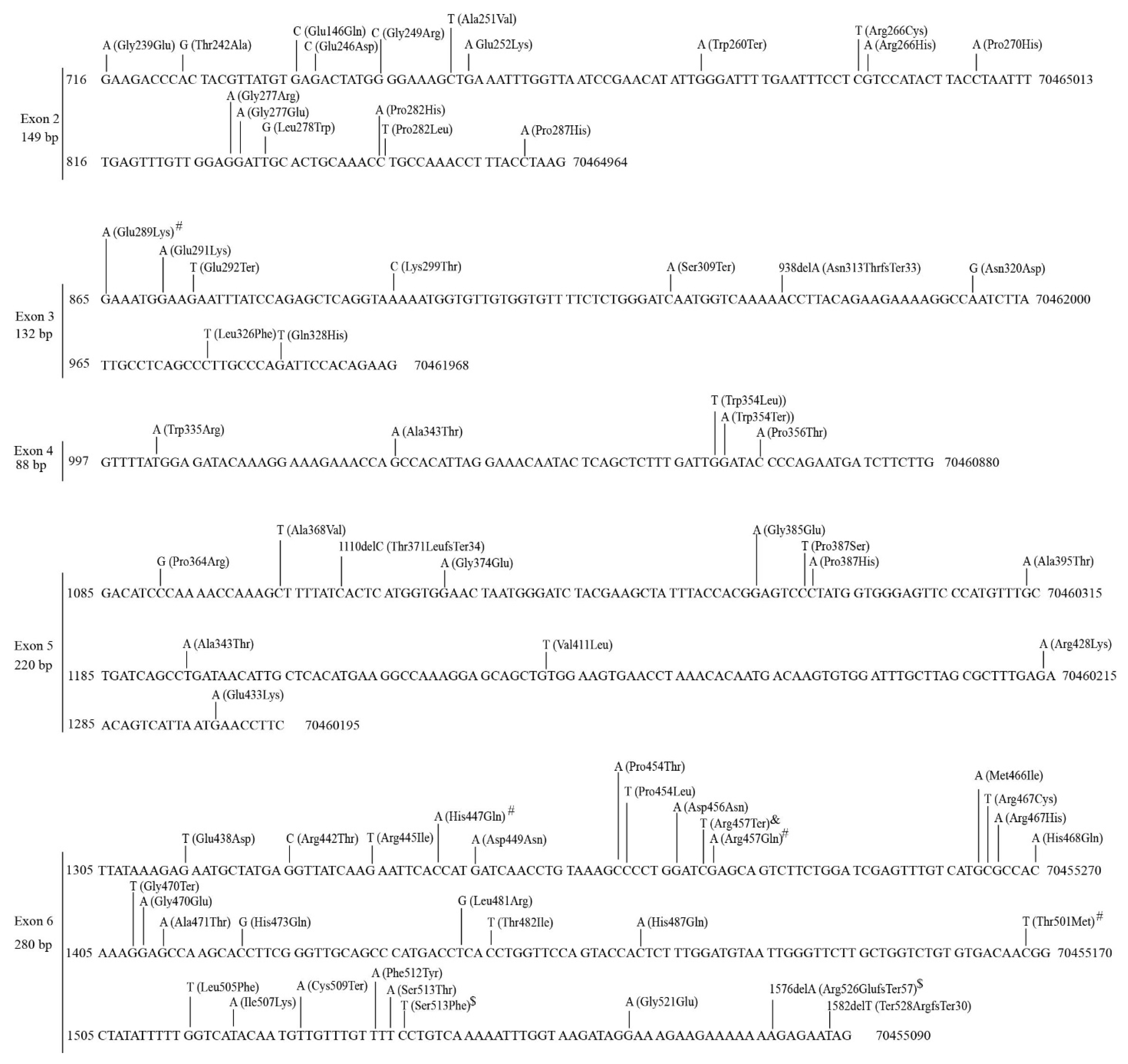
3.2.2. Mutations in the UGT1A Subfamily Genes
3.2.3. Mutations in the UGT2A Subfamily of Genes
3.2.4. Mutations in the UGT2B Subfamily Genes
3.2.5. Mutations in the UGT3 Subfamily Genes
3.2.6. Mutations in the UGT8 Gene
3.2.7. Mutations in the 5′ UTRs of UGT Genes
3.2.8. Mutations in the 3′ UTRs of UGT Genes
3.2.9. Mutations in the Splice Sites of UGT Genes
3.2.10. Recurrent Mutations in UGT Genes
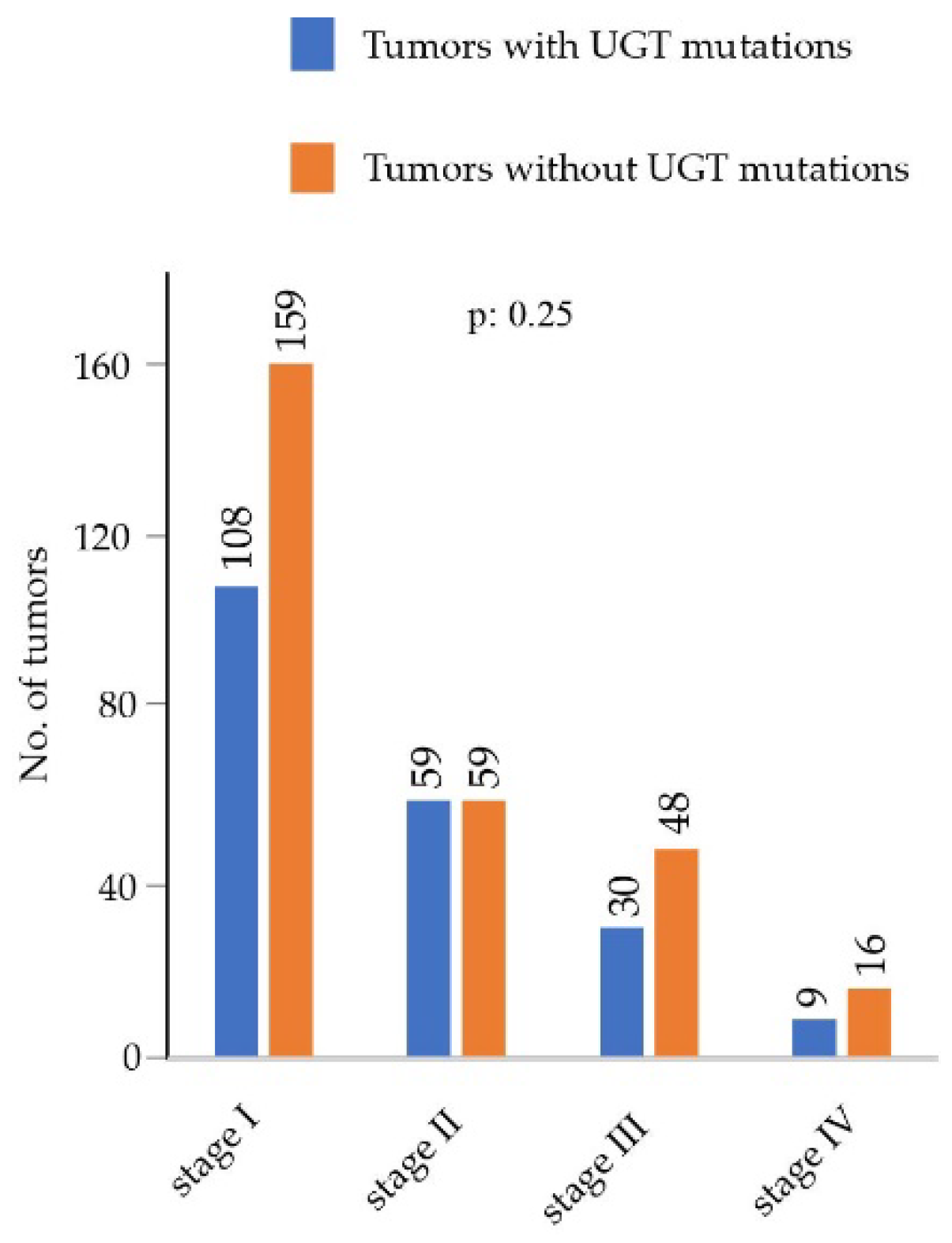
3.3. Assesment of Associations of UGT Mutations with Clinicopathological Parameters Using the LUAD Cohort
3.4. Mutations in UGT Genes in Human Cancer Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mackenzie, P.I.; Bock, K.W.; Burchell, B.; Guillemette, C.; Ikushiro, S.; Iyanagi, T.; Miners, J.O.; Owens, I.S.; Nebert, D.W. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genom. 2005, 15, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, P.I.; Owens, I.S.; Burchell, B.; Bock, K.W.; Bairoch, A.; Belanger, A.; Fournel-Gigleux, S.; Green, M.; Hum, D.W.; Iyanagi, T.; et al. The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 1997, 7, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Meech, R.; Hu, D.G.; McKinnon, R.A.; Mubarokah, S.N.; Haines, A.Z.; Nair, P.C.; Rowland, A.; Mackenzie, P.I. The UDP-Glycosyltransferase (UGT) Superfamily: New Members, New Functions, and Novel Paradigms. Physiol. Rev. 2019, 99, 1153–1222. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Hulin, J.U.; Nair, P.C.; Haines, A.Z.; McKinnon, R.A.; Mackenzie, P.I.; Meech, R. The UGTome: The expanding diversity of UDP glycosyltransferases and its impact on small molecule metabolism. Pharmacol. Ther. 2019, 204, 107414. [Google Scholar] [CrossRef]
- Hu, D.G.; Meech, R.; McKinnon, R.A.; Mackenzie, P.I. Transcriptional regulation of human UDP-glucuronosyltransferase genes. Drug. Metab. Rev. 2014, 46, 421–458. [Google Scholar] [CrossRef]
- MacKenzie, P.I.; Rogers, A.; Elliot, D.J.; Chau, N.; Hulin, J.A.; Miners, J.O.; Meech, R. The novel UDP glycosyltransferase 3A2: Cloning, catalytic properties, and tissue distribution. Mol. Pharmacol. 2011, 79, 472–478. [Google Scholar] [CrossRef]
- Mackenzie, P.I.; Rogers, A.; Treloar, J.; Jorgensen, B.R.; Miners, J.O.; Meech, R. Identification of UDP glycosyltransferase 3A1 as a UDP N-acetylglucosaminyltransferase. J. Biol. Chem. 2008, 283, 36205–36210. [Google Scholar] [CrossRef]
- Meech, R.; Mubarokah, N.; Shivasami, A.; Rogers, A.; Nair, P.C.; Hu, D.G.; McKinnon, R.A.; Mackenzie, P.I. A novel function for UDP glycosyltransferase 8: Galactosidation of bile acids. Mol. Pharmacol. 2015, 87, 442–450. [Google Scholar] [CrossRef]
- Vergara, A.G.; Watson, C.J.W.; Chen, G.; Lazarus, P. UDP-Glycosyltransferase 3A Metabolism of Polycyclic Aromatic Hydrocarbons: Potential Importance in Aerodigestive Tract Tissues. Drug Metab. Dispos. 2020, 48, 160–168. [Google Scholar] [CrossRef]
- Takano, M.; Sugiyama, T. UGT1A1 polymorphisms in cancer: Impact on irinotecan treatment. Pharmgenomics Pers. Med. 2017, 10, 61–68. [Google Scholar] [CrossRef]
- Minotti, G.; Menna, P.; Salvatorelli, E.; Cairo, G.; Gianni, L. Anthracyclines: Molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Mackenzie, P.I.; Lu, L.; Meech, R.; McKinnon, R.A. Induction of human UDP-Glucuronosyltransferase 2B7 gene expression by cytotoxic anticancer drugs in liver cancer HepG2 cells. Drug Metab. Dispos. 2015, 43, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Rogers, A.; Mackenzie, P.I. Epirubicin upregulates UDP glucuronosyltransferase 2B7 expression in liver cancer cells via the p53 pathway. Mol. Pharmacol. 2014, 85, 887–897. [Google Scholar] [CrossRef]
- Joy, A.A.; Vos, L.J.; Pituskin, E.; Cook, S.F.; Bies, R.R.; Vlahadamis, A.; King, K.; Basi, S.K.; Meza-Junco, J.; Mackey, J.R.; et al. Uridine Glucuronosyltransferase 2B7 Polymorphism-Based Pharmacogenetic Dosing of Epirubicin in FEC Chemotherapy for Early-Stage Breast Cancer. Clin. Breast Cancer 2021, 21, e584–e593. [Google Scholar] [CrossRef] [PubMed]
- Parmar, S.; Stingl, J.C.; Huber-Wechselberger, A.; Kainz, A.; Renner, W.; Langsenlehner, U.; Krippl, P.; Brockmoller, J.; Haschke-Becher, E. Impact of UGT2B7 His268Tyr polymorphism on the outcome of adjuvant epirubicin treatment in breast cancer. Breast Cancer Res. 2011, 13, R57. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.; King, C.D.; Whitington, P.F.; Green, M.D.; Roy, S.K.; Tephly, T.R.; Coffman, B.L.; Ratain, M.J. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J. Clin. Investig. 1998, 101, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Strassburg, C.P. Hyperbilirubinemia syndromes (Gilbert-Meulengracht, Crigler-Najjar, Dubin-Johnson, and Rotor syndrome). Best Pract. Res. Clin. Gastroenterol. 2010, 24, 555–571. [Google Scholar] [CrossRef]
- Di Paolo, A.; Bocci, G.; Polillo, M.; Del Re, M.; Di Desidero, T.; Lastella, M.; Danesi, R. Pharmacokinetic and pharmacogenetic predictive markers of irinotecan activity and toxicity. Curr. Drug Metab. 2011, 12, 932–943. [Google Scholar] [CrossRef]
- Hoskins, J.M.; Goldberg, R.M.; Qu, P.; Ibrahim, J.G.; McLeod, H.L. UGT1A1*28 genotype and irinotecan-induced neutropenia: Dose matters. J. Natl. Cancer Inst. 2007, 99, 1290–1295. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, D.; Kuang, Q.; Liu, G.; Xu, W. Association of UGT1A1*28 polymorphisms with irinotecan-induced toxicities in colorectal cancer: A meta-analysis in Caucasians. Pharmacogenomics J. 2014, 14, 120–129. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, J.F.; Zhang, J.; Kong, S.J.; Zhang, H.Y.; Chen, X.M. UGT1A1*6 polymorphisms are correlated with irinotecan-induced neutropenia: A systematic review and meta-analysis. Cancer Chemother. Pharmacol. 2017, 80, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Allain, E.P.; Rouleau, M.; Levesque, E.; Guillemette, C. Emerging roles for UDP-glucuronosyltransferases in drug resistance and cancer progression. Br. J. Cancer 2020, 122, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Mackenzie, P.I.; McKinnon, R.A.; Meech, R. Genetic polymorphisms of human UDP-glucuronosyltransferase (UGT) genes and cancer risk. Drug Metab. Rev. 2016, 48, 47–69. [Google Scholar] [CrossRef] [PubMed]
- Mazaris, E.; Tsiotras, A. Molecular pathways in prostate cancer. Nephrourol. Mon. 2013, 5, 792–800. [Google Scholar] [CrossRef]
- Chouinard, S.; Yueh, M.F.; Tukey, R.H.; Giton, F.; Fiet, J.; Pelletier, G.; Barbier, O.; Belanger, A. Inactivation by UDP-glucuronosyltransferase enzymes: The end of androgen signaling. J. Steroid. Biochem. Mol. Biol. 2008, 109, 247–253. [Google Scholar] [CrossRef]
- Turgeon, D.; Carrier, J.S.; Levesque, E.; Hum, D.W.; Belanger, A. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology 2001, 142, 778–787. [Google Scholar] [CrossRef]
- Cai, L.; Huang, W.; Chou, K.C. Prostate cancer with variants in CYP17 and UGT2B17 genes: A meta-analysis. Protein. Pept. Lett. 2012, 19, 62–69. [Google Scholar] [CrossRef]
- Kpoghomou, M.A.; Soatiana, J.E.; Kalembo, F.W.; Bishwajit, G.; Sheng, W. UGT2B17 Polymorphism and Risk of Prostate Cancer: A Meta-Analysis. ISRN Oncol. 2013, 2013, 465916. [Google Scholar] [CrossRef]
- Li, H.; Xie, N.; Chen, R.; Verreault, M.; Fazli, L.; Gleave, M.E.; Barbier, O.; Dong, X. UGT2B17 Expedites Progression of Castration-Resistant Prostate Cancers by Promoting Ligand-Independent AR Signaling. Cancer Res. 2016, 76, 6701–6711. [Google Scholar] [CrossRef]
- Grant, D.J.; Chen, Z.; Howard, L.E.; Wiggins, E.; De Hoedt, A.; Vidal, A.C.; Carney, S.T.; Squires, J.; Magyar, C.E.; Huang, J.; et al. UDP-glucuronosyltransferases and biochemical recurrence in prostate cancer progression. BMC Cancer 2017, 17, 463. [Google Scholar] [CrossRef]
- Margaillan, G.; Rouleau, M.; Fallon, J.K.; Caron, P.; Villeneuve, L.; Turcotte, V.; Smith, P.C.; Joy, M.S.; Guillemette, C. Quantitative profiling of human renal UDP-glucuronosyltransferases and glucuronidation activity: A comparison of normal and tumoral kidney tissues. Drug Metab. Dispos. 2015, 43, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.B.; Mostaghel, E.A.; Vessella, R.; Hess, D.L.; Kalhorn, T.F.; Higano, C.S.; True, L.D.; Nelson, P.S. Maintenance of intratumoral androgens in metastatic prostate cancer: A mechanism for castration-resistant tumor growth. Cancer Res. 2008, 68, 4447–4454. [Google Scholar] [CrossRef] [PubMed]
- Paquet, S.; Fazli, L.; Grosse, L.; Verreault, M.; Tetu, B.; Rennie, P.S.; Belanger, A.; Barbier, O. Differential expression of the androgen-conjugating UGT2B15 and UGT2B17 enzymes in prostate tumor cells during cancer progression. J. Clin. Endocrinol. Metab. 2012, 97, E428–E432. [Google Scholar] [CrossRef] [PubMed]
- Stanbrough, M.; Bubley, G.J.; Ross, K.; Golub, T.R.; Rubin, M.A.; Penning, T.M.; Febbo, P.G.; Balk, S.P. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006, 66, 2815–2825. [Google Scholar] [CrossRef]
- Court, M.H.; Zhang, X.; Ding, X.; Yee, K.K.; Hesse, L.M.; Finel, M. Quantitative distribution of mRNAs encoding the 19 human UDP-glucuronosyltransferase enzymes in 26 adult and 3 fetal tissues. Xenobiotica 2012, 42, 266–277. [Google Scholar] [CrossRef]
- Nakamura, A.; Nakajima, M.; Yamanaka, H.; Fujiwara, R.; Yokoi, T. Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab. Dispos. 2008, 36, 1461–1464. [Google Scholar] [CrossRef]
- Nishimura, M.; Naito, S. Tissue-specific mRNA expression profiles of human phase I metabolizing enzymes except for cytochrome P450 and phase II metabolizing enzymes. Drug Metab. Pharmacokinet. 2006, 21, 357–374. [Google Scholar] [CrossRef]
- Ohno, S.; Nakajin, S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab. Dispos. 2009, 37, 32–40. [Google Scholar] [CrossRef]
- Hu, D.G.; Marri, S.; Mackenzie, P.I.; Hulin, J.A.; McKinnon, R.A.; Meech, R. The Expression Profiles and Deregulation of UDP-Glycosyltransferase (UGT) Genes in Human Cancers and Their Association with Clinical Outcomes. Cancers 2021, 13, 4491. [Google Scholar] [CrossRef]
- Beyerle, J.; Holowatyj, A.N.; Haffa, M.; Frei, E.; Gigic, B.; Schrotz-King, P.; Boehm, J.; Habermann, N.; Stiborova, M.; Scherer, D.; et al. Expression Patterns of Xenobiotic-Metabolizing Enzymes in Tumor and Adjacent Normal Mucosa Tissues among Patients with Colorectal Cancer: The ColoCare Study. Cancer Epidemiol. Biomark. Prev. 2020, 29, 460–469. [Google Scholar] [CrossRef]
- Cengiz, B.; Yumrutas, O.; Bozgeyik, E.; Borazan, E.; Igci, Y.Z.; Bozgeyik, I.; Oztuzcu, S. Differential expression of the UGT1A family of genes in stomach cancer tissues. Tumour. Biol. 2015, 36, 5831–5837. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Marri, S.; McKinnon, R.A.; Mackenzie, P.I.; Meech, R. Deregulation of the Genes that Are Involved in Drug Absorption, Distribution, Metabolism, and Excretion in Hepatocellular Carcinoma. J. Pharmacol. Exp. Ther. 2019, 368, 363–381. [Google Scholar] [CrossRef] [PubMed]
- Strassburg, C.P.; Manns, M.P.; Tukey, R.H. Differential down-regulation of the UDP-glucuronosyltransferase 1A locus is an early event in human liver and biliary cancer. Cancer Res. 1997, 57, 2979–2985. [Google Scholar] [PubMed]
- Strassburg, C.P.; Nguyen, N.; Manns, M.P.; Tukey, R.H. Polymorphic expression of the UDP-glucuronosyltransferase UGT1A gene locus in human gastric epithelium. Mol. Pharmacol. 1998, 54, 647–654. [Google Scholar] [PubMed]
- Allain, E.P.; Rouleau, M.; Vanura, K.; Tremblay, S.; Vaillancourt, J.; Bat, V.; Caron, P.; Villeneuve, L.; Labriet, A.; Turcotte, V.; et al. UGT2B17 modifies drug response in chronic lymphocytic leukaemia. Br. J. Cancer 2020, 123, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Gruber, M.; Bellemare, J.; Hoermann, G.; Gleiss, A.; Porpaczy, E.; Bilban, M.; Le, T.; Zehetmayer, S.; Mannhalter, C.; Gaiger, A.; et al. Overexpression of uridine diphospho glucuronosyltransferase 2B17 in high-risk chronic lymphocytic leukemia. Blood 2013, 121, 1175–1183. [Google Scholar] [CrossRef]
- Giuliani, L.; Ciotti, M.; Stoppacciaro, A.; Pasquini, A.; Silvestri, I.; De Matteis, A.; Frati, L.; Agliano, A.M. UDP-glucuronosyltransferases 1A expression in human urinary bladder and colon cancer by immunohistochemistry. Oncol. Rep. 2005, 13, 185–191. [Google Scholar]
- Yang, W.; Ma, J.; Zhou, W.; Li, Z.; Zhou, X.; Cao, B.; Zhang, Y.; Liu, J.; Yang, Z.; Zhang, H.; et al. Identification of hub genes and outcome in colon cancer based on bioinformatics analysis. Cancer Manag. Res. 2019, 11, 323–338. [Google Scholar] [CrossRef]
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018, 6, 271–281.e7. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 173, 371–385.e318. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jane-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.A.; Lawrence, M.S.; Keats, J.J.; Cibulskis, K.; Sougnez, C.; Schinzel, A.C.; Harview, C.L.; Brunet, J.P.; Ahmann, G.J.; Adli, M.; et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011, 471, 467–472. [Google Scholar] [CrossRef]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Fan, Y.; Xi, L.; Hughes, D.S.; Zhang, J.; Zhang, J.; Futreal, P.A.; Wheeler, D.A.; Wang, W. MuSE: Accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol. 2016, 17, 178. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef]
- Larson, D.E.; Harris, C.C.; Chen, K.; Koboldt, D.C.; Abbott, T.E.; Dooling, D.J.; Ley, T.J.; Mardis, E.R.; Wilson, R.K.; Ding, L. SomaticSniper: Identification of somatic point mutations in whole genome sequencing data. Bioinformatics 2012, 28, 311–317. [Google Scholar] [CrossRef]
- Radenbaugh, A.J.; Ma, S.; Ewing, A.; Stuart, J.M.; Collisson, E.A.; Zhu, J.; Haussler, D. RADIA: RNA and DNA integrated analysis for somatic mutation detection. PLoS ONE 2014, 9, e111516. [Google Scholar] [CrossRef]
- Edwards, L.; Gupta, R.; Filipp, F.V. Hypermutation of DPYD Deregulates Pyrimidine Metabolism and Promotes Malignant Progression. Mol. Cancer Res. 2016, 14, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Mackenzie, P.I.; Nair, P.C.; McKinnon, R.A.; Meech, R. The Expression Profiles of ADME Genes in Human Cancers and Their Associations with Clinical Outcomes. Cancers 2020, 12, 3369. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, R.; Raffenne, J.; Paradis, V.; Couvelard, A.; de Reynies, A.; Blum, Y.; Cros, J. Prognostic Biomarkers in Pancreatic Cancer: Avoiding Errata When Using the TCGA Dataset. Cancers 2019, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Peran, I.; Madhavan, S.; Byers, S.W.; McCoy, M.D. Curation of the Pancreatic Ductal Adenocarcinoma Subset of the Cancer Genome Atlas Is Essential for Accurate Conclusions about Survival-Related Molecular Mechanisms. Clin. Cancer Res. 2018, 24, 3813–3819. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.B.; Light, N.; Fabrizio, D.; Zatzman, M.; Fuligni, F.; de Borja, R.; Davidson, S.; Edwards, M.; Elvin, J.A.; Hodel, K.P.; et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017, 171, 1042–1056.e1010. [Google Scholar] [CrossRef] [PubMed]
- Akbani, R.; Ng, P.K.; Werner, H.M.; Shahmoradgoli, M.; Zhang, F.; Ju, Z.; Liu, W.; Yang, J.Y.; Yoshihara, K.; Li, J.; et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 2014, 5, 3887. [Google Scholar] [CrossRef]
- Govindan, R.; Ding, L.; Griffith, M.; Subramanian, J.; Dees, N.D.; Kanchi, K.L.; Maher, C.A.; Fulton, R.; Fulton, L.; Wallis, J.; et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012, 150, 1121–1134. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Pfeifer, G.P.; You, Y.H.; Besaratinia, A. Mutations induced by ultraviolet light. Mutat. Res. 2005, 571, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Pleasance, E.D.; Stephens, P.J.; O’Meara, S.; McBride, D.J.; Meynert, A.; Jones, D.; Lin, M.L.; Beare, D.; Lau, K.W.; Greenman, C.; et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 2010, 463, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Bouffet, E.; Larouche, V.; Campbell, B.B.; Merico, D.; de Borja, R.; Aronson, M.; Durno, C.; Krueger, J.; Cabric, V.; Ramaswamy, V.; et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J. Clin. Oncol. 2016, 34, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- Johanns, T.M.; Miller, C.A.; Dorward, I.G.; Tsien, C.; Chang, E.; Perry, A.; Uppaluri, R.; Ferguson, C.; Schmidt, R.E.; Dahiya, S.; et al. Immunogenomics of Hypermutated Glioblastoma: A Patient with Germline POLE Deficiency Treated with Checkpoint Blockade Immunotherapy. Cancer Discov. 2016, 6, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Santin, A.D.; Bellone, S.; Buza, N.; Choi, J.; Schwartz, P.E.; Schlessinger, J.; Lifton, R.P. Regression of Chemotherapy-Resistant Polymerase epsilon (POLE) Ultra-Mutated and MSH6 Hyper-Mutated Endometrial Tumors with Nivolumab. Clin. Cancer Res. 2016, 22, 5682–5687. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002, 12, 436–446. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R., 3rd; Kalocsay, M.; Jane-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e316. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Donehower, L.A.; Soussi, T.; Korkut, A.; Liu, Y.; Schultz, A.; Cardenas, M.; Li, X.; Babur, O.; Hsu, T.K.; Lichtarge, O.; et al. Integrated analysis of TP53 gene and pathway alternations in the Cancer Genome Atlas. Cell Rep. 2019, 28, 1370–1384.e5. [Google Scholar] [CrossRef] [PubMed]
- Girard, H.; Levesque, E.; Bellemare, J.; Journault, K.; Caillier, B.; Guillemette, C. Genetic diversity at the UGT1 locus is amplified by a novel 3’ alternative splicing mechanism leading to nine additional UGT1A proteins that act as regulators of glucuronidation activity. Pharmacogenet. Genom. 2007, 17, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Hulin, J.A.; Wijayakumara, D.D.; McKinnon, R.A.; Mackenzie, P.I.; Meech, R. Intergenic Splicing between Four Adjacent UGT Genes (2B15, 2B29P2, 2B17, 2B29P1) Gives Rise to Variant UGT Proteins That Inhibit Glucuronidation via Protein-Protein Interactions. Mol. Pharmacol. 2018, 94, 938–952. [Google Scholar] [CrossRef] [PubMed]
- Menard, V.; Eap, O.; Roberge, J.; Harvey, M.; Levesque, E.; Guillemette, C. Transcriptional diversity at the UGT2B7 locus is dictated by extensive pre-mRNA splicing mechanisms that give rise to multiple mRNA splice variants. Pharmacogenet. Genom. 2011, 21, 631–641. [Google Scholar] [CrossRef]
- Kozak, M. An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987, 15, 8125–8148. [Google Scholar] [CrossRef]
- Kozak, M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997, 16, 2482–2492. [Google Scholar] [CrossRef]
- Kozak, M. Initiation of translation in prokaryotes and eukaryotes. Gene 1999, 234, 187–208. [Google Scholar] [CrossRef]
- Nakagawa, S.; Niimura, Y.; Gojobori, T.; Tanaka, H.; Miura, K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008, 36, 861–871. [Google Scholar] [CrossRef]
- Hu, D.G.; Mackenzie, P.I.; Hulin, J.A.; McKinnon, R.A.; Meech, R. Regulation of human UDP-glycosyltransferase (UGT) genes by miRNAs. Drug Metab. Rev. 2022, 54, 120–140. [Google Scholar] [CrossRef]
- Mount, S.M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982, 10, 459–472. [Google Scholar] [CrossRef]
- Bozic, I.; Antal, T.; Ohtsuki, H.; Carter, H.; Kim, D.; Chen, S.; Karchin, R.; Kinzler, K.W.; Vogelstein, B.; Nowak, M.A. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 18545–18550. [Google Scholar] [CrossRef] [PubMed]
- Merid, S.K.; Goranskaya, D.; Alexeyenko, A. Distinguishing between driver and passenger mutations in individual cancer genomes by network enrichment analysis. BMC Bioinform. 2014, 15, 308. [Google Scholar] [CrossRef] [PubMed]
- Pon, J.R.; Marra, M.A. Driver and passenger mutations in cancer. Annu. Rev. Pathol. 2015, 10, 25–50. [Google Scholar] [CrossRef] [PubMed]
- Wodarz, D.; Newell, A.C.; Komarova, N.L. Passenger mutations can accelerate tumour suppressor gene inactivation in cancer evolution. J. R. Soc. Interface 2018, 15, 20170967. [Google Scholar] [CrossRef] [PubMed]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef]
- Consortium, I.T.P.-C.A.o.W.G. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Innocenti, F.; Iyer, L.; Ramirez, J.; Green, M.D.; Ratain, M.J. Epirubicin glucuronidation is catalyzed by human UDP-glucuronosyltransferase 2B7. Drug Metab. Dispos. 2001, 29, 686–692. [Google Scholar]
- Wen, Z.; Tallman, M.N.; Ali, S.Y.; Smith, P.C. UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human liver and intestinal microsomes: Structural characterization of phenolic and alcoholic glucuronides of etoposide and estimation of enzyme kinetics. Drug Metab. Dispos. 2007, 35, 371–380. [Google Scholar] [CrossRef]
- Hanioka, N.; Ozawa, S.; Jinno, H.; Ando, M.; Saito, Y.; Sawada, J. Human liver UDP-glucuronosyltransferase isoforms involved in the glucuronidation of 7-ethyl-10-hydroxycamptothecin. Xenobiotica 2001, 31, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Oguri, T.; Takahashi, T.; Miyazaki, M.; Isobe, T.; Kohno, N.; Mackenzie, P.I.; Fujiwara, Y. UGT1A10 is responsible for SN-38 glucuronidation and its expression in human lung cancers. Anticancer Res. 2004, 24, 2893–2896. [Google Scholar] [PubMed]
- Tallman, M.N.; Ritter, J.K.; Smith, P.C. Differential rates of glucuronidation for 7-ethyl-10-hydroxy-camptothecin (SN-38) lactone and carboxylate in human and rat microsomes and recombinant UDP-glucuronosyltransferase isoforms. Drug Metab. Dispos. 2005, 33, 977–983. [Google Scholar] [CrossRef]
- Xiao, L.; Zhu, L.; Li, W.; Li, C.; Cao, Y.; Ge, G.; Sun, X. New Insights into SN-38 Glucuronidation: Evidence for the Important Role of UDP Glucuronosyltransferase 1A9. Basic Clin. Pharmacol. Toxicol. 2018, 122, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Takeshima, N.; Hayasaka, Y.; Notsu, A.; Yamazaki, M.; Kawabata, T.; Yamazaki, K.; Mori, K.; Yasui, H. Comparison of irinotecan and oxaliplatin as the first-line therapies for metastatic colorectal cancer: A meta-analysis. BMC Cancer 2021, 21, 116. [Google Scholar] [CrossRef]
- Erdem, G.U.; Bozkaya, Y.; Ozdemir, N.Y.; Demirci, N.S.; Yazici, O.; Zengin, N. 5-fluorouracil, leucovorin, and irinotecan (FOLFIRI) as a third-line chemotherapy treatment in metastatic gastric cancer, after failure of fluoropyrimidine, platinum, anthracycline, and taxane. Bosn. J. Basic Med. Sci. 2018, 18, 170–177. [Google Scholar] [CrossRef]
- Kondo, R.; Watanabe, S.; Shoji, S.; Ichikawa, K.; Abe, T.; Baba, J.; Tanaka, J.; Tsukada, H.; Terada, M.; Sato, K.; et al. A Phase II Study of Irinotecan for Patients with Previously Treated Small-Cell Lung Cancer. Oncology 2018, 94, 223–232. [Google Scholar] [CrossRef]
- Miyamoto, M.; Takano, M.; Kuwahara, M.; Soyama, H.; Kato, K.; Matuura, H.; Sakamoto, T.; Takasaki, K.; Aoyama, T.; Yoshikawa, T.; et al. Efficacy of combination chemotherapy using irinotecan and nedaplatin for patients with recurrent and refractory endometrial carcinomas: Preliminary analysis and literature review. Cancer Chemother. Pharmacol. 2018, 81, 111–117. [Google Scholar] [CrossRef]
- Pasquini, G.; Vasile, E.; Caparello, C.; Vivaldi, C.; Musettini, G.; Lencioni, M.; Petrini, I.; Fornaro, L.; Falcone, A. Third-Line Chemotherapy with Irinotecan plus 5-Fluorouracil in Caucasian Metastatic Gastric Cancer Patients. Oncology 2016, 91, 311–316. [Google Scholar] [CrossRef]
- Tanaka, T.; Tanaka, M.; Furusawa, H.; Kamada, Y.; Sagara, Y.; Anan, K.; Miyara, K.; Kai, Y.; Uga, T.; Tamura, K.; et al. Pilot Study of Irinotecan and S-1 (IRIS) for Advanced and Metastatic Breast Cancer. Anticancer Res. 2020, 40, 4779–4785. [Google Scholar] [CrossRef]
- McAndrew, N.P.; Finn, R.S. Clinical Review on the Management of Hormone Receptor-Positive Metastatic Breast Cancer. JCO Oncol. Pract. 2022, 18, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Ogura, K.; Nakano, H.; Ohnuma, T.; Kaku, T.; Hiratsuka, A.; Muro, K.; Watabe, T. Reverse geometrical selectivity in glucuronidation and sulfation of cis- and trans-4-hydroxytamoxifens by human liver UDP-glucuronosyltransferases and sulfotransferases. Biochem. Pharmacol. 2002, 63, 1817–1830. [Google Scholar] [CrossRef]
- Sun, D.; Chen, G.; Dellinger, R.W.; Sharma, A.K.; Lazarus, P. Characterization of 17-dihydroexemestane glucuronidation: Potential role of the UGT2B17 deletion in exemestane pharmacogenetics. Pharmacogenet. Genom. 2010, 20, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Kohn, G.E.; Rodriguez, K.M.; Hotaling, J.; Pastuszak, A.W. The History of Estrogen Therapy. Sex. Med. Rev. 2019, 7, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Guillemette, C.; Belanger, A.; Lepine, J. Metabolic inactivation of estrogens in breast tissue by UDP-glucuronosyltransferase enzymes: An overview. Breast Cancer Res. 2004, 6, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Need, E.F.; Selth, L.A.; Harris, T.J.; Birrell, S.N.; Tilley, W.D.; Buchanan, G. Research resource: Interplay between the genomic and transcriptional networks of androgen receptor and estrogen receptor alpha in luminal breast cancer cells. Mol. Endocrinol. 2012, 26, 1941–1952. [Google Scholar] [CrossRef]





| Cancer types | Description | No. of Tumors | Total No. of Mutations | No. of Mutations per Tumor | No. of Tumors with UGT Mutations | Total No. of UGT Mutations | No. of Missense Frameshift Nonsense | Percentage of Tumors with UGT Mutations |
|---|---|---|---|---|---|---|---|---|
| ACC | Adrenocortical Carcinoma | 92 | 11,981 | 130 | 9 | 14 | 8 | 0.097 |
| BLCA | Bladder Urothelial Carcinoma | 411 | 155,233 | 377 | 102 | 145 | 90 | 0.248 |
| BRCA | Breast Invasive Carcinoma | 1019 | 135,026 | 132 | 86 | 125 | 78 | 0.084 |
| CESC | Cervical Squamous Cell Carcinoma and | |||||||
| Endocervical Adenocarcinoma | 289 | 83,232 | 288 | 48 | 71 | 47 | 0.168 | |
| CHOL | Cholangiocarcinoma | 36 | 4500 | 125 | 3 | 3 | 3 | 0.083 |
| COAD | Colon Adenocarcinoma | 402 | 242,404 | 602 | 102 | 218 | 159 | 0.253 |
| DLBC | Lymphoid Neoplasm Diffuse Large | |||||||
| B-cell Lymphoma | 37 | 7,784 | 210 | 5 | 5 | 3 | 0.142 | |
| ESCA | Esophageal Carcinoma | 183 | 48,196 | 263 | 27 | 31 | 20 | 0.147 |
| GBM | Glioblastoma Multiforme | 380 | 40,617 | 106 | 82 | 96 | 66 | 0.215 |
| HNSC | Head and Neck Squamous Cell Carcinoma | 507 | 125,417 | 247 | 106 | 147 | 83 | 0.209 |
| KICH | Kidney Chromophobe | 66 | 3324 | 50 | 4 | 4 | 3 | 0.060 |
| KIRC | Kidney Renal Clear Cell Carcinoma | 371 | 32,001 | 86 | 25 | 28 | 20 | 0.067 |
| KIRP | Kidney Renal Papillary Cell Carcinoma | 281 | 35,445 | 126 | 32 | 33 | 24 | 0.113 |
| LAML | Acute Myeloid Leukemia | 140 | 8332 | 59 | 6 | 10 | 5 | 0.042 |
| LGG | Brain Lower Grade Glioma | 512 | 24,000 | 46 | 34 | 37 | 30 | 0.066 |
| LIHC | Liver Hepatocellular Carcinoma | 363 | 60,432 | 166 | 45 | 50 | 32 | 0.123 |
| LUAD | Lung Adenocarcinoma | 512 | 243,687 | 475 | 235 | 412 | 287 | 0.458 |
| LUSC | Lung Squamous Cell Carcinoma | 484 | 204,623 | 422 | 166 | 253 | 182 | 0.342 |
| MESO | Mesothelioma | 82 | 3979 | 48 | 1 | 1 | 1 | 0.012 |
| OV | Ovary Serous Cystadenocarcinoma | 406 | 53,115 | 130 | 60 | 71 | 44 | 0.147 |
| PCPG | Pheochromocytoma and Paraganglioma | 179 | 2726 | 15 | 4 | 4 | 1 | 0.022 |
| PAAD | Pancreatic Adenocarcinoma | 155 | 8728 | 56 | 3 | 3 | 2 | 0.019 |
| PRAD | Prostate Adenocarcinoma | 495 | 24,778 | 50 | 19 | 22 | 15 | 0.038 |
| READ | Rectum Adenocarcinoma | 146 | 30,380 | 208 | 19 | 26 | 18 | 0.13 |
| SARC | Sarcoma | 236 | 31,678 | 134 | 33 | 41 | 26 | 0.139 |
| SKCM | Skin Cutaneous Melanoma (primary) | 103 | 68,991 | 669 | 56 | 160 | 94 | 0.543 |
| SKCM | Skin Cutaneous Melanoma (Metastatic) | 364 | 438,405 | 1204 | 246 | 926 | 592 | 0.675 |
| STAD | Stomach Adenocarcinoma | 437 | 221,714 | 507 | 105 | 175 | 120 | 0.240 |
| TGCT | Testicular Germ Cell Tumor | 144 | 3588 | 24 | 0 | 0 | 0 | 0.000 |
| THCA | Thyroid Carcinoma | 491 | 12,050 | 24 | 9 | 10 | 8 | 0.018 |
| THYM | Thymoma | 122 | 4888 | 40 | 2 | 3 | 2 | 0.016 |
| UCEC | Uterine Corpus Endometrial Carcinoma | 487 | 307,642 | 631 | 122 | 297 | 193 | 0.250 |
| UCS | Uterine Carcinoma | 57 | 5261 | 92 | 5 | 5 | 3 | 0.087 |
| UVM | Uveal Melanoma | 80 | 1935 | 24 | 1 | 1 | 1 | 0.012 |
| SUM | 10,069 | 2,686,092 | 266 | 1802 | 3427 | 2260 | 0.178 |
| UGT Gene | 5′UTR | Missense | Translation Start Site | Nonsense | Silent | Frame Shift Del | Frame Shift Ins | In Frame Del | In Frame Ins | Non Stop | Intron | Splice Site | 3′UTR | Sum | RefSeq Transcript |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A8 | 3 | 47 (26) | 8 | 19 | 4 | 2 | 83 | ENST00000373450 | |||||||
| 1A10 | 3 | 59 (31) | 3 | 24 | 4 | 1 | 94 | ENST00000344644 | |||||||
| 1A9 | 2 | 56 (27) | 7 | 19 | 5 | 7 | 96 | ENST00000354728 | |||||||
| 1A7 | 46 (25) | 3 | 12 | 4 | 3 | 68 | ENST00000373426 | ||||||||
| 1A6 | 14 | 43 (18) | 1 | 5 | 19 | 1 | 83 | ENST00000305139 | |||||||
| 1A5 | 48 (18) | 2 | 24 | 1 | 1 | 76 | ENST00000373414 | ||||||||
| 1A4 | 1 | 49 (17) | 4 | 20 | 10 | 2 | 86 | ENST00000373409 | |||||||
| 1A3 | 1 | 47 (18) | 2 | 24 | 4 | 1 | 79 | ENST00000482026 | |||||||
| 1A1 | 2 | 42 * | 32 | 2 | 1 | 79 | ENST00000609767 | ||||||||
| 1A E2–5 | 43 (29) | 1 | 25 | 7 | 2 | 1 | 8 | 87 | ENST00000344644 ENST00000373409 | ||||||
| 2A1 | 5 | 44 (29) | 5 | 15 | 3 | 1 | 2 | 3 | 78 | ENST00000514019 | |||||
| 2A2 | 49 (15) | 7 | 15 | 2 | 73 | ENST00000457664 | |||||||||
| 2A1/2A2 E2–6 | 70 (53) | 11 | 22 | 6 | 1 | 5 | 6 | 121 | ENST00000514019 ENST00000503640 | ||||||
| 2A3 | 2 | 127 (84) | 10 | 33 | 7 | 3 | 3 | 11 | 196 | ENST00000251566 | |||||
| 2B4 | 7 | 151 (93) | 21 | 74 | 5 | 1 | 2 | 6 | 22 | 289 | ENST00000305107 | ||||
| 2B7 | 2 | 106 (65) | 4 | 37 | 4 | 1 | 3 | 15 | 172 | ENST00000305231 | |||||
| 2B10 | 3 | 116 (77) | 11 | 27 | 4 | 2 | 3 | 7 | 20 | 193 | ENST00000265403 | ||||
| 2B11 | 20 | 115 (79) | 16 | 38 | 3 | 1 | 4 | 8 | 205 | ENST00000446444 | |||||
| 2B15 | 2 | 122 (78) | 14 | 38 | 3 | 1 | 1 | 1 | 1 | 27 | 210 | ENST00000338206 | |||
| 2B17 | 1 | 100 (61) | 10 | 29 | 5 | 2 | 147 | ENST00000317746 | |||||||
| 2B28 | 1 | 133 (80) | 15 | 47 | 1 | 4 | 9 | 210 | ENST00000335568 | ||||||
| 3A1 | 20 | 153 (64) | 9 | 68 | 2 | 1 | 19 | 6 | 29 | 307 | ENST00000274278 | ||||
| 3A2 | 10 | 143 (65) | 12 | 64 | 3 | 1 | 1 | 21 | 255 | ENST00000282507 | |||||
| UGT8 | 89 (47) | 4 | 29 | 3 | 1 | 8 | 6 | 140 | ENST00000310836 | ||||||
| Total | 99 | 1998 (1099) | 1 | 184 | 754 | 93 | 29 | 5 | 1 | 36 | 45 | 182 | 3427 |
| UGT Genes | CCLE Cell Lines with UGT Mutations | Translation Start Site | Missense | Nonsense | Frame Shift Del | Frame Shift Ins | In Frame Del | Nonstop | Splice Site | SUM |
|---|---|---|---|---|---|---|---|---|---|---|
| 1A8 | 28 | 20 | 2 | 8 | 4 | 34 (5) | ||||
| 1A10 | 35 | 36 | 2 | 38 (1) | ||||||
| 1A9 | 31 | 28 | 4 | 6 | 39 (4) | |||||
| 1A7 | 35 | 1 | 32 | 1 | 2 | 1 | 36 (10) | |||
| 1A6 | 16 | 12 | 1 | 2 | 1 | 16 (3) | ||||
| 1A5 | 17 | 13 | 1 | 2 | 1 | 17 (2) | ||||
| 1A4 | 30 | 21 | 7 | 2 | 1 | 31 (2) | ||||
| 1A3 | 21 | 11 | 6 | 3 | 1 | 21 | ||||
| 1A1 | 14 | 15 | 15 (1) | |||||||
| 1A E2–5 | 22 | 19 | 6 | 1 | 26 (6) | |||||
| 2A1 | 15 | 13 | 2 | 15 | ||||||
| 2A2 | 22 | 22 | 22 (2) | |||||||
| 2A1/2A2 E2–6 | 30 | 13 | 3 | 3 | 19 (1) | |||||
| 2A3 | 45 | 42 | 4 | 4 | 1 | 51 (7) | ||||
| 2B4 | 50 | 49 | 6 | 3 | 1 | 59 (10) | ||||
| 2B7 | 42 | 33 | 4 | 2 | 1 | 2 | 42 (6) | |||
| 2B10 | 60 | 49 | 5 | 4 | 1 | 3 | 62 (11) | |||
| 2B11 | 54 | 51 | 3 | 2 | 56 (7) | |||||
| 2B15 | 46 | 45 | 3 | 1 | 49 (8) | |||||
| 2B17 | 18 | 16 | 1 | 1 | 1 | 19 (1) | ||||
| 2B28 | 66 | 64 | 7 | 2 | 1 | 2 | 76 (10) | |||
| 3A1 | 50 | 49 | 4 | 2 | 3 | 58 (6) | ||||
| 3A2 | 55 | 1 | 47 | 3 | 6 | 1 | 2 | 60 (7) | ||
| UGT8 | 29 | 28 | 2 | 3 | 1 | 34 (4) | ||||
| Total | 502 | 2 | 728 | 51 | 65 | 28 | 2 | 2 | 17 | 895 (114) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, D.G.; Marri, S.; Hulin, J.-A.; McKinnon, R.A.; Mackenzie, P.I.; Meech, R. The Somatic Mutation Landscape of UDP-Glycosyltransferase (UGT) Genes in Human Cancers. Cancers 2022, 14, 5708. https://doi.org/10.3390/cancers14225708
Hu DG, Marri S, Hulin J-A, McKinnon RA, Mackenzie PI, Meech R. The Somatic Mutation Landscape of UDP-Glycosyltransferase (UGT) Genes in Human Cancers. Cancers. 2022; 14(22):5708. https://doi.org/10.3390/cancers14225708
Chicago/Turabian StyleHu, Dong Gui, Shashikanth Marri, Julie-Ann Hulin, Ross A. McKinnon, Peter I. Mackenzie, and Robyn Meech. 2022. "The Somatic Mutation Landscape of UDP-Glycosyltransferase (UGT) Genes in Human Cancers" Cancers 14, no. 22: 5708. https://doi.org/10.3390/cancers14225708
APA StyleHu, D. G., Marri, S., Hulin, J.-A., McKinnon, R. A., Mackenzie, P. I., & Meech, R. (2022). The Somatic Mutation Landscape of UDP-Glycosyltransferase (UGT) Genes in Human Cancers. Cancers, 14(22), 5708. https://doi.org/10.3390/cancers14225708






