Signatures of Breast Cancer Progression in the Blood: What Could Be Learned from Circulating Tumor Cell Transcriptomes
Abstract
Simple Summary
Abstract
1. Introduction
2. Breast Cancer Gene Signatures
3. Circulating Tumor Cells: The Kinetic Phase of Metastasis
4. CTC Detection and Clinical Significance of CTC Count in Breast Cancer
5. CTC Gene Expression Profile Studies in Breast Cancer
5.1. Experimental Strategies to Identify CTC Detection-Specific Genes in Breast Cancer
5.2. Clinical Significance of EMT, Metastasis and Chemoresistance-Related Gene Expression in Circulating Breast Cancer Cells
5.3. Sources of Breast Cancer Molecular Signatures from Ex Vivo Expanded CTCs and Circulating Cell-Free Tumor-Derived Biomolecules
6. Challenges to the Integration of CTC Molecular Analysis in the Clinical Practice and Future Perspective
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Laversanne, M.; Brewster, D.H.; Gombe Mbalawa, C.; Kohler, B.; Piñeros, M.; Steliarova-Foucher, E.; Swaminathan, R.; Antoni, S.; et al. Cancer Incidence in Five Continents: Inclusion criteria, highlights from Volume X and the global status of cancer registration. Int. J. Cancer 2015, 137, 2060–2071. [Google Scholar] [CrossRef] [PubMed]
- Independent UK Panel on Breast Cancer Screening. The benefits and harms of breast cancer screening: An independent review. Lancet 2012, 380, 1778–1786. [Google Scholar] [CrossRef]
- Oeffinger, K.C.; Fontham, E.T.; Etzioni, R.; Herzig, A.; Michaelson, J.S.; Shih, Y.C.; Walter, L.C.; Church, T.R.; Flowers, C.R.; LaMonte, S.J.; et al. Breast Cancer Screening for Women at Average Risk: 2015 Guideline Update from the American Cancer Society. JAMA 2015, 314, 1599–1614, Erratum in: JAMA 2016, 315, 1406. [Google Scholar] [CrossRef] [PubMed]
- Saphner, T.; Tormey, D.C.; Gray, R. Annual hazard rates of recurrence for breast cancer after primary therapy. J. Clin. Oncol. 1996, 14, 2738–2746. [Google Scholar] [CrossRef]
- Karrison, T.G.; Ferguson, D.J.; Meier, P. Dormancy of mammary carcinoma after mastectomy. J. Natl. Cancer Inst. 1999, 91, 80–85. [Google Scholar] [CrossRef]
- Colleoni, M.; Sun, Z.; Price, K.N.; Karlsson, P.; Forbes, J.F.; Thürlimann, B.; Gianni, L.; Castiglione, M.; Gelber, R.D.; Coates, A.S.; et al. Annual Hazard Rates of Recurrence for Breast Cancer During 24 Years of Follow-Up: Results from the International Breast Cancer Study Group Trials I to, V. J. Clin. Oncol. 2016, 34, 927–935. [Google Scholar] [CrossRef]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334. [Google Scholar] [CrossRef]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Yates, L.R.; Knappskog, S.; Wedge, D.; Farmery, J.H.R.; Gonzalez, S.; Martincorena, I.; Alexandrov, L.B.; Van Loo, P.; Haugland, H.K.; Lilleng, P.K.; et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 2017, 32, 169–184.e7. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. (Eds.) SEER Cancer Statistics Review, 1975–2018; National Cancer Institute: Bethesda, MD, USA, 2021. Available online: https://seer.cancer.gov/archive/csr/1975_2018/ (accessed on 28 July 2022).
- Pantel, K.; Cote, R.J.; Fodstad, O. Detection and clinical importance of micrometastatic disease. J. Natl. Cancer Inst. 1999, 91, 1113–1124. [Google Scholar] [CrossRef]
- Klein, C.A.; Blankenstein, T.J.; Schmidt-Kittler, O.; Petronio, M.; Polzer, B.; Stoecklein, N.H.; Riethmüller, G. Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet 2002, 360, 683–689. [Google Scholar] [CrossRef]
- Schmidt-Kittler, O.; Ragg, T.; Daskalakis, A.; Granzow, M.; Ahr, A.; Blankenstein, T.J.; Kaufmann, M.; Diebold, J.; Arnholdt, H.; Muller, P.; et al. From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proc. Natl. Acad. Sci. USA 2003, 100, 7737–7742. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 2009, 9, 302–312. [Google Scholar] [CrossRef]
- Hu, Z.; Li, Z.; Ma, Z.; Curtis, C. Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases. Nat. Genet. 2020, 52, 701–708. [Google Scholar] [CrossRef]
- Hüsemann, Y.; Geigl, J.B.; Schubert, F.; Musiani, P.; Meyer, M.; Burghart, E.; Forni, G.; Eils, R.; Fehm, T.; Riethmüller, G.; et al. Systemic spread is an early step in breast cancer. Cancer Cell 2008, 13, 58–68. [Google Scholar] [CrossRef]
- Sänger, N.; Effenberger, K.E.; Riethdorf, S.; Van Haasteren, V.; Gauwerky, J.; Wiegratz, I.; Strebhardt, K.; Kaufmann, M.; Pantel, K. Disseminated tumor cells in the bone marrow of patients with ductal carcinoma in situ. Int. J. Cancer 2011, 129, 2522–2526. [Google Scholar] [CrossRef]
- Hosseini, H.; Obradović, M.M.S.; Hoffmann, M.; Harper, K.L.; Sosa, M.S.; Werner-Klein, M.; Nanduri, L.K.; Werno, C.; Ehrl, C.; Maneck, M.; et al. Early dissemination seeds metastasis in breast cancer. Nature 2016, 540, 552–558. [Google Scholar] [CrossRef]
- Bertos, N.R.; Park, M. Breast cancer—One term, many entities? J. Clin. Investig. 2011, 121, 3789–3796. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Wirapati, P.; Sotiriou, C.; Kunkel, S.; Farmer, P.; Pradervand, S.; Haibe-Kains, B.; Desmedt, C.; Ignatiadis, M.; Sengstag, T.; Schütz, F.; et al. Meta-analysis of gene expression profiles in breast cancer: Toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008, 10, R65. [Google Scholar] [CrossRef] [PubMed]
- Reis-Filho, J.S.; Weigelt, B.; Fumagalli, D.; Sotiriou, C. Molecular profiling: Moving away from tumor philately. Sci. Transl. Med. 2010, 2, 47ps43. [Google Scholar] [CrossRef]
- Nielsen, T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef]
- Bertucci, F.; Finetti, P.; Cervera, N.; Esterni, B.; Hermitte, F.; Viens, P.; Birnbaum, D. How basal are triple-negative breast cancers? Int. J. Cancer 2008, 123, 236–240. [Google Scholar] [CrossRef]
- Toft, D.J.; Cryns, V.L. Minireview: Basal-like breast cancer: From molecular profiles to targeted therapies. Mol. Endocrinol. 2011, 25, 199–211. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Herschkowitz, J.I.; Simin, K.; Weigman, V.J.; Mikaelian, I.; Usary, J.; Hu, Z.; Rasmussen, K.E.; Jones, L.P.; Assefnia, S.; Chandrasekharan, S.; et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007, 8, R76. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef]
- Bruna, A.; Greenwood, W.; Le Quesne, J.; Teschendorff, A.; Miranda-Saavedra, D.; Rueda, O.M.; Sandoval, J.L.; Vidakovic, A.T.; Saadi, A.; Pharoah, P.; et al. TGFβ induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat. Commun. 2012, 3, 1055. [Google Scholar] [CrossRef]
- Dias, K.; Dvorkin-Gheva, A.; Hallett, R.M.; Wu, Y.; Hassell, J.; Pond, G.R.; Levine, M.; Whelan, T.; Bane, A.L. Claudin-Low Breast Cancer; Clinical & Pathological Characteristics. PLoS ONE 2017, 12, e0168669. [Google Scholar] [CrossRef]
- Fougner, C.; Bergholtz, H.; Norum, J.H.; Sørlie, T. Re-definition of claudin-low as a breast cancer phenotype. Nat. Commun. 2020, 11, 1787. [Google Scholar] [CrossRef] [PubMed]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef] [PubMed]
- van’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Wallden, B.; Storhoff, J.; Nielsen, T.; Dowidar, N.; Schaper, C.; Ferree, S.; Liu, S.; Leung, S.; Geiss, G.; Snider, J.; et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med. Genom. 2015, 8, 54. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- van de Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef]
- Filipits, M.; Rudas, M.; Jakesz, R.; Dubsky, P.; Fitzal, F.; Singer, C.F.; Dietze, O.; Greil, R.; Jelen, A.; Sevelda, P.; et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 2011, 17, 6012–6020. [Google Scholar] [CrossRef]
- Giuliano, A.E.; Connolly, J.L.; Edge, S.B.; Mittendorf, E.A.; Rugo, H.S.; Solin, L.J.; Weaver, D.L.; Winchester, D.J.; Hortobagyi, G.N. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 290–303, Erratum in: CA Cancer J. Clin. 2017, 67, 345. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.A.; Marmor, S.; Hui, J.Y.C.; Beckwith, H.; Blaes, A.H.; Potter, D.; Tuttle, T.M. The 70-gene signature test as a prognostic and predictive biomarker in patients with invasive lobular breast cancer. Breast Cancer Res. Treat. 2022, 191, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Groenendijk, F.H.; Treece, T.; Yoder, E.; Baron, P.; Beitsch, P.; Audeh, W.; Dinjens, W.N.M.; Bernards, R.; Whitworth, P. Estrogen receptor variants in ER-positive basal-type breast cancers responding to therapy like ER-negative breast cancers. NPJ Breast Cancer 2019, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Végran, F.; Boidot, R.; Coudert, B.; Fumoleau, P.; Arnould, L.; Garnier, J.; Causeret, S.; Fraise, J.; Dembélé, D.; Lizard-Nacol, S. Gene expression profile and response to trastuzumab-docetaxel-based treatment in breast carcinoma. Br. J. Cancer 2009, 101, 1357–1364. [Google Scholar] [CrossRef]
- Minuti, G.; Cappuzzo, F.; Duchnowska, R.; Jassem, J.; Fabi, A.; O’Brien, T.; Mendoza, A.D.; Landi, L.; Biernat, W.; Czartoryska-Arłukowicz, B.; et al. Increased MET and HGF gene copy numbers are associated with trastuzumab failure in HER2-positive metastatic breast cancer. Br. J. Cancer 2012, 107, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Seshadri, R.; Firgaira, F.A.; Horsfall, D.J.; McCaul, K.; Setlur, V.; Kitchen, P. Clinical significance of HER-2/neu oncogene amplification in primary breast cancer. The South Australian Breast Cancer Study Group. J. Clin. Oncol. 1993, 11, 1936–1942. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef]
- Banerji, U.; van Herpen, C.M.L.; Saura, C.; Thistlethwaite, F.; Lord, S.; Moreno, V.; Macpherson, I.R.; Boni, V.; Rolfo, C.; de Vries, E.G.E.; et al. Trastuzumab duocarmazine in locally advanced and metastatic solid tumours and HER2-expressing breast cancer: A phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 2019, 20, 1124–1135. [Google Scholar] [CrossRef]
- Modi, S.; Park, H.; Murthy, R.K.; Iwata, H.; Tamura, K.; Tsurutani, J.; Moreno-Aspitia, A.; Doi, T.; Sagara, Y.; Redfern, C.; et al. Antitumor Activity and Safety of Trastuzumab Deruxtecan in Patients with HER2-Low-Expressing Advanced Breast Cancer: Results from a Phase Ib Study. J. Clin. Oncol. 2020, 38, 1887–1896. [Google Scholar] [CrossRef]
- Dieci, M.V.; Barbieri, E.; Piacentini, F.; Ficarra, G.; Bettelli, S.; Dominici, M.; Conte, P.F.; Guarneri, V. Discordance in receptor status between primary and recurrent breast cancer has a prognostic impact: A single-institution analysis. Ann Oncol. 2013, 24, 101–108. [Google Scholar] [CrossRef]
- Guarneri, V.; Giovannelli, S.; Ficarra, G.; Bettelli, S.; Maiorana, A.; Piacentini, F.; Barbieri, E.; Dieci, M.V.; D’Amico, R.; Jovic, G.; et al. Comparison of HER-2 and hormone receptor expression in primary breast cancers and asynchronous paired metastases: Impact on patient management. Oncologist 2008, 13, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Grinda, T.; Joyon, N.; Lusque, A.; Lefèvre, S.; Arnould, L.; Penault-Llorca, F.; Macgrogan, G.; Treilleux, I.; Vincent-Salomon, A.; Haudebourg, J.; et al. Phenotypic discordance between primary and metastatic breast cancer in the large-scale real-life multicenter French ESME cohort. NPJ Breast Cancer 2021, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Yardley, D.A.; Tripathy, D.; Brufsky, A.M.; Rugo, H.S.; Kaufman, P.A.; Mayer, M.; Magidson, J.; Yoo, B.; Quah, C.; Ulcickas Yood, M. Long-term survivor characteristics in HER2-positive metastatic breast cancer from registHER. Br. J. Cancer 2014, 110, 2756–2764. [Google Scholar] [CrossRef] [PubMed]
- Yeo, B.; Kotsori, K.; Mohammed, K.; Walsh, G.; Smith, I.E. Long-term outcome of HER2 positive metastatic breast cancer patients treated with first-line trastuzumab. Breast 2015, 24, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Miglietta, F.; Griguolo, G.; Bottosso, M.; Giarratano, T.; Lo Mele, M.; Fassan, M.; Cacciatore, M.; Genovesi, E.; De Bartolo, D.; Vernaci, G.; et al. Evolution of HER2-low expression from primary to recurrent breast cancer. NPJ Breast Cancer 2021, 7, 137, Erratum in: NPJ Breast Cancer 2021, 7, 149. [Google Scholar] [CrossRef]
- Bruzas, S.; Gluz, O.; Harbeck, N.; Schmid, P.; Cortés, J.; Blohmer, J.; Seiberling, C.; Chiari, O.; Harrach, H.; Ataseven, B.; et al. Gene signatures in patients with early breast cancer and relapse despite pathologic complete response. NPJ Breast Cancer 2022, 8, 42. [Google Scholar] [CrossRef]
- Carey, L.A.; Dees, E.C.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Adamo, B.; Anders, C.K. Stratifying triple-negative breast cancer: Which definition(s) to use? Breast Cancer Res. 2011, 13, 105. [Google Scholar] [CrossRef]
- Bianchini, G.; De Angelis, C.; Licata, L.; Gianni, L. Treatment landscape of triple-negative breast cancer—Expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022, 19, 91–113. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bäuerle, T.; Wallwiener, M.; et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013, 5, 180ra48, Erratum in: Sci. Transl. Med. 2013, 5, 189er5. [Google Scholar] [CrossRef] [PubMed]
- Weiss, L. Metastatic inefficiency. Adv. Cancer Res. 1990, 54, 159–211. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef]
- Strilic, B.; Offermanns, S. Intravascular Survival and Extravasation of Tumor Cells. Cancer Cell 2017, 32, 282–293. [Google Scholar] [CrossRef]
- Psaila, B.; Lyden, D. The metastatic niche: Adapting the foreign soil. Nat. Rev. Cancer 2009, 9, 285–293. [Google Scholar] [CrossRef]
- Baserga, R.; Kisieleski, W.E.; Halvorsen, K. A study on the establishment and growth of tumor metastases with tritiated thymidine. Cancer Res. 1960, 20, 910–917. [Google Scholar]
- Fidler, I.J. Metastasis: Quantitative analysis of distribution and fate of tumor emboli labeled with 125 I-5-iodo-2′-deoxyuridine. J. Natl. Cancer Inst. 1970, 45, 773–782. [Google Scholar]
- Weiss, L. Biomechanical destruction of cancer cells in skeletal muscle: A rate-regulator for hematogenous metastasis. Clin. Exp. Metastasis 1989, 7, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Fujikawa, T.; Tanabe, A.; Sakurai, K. Quantitative analysis of distribution and fate of human lung cancer emboli labeled with 125I-5-iodo-2′-deoxyuridine in nude mice. Surg. Today 1993, 23, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Fidler, I.J.; Kumar, R.; Bielenberg, D.R.; Ellis, L.M. Molecular determinants of angiogenesis in cancer metastasis. Cancer J. Sci. Am. 1998, 4 (Suppl. 1), S58–S66. [Google Scholar] [PubMed]
- Koop, S.; MacDonald, I.C.; Luzzi, K.; Schmidt, E.E.; Morris, V.L.; Grattan, M.; Khokha, R.; Chambers, A.F.; Groom, A.C. Fate of melanoma cells entering the microcirculation: Over 80% survive and extravasate. Cancer Res. 1995, 55, 2520–2523. [Google Scholar]
- Luzzi, K.J.; MacDonald, I.C.; Schmidt, E.E.; Kerkvliet, N.; Morris, V.L.; Chambers, A.F.; Groom, A.C. Multistep nature of metastatic inefficiency: Dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998, 153, 865–873. [Google Scholar] [CrossRef]
- Liotta, L.A.; Steeg, P.S.; Stetler-Stevenson, W.G. Cancer metastasis and angiogenesis: An imbalance of positive and negative regulation. Cell 1991, 64, 327–336. [Google Scholar] [CrossRef]
- Cameron, M.D.; Schmidt, E.E.; Kerkvliet, N.; Nadkarni, K.V.; Morris, V.L.; Groom, A.C.; Chambers, A.F.; MacDonald, I.C. Temporal progression of metastasis in lung: Cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000, 60, 2541–2546. [Google Scholar]
- Butler, T.P.; Gullino, P.M. Quantitation of cell shedding into efferent blood of mammary adenocarcinoma. Cancer Res. 1975, 35, 512–516. [Google Scholar]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef]
- Cleris, L.; Daidone, M.G.; Fina, E.; Cappelletti, V. The Detection and Morphological Analysis of Circulating Tumor and Host Cells in Breast Cancer Xenograft Models. Cells 2019, 8, 683. [Google Scholar] [CrossRef]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Follain, G.; Osmani, N.; Azevedo, A.S.; Allio, G.; Mercier, L.; Karreman, M.A.; Solecki, G.; Garcia Leòn, M.J.; Lefebvre, O.; Fekonja, N.; et al. Hemodynamic Forces Tune the Arrest, Adhesion, and Extravasation of Circulating Tumor Cells. Dev. Cell 2018, 45, 33–52.e12. [Google Scholar] [CrossRef] [PubMed]
- Hamza, B.; Miller, A.B.; Meier, L.; Stockslager, M.; Ng, S.R.; King, E.M.; Lin, L.; DeGouveia, K.L.; Mulugeta, N.; Calistri, N.L.; et al. Measuring kinetics and metastatic propensity of CTCs by blood exchange between mice. Nat. Commun. 2021, 12, 5680. [Google Scholar] [CrossRef] [PubMed]
- Borriello, L.; Coste, A.; Traub, B.; Sharma, V.P.; Karagiannis, G.S.; Lin, Y.; Wang, Y.; Ye, X.; Duran, C.L.; Chen, X.; et al. Primary tumor associated macrophages activate programs of invasion and dormancy in disseminating tumor cells. Nat. Commun. 2022, 13, 626. [Google Scholar] [CrossRef]
- Linde, N.; Casanova-Acebes, M.; Sosa, M.S.; Mortha, A.; Rahman, A.; Farias, E.; Harper, K.; Tardio, E.; Reyes Torres, I.; Jones, J.; et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat. Commun. 2018, 9, 21. [Google Scholar] [CrossRef]
- Aceto, N. Bring along your friends: Homotypic and heterotypic circulating tumor cell clustering to accelerate metastasis. Biomed. J. 2020, 43, 18–23. [Google Scholar] [CrossRef]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef]
- Dujon, A.M.; Capp, J.P.; Brown, J.S.; Pujol, P.; Gatenby, R.A.; Ujvari, B.; Alix-Panabières, C.; Thomas, F. Is There One Key Step in the Metastatic Cascade? Cancers 2021, 13, 3693. [Google Scholar] [CrossRef]
- Allan, A.L.; Keeney, M. Circulating tumor cell analysis: Technical and statistical considerations for application to the clinic. J. Oncol. 2010, 2010, 426218. [Google Scholar] [CrossRef]
- Tibbe, A.G.; Miller, M.C.; Terstappen, L.W. Statistical considerations for enumeration of circulating tumor cells. Cytom. A. 2007, 71, 154–162. [Google Scholar] [CrossRef]
- Coumans, F.A.; Ligthart, S.T.; Terstappen, L.W. Interpretation of changes in circulating tumor cell counts. Transl. Oncol. 2012, 5, 486–491. [Google Scholar] [CrossRef]
- Juratli, M.A.; Sarimollaoglu, M.; Nedosekin, D.A.; Melerzanov, A.V.; Zharov, V.P.; Galanzha, E.I. Dynamic Fluctuation of Circulating Tumor Cells during Cancer Progression. Cancers 2014, 6, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Suo, Y.; Fu, Y.; Zhang, F.; Ding, N.; Pang, K.; Xie, C.; Weng, X.; Tian, M.; He, H.; et al. In vivo flow cytometry reveals a circadian rhythm of circulating tumor cells. Light Sci. Appl. 2021, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.L.; Fitzgerald, J.E.; Ivich, F.; Sontag, E.D.; Niedre, M. Short-Term Circulating Tumor Cell Dynamics in Mouse Xenograft Models and Implications for Liquid Biopsy. Front. Oncol. 2020, 10, 601085. [Google Scholar] [CrossRef] [PubMed]
- Diamantopoulou, Z.; Castro-Giner, F.; Schwab, F.D.; Foerster, C.; Saini, M.; Budinjas, S.; Strittmatter, K.; Krol, I.; Seifert, B.; Heinzelmann-Schwarz, V.; et al. The metastatic spread of breast cancer accelerates during sleep. Nature 2022, 607, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, T.R. A Case of Cancer in Which Cells Similar to Those in the Tumours Were Seen in the Blood after Death. Med. J. Aust. 1869, 14, 146–147. [Google Scholar]
- Gross, H.J.; Verwer, B.; Houck, D.; Hoffman, R.A.; Recktenwald, D. Model study detecting breast cancer cells in peripheral blood mononuclear cells at frequencies as low as 10(-7). Proc. Natl. Acad. Sci. USA 1995, 92, 537–541. [Google Scholar] [CrossRef]
- Mori, M.; Mimori, K.; Ueo, H.; Karimine, N.; Barnard, G.F.; Sugimachi, K.; Akiyoshi, T. Molecular detection of circulating solid carcinoma cells in the peripheral blood: The concept of early systemic disease. Int. J. Cancer 1996, 68, 739–743. [Google Scholar] [CrossRef]
- Neumaier, M.; Gerhard, M.; Wagener, C. Diagnosis of micrometastases by the amplification of tissue-specific genes. Gene 1995, 159, 43–47. [Google Scholar] [CrossRef]
- Datta, Y.H.; Adams, P.T.; Drobyski, W.R.; Ethier, S.P.; Terry, V.H.; Roth, M.S. Sensitive detection of occult breast cancer by the reverse-transcriptase polymerase chain reaction. J. Clin. Oncol. 1994, 12, 475–482. [Google Scholar] [CrossRef]
- Schmid-Schönbein, G.W.; Shih, Y.Y.; Chien, S. Morphometry of human leukocytes. Blood 1980, 56, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Mendelaar, P.A.J.; Kraan, J.; Van, M.; Zeune, L.L.; Terstappen, L.W.M.M.; Oomen-de Hoop, E.; Martens, J.W.M.; Sleijfer, S. Defining the dimensions of circulating tumor cells in a large series of breast, prostate, colon, and bladder cancer patients. Mol. Oncol. 2021, 15, 116–125. [Google Scholar] [CrossRef]
- Werner, S.L.; Graf, R.P.; Landers, M.; Valenta, D.T.; Schroeder, M.; Greene, S.B.; Bales, N.; Dittamore, R.; Marrinucci, D. Analytical Validation and Capabilities of the Epic CTC Platform: Enrichment-Free Circulating Tumour Cell Detection and Characterization. J. Circ. Biomark. 2015, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Kaldjian, E.P.; Ramirez, A.B.; Sun, Y.; Campton, D.E.; Werbin, J.L.; Varshavskaya, P.; Quarre, S.; George, T.; Madan, A.; Blau, C.A.; et al. The RareCyte® platform for next-generation analysis of circulating tumor cells. Cytom. A 2018, 93, 1220–1225. [Google Scholar] [CrossRef] [PubMed]
- Krol, I.; Schwab, F.D.; Carbone, R.; Ritter, M.; Picocci, S.; De Marni, M.L.; Stepien, G.; Franchi, G.M.; Zanardi, A.; Rissoglio, M.D.; et al. Detection of clustered circulating tumour cells in early breast cancer. Br. J. Cancer 2021, 125, 23–27. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12 Pt 1, 4218–4224. [Google Scholar] [CrossRef]
- Bidard, F.C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Pierga, J.Y.; Reuben, J.; Rademaker, A.; Davis, A.A.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit. Rev. Oncol. Hematol. 2019, 134, 39–45. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Hendrix, L.H.; Hyslop, T.; Barry, W.T.; Winer, E.P.; Hudis, C.; Toppmeyer, D.; Carey, L.A.; Partridge, A.H.; Pierga, J.Y.; et al. Serial Analysis of Circulating Tumor Cells in Metastatic Breast Cancer Receiving First-Line Chemotherapy. J. Natl. Cancer Inst. 2021, 113, 443–452. [Google Scholar] [CrossRef]
- Bidard, F.C.; Jacot, W.; Kiavue, N.; Dureau, S.; Kadi, A.; Brain, E.; Bachelot, T.; Bourgeois, H.; Gonçalves, A.; Ladoire, S.; et al. Efficacy of Circulating Tumor Cell Count-Driven vs Clinician-Driven First-line Therapy Choice in Hormone Receptor-Positive, ERBB2-Negative Metastatic Breast Cancer: The STIC CTC Randomized Clinical Trial. JAMA Oncol. 2021, 7, 34–41. [Google Scholar] [CrossRef]
- Cabel, L.; Berger, F.; Cottu, P.; Loirat, D.; Rampanou, A.; Brain, E.; Cyrille, S.; Bourgeois, H.; Kiavue, N.; Deluche, E.; et al. Clinical utility of circulating tumour cell-based monitoring of late-line chemotherapy for metastatic breast cancer: The randomised CirCe01 trial. Br. J. Cancer 2021, 124, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Müller, V.; Zhang, L.; Rau, T.; Loibl, S.; Komor, M.; Roller, M.; Huober, J.; Fehm, T.; Schrader, I.; et al. Detection and HER2 expression of circulating tumor cells: Prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin. Cancer Res. 2010, 16, 2634–2645. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.C.; Mathiot, C.; Delaloge, S.; Brain, E.; Giachetti, S.; de Cremoux, P.; Marty, M.; Pierga, J.Y. Single circulating tumor cell detection and overall survival in nonmetastatic breast cancer. Ann Oncol. 2010, 21, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Mathiesen, R.R.; Borgen, E.; Renolen, A.; Løkkevik, E.; Nesland, J.M.; Anker, G.; Ostenstad, B.; Lundgren, S.; Risberg, T.; Mjaaland, I.; et al. Persistence of disseminated tumor cells after neoadjuvant treatment for locally advanced breast cancer predicts poor survival. Breast Cancer Res. 2012, 14, R117. [Google Scholar] [CrossRef]
- Lucci, A.; Hall, C.S.; Lodhi, A.K.; Bhattacharyya, A.; Anderson, A.E.; Xiao, L.; Bedrosian, I.; Kuerer, H.M.; Krishnamurthy, S. Circulating tumour cells in non-metastatic breast cancer: A prospective study. Lancet Oncol. 2012, 13, 688–695. [Google Scholar] [CrossRef]
- Sandri, M.T.; Zorzino, L.; Cassatella, M.C.; Bassi, F.; Luini, A.; Casadio, C.; Botteri, E.; Rotmensz, N.; Adamoli, L.; Nolè, F. Changes in circulating tumor cell detection in patients with localized breast cancer before and after surgery. Ann. Surg. Oncol. 2010, 17, 1539–1545. [Google Scholar] [CrossRef]
- Pierga, J.Y.; Bidard, F.C.; Mathiot, C.; Brain, E.; Delaloge, S.; Giachetti, S.; de Cremoux, P.; Salmon, R.; Vincent-Salomon, A.; Marty, M. Circulating tumor cell detection predicts early metastatic relapse after neoadjuvant chemotherapy in large operable and locally advanced breast cancer in a phase II randomized trial. Clin. Cancer Res. 2008, 14, 7004–7010. [Google Scholar] [CrossRef]
- Janni, W.J.; Rack, B.; Terstappen, L.W.; Pierga, J.Y.; Taran, F.A.; Fehm, T.; Hall, C.; de Groot, M.R.; Bidard, F.C.; Friedl, T.W.; et al. Pooled Analysis of the Prognostic Relevance of Circulating Tumor Cells in Primary Breast Cancer. Clin. Cancer Res. 2016, 22, 2583–2593. [Google Scholar] [CrossRef]
- Pachmann, K.; Camara, O.; Kavallaris, A.; Krauspe, S.; Malarski, N.; Gajda, M.; Kroll, T.; Jörke, C.; Hammer, U.; Altendorf-Hofmann, A.; et al. Monitoring the response of circulating epithelial tumor cells to adjuvant chemotherapy in breast cancer allows detection of patients at risk of early relapse. J. Clin. Oncol. 2008, 26, 1208–1215. [Google Scholar] [CrossRef]
- Fischer, J.C.; Niederacher, D.; Topp, S.A.; Honisch, E.; Schumacher, S.; Schmitz, N.; Zacarias Föhrding, L.; Vay, C.; Hoffmann, I.; Kasprowicz, N.S.; et al. Diagnostic leukapheresis enables reliable detection of circulating tumor cells of nonmetastatic cancer patients. Proc. Natl. Acad. Sci. USA 2013, 110, 16580–16585. [Google Scholar] [CrossRef]
- Lang, J.E.; Ring, A.; Porras, T.; Kaur, P.; Forte, V.A.; Mineyev, N.; Tripathy, D.; Press, M.F.; Campo, D. RNA-Seq of Circulating Tumor Cells in Stage II-III Breast Cancer. Ann. Surg. Oncol. 2018, 25, 2261–2270. [Google Scholar] [CrossRef] [PubMed]
- Fina, E.; Reduzzi, C.; Motta, R.; Di Cosimo, S.; Bianchi, G.; Martinetti, A.; Wechsler, J.; Cappelletti, V.; Daidone, M.G. Did circulating tumor cells tell us all they could? The missed circulating tumor cell message in breast cancer. Int. J. Biol. Markers. 2015, 30, e429–e433. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, D.A.; Zweitzig, D.R.; Foulk, B.W.; Miller, M.C.; Doyle, G.V.; Pienta, K.J.; Meropol, N.J.; Weiner, L.M.; Cohen, S.J.; Moreno, J.G.; et al. Global gene expression profiling of circulating tumor cells. Cancer Res. 2005, 65, 4993–4997. [Google Scholar] [CrossRef] [PubMed]
- Fina, E.; Callari, M.; Reduzzi, C.; D’Aiuto, F.; Mariani, G.; Generali, D.; Pierotti, M.A.; Daidone, M.G.; Cappelletti, V. Gene expression profiling of circulating tumor cells in breast cancer. Clin. Chem. 2015, 61, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Obermayr, E.; Sanchez-Cabo, F.; Tea, M.K.; Singer, C.F.; Krainer, M.; Fischer, M.B.; Sehouli, J.; Reinthaller, A.; Horvat, R.; Heinze, G.; et al. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer 2010, 10, 666. [Google Scholar] [CrossRef]
- Lu, J.; Fan, T.; Zhao, Q.; Zeng, W.; Zaslavsky, E.; Chen, J.J.; Frohman, M.A.; Golightly, M.G.; Madajewicz, S.; Chen, W.T. Isolation of circulating epithelial and tumor progenitor cells with an invasive phenotype from breast cancer patients. Int. J. Cancer 2010, 126, 669–683. [Google Scholar] [CrossRef]
- Kwan, T.T.; Bardia, A.; Spring, L.M.; Giobbie-Hurder, A.; Kalinich, M.; Dubash, T.; Sundaresan, T.; Hong, X.; LiCausi, J.A.; Ho, U.; et al. A Digital RNA Signature of Circulating Tumor Cells Predicting Early Therapeutic Response in Localized and Metastatic Breast Cancer. Cancer Discov. 2018, 8, 1286–1299. [Google Scholar] [CrossRef]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Rugo, H.S.; Wolf, D.M.; Hauranieh, L.; Roy, R.; Pendyala, P.; Sosa, E.V.; Scott, J.H.; Lee, J.S.; Pitcher, B.; et al. Expanded Genomic Profiling of Circulating Tumor Cells in Metastatic Breast Cancer Patients to Assess Biomarker Status and Biology Over Time (CALGB 40502 and CALGB 40503, Alliance). Clin. Cancer Res. 2018, 24, 1486–1499. [Google Scholar] [CrossRef]
- Wang, H.; Molina, J.; Jiang, J.; Ferber, M.; Pruthi, S.; Jatkoe, T.; Derecho, C.; Rajpurohit, Y.; Zheng, J.; Wang, Y. Gene expression markers in circulating tumor cells may predict bone metastasis and response to hormonal treatment in breast cancer. Mol. Clin. Oncol. 2013, 1, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Pauken, C.M.; Kenney, S.R.; Brayer, K.J.; Guo, Y.; Brown-Glaberman, U.A.; Marchetti, D. Heterogeneity of Circulating Tumor Cell Neoplastic Subpopulations Outlined by Single-Cell Transcriptomics. Cancers 2021, 13, 4885. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, A.M.; Mostert, B.; Bolt-de Vries, J.; Peeters, D.; de Jongh, F.E.; Stouthard, J.M.; Dirix, L.Y.; van Dam, P.A.; Van Galen, A.; de Weerd, V.; et al. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin. Cancer Res. 2011, 17, 3600–3618. [Google Scholar] [CrossRef] [PubMed]
- Fina, E.; Cleris, L.; Dugo, M.; Lecchi, M.; Ciniselli, C.M.; Lecis, D.; Bianchi, G.V.; Verderio, P.; Daidone, M.G.; Cappelletti, V. Gene signatures of circulating breast cancer cell models are a source of novel molecular determinants of metastasis and improve circulating tumor cell detection in patients. J. Exp. Clin. Cancer Res. 2022, 41, 78. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.; Cabezas-Sainz, P.; Pereira-Veiga, T.; Falo, C.; Abalo, A.; Morilla, I.; Curiel, T.; Cueva, J.; Rodríguez, C.; Varela-Pose, V.; et al. Looking for a Better Characterization of Triple-Negative Breast Cancer by Means of Circulating Tumor Cells. J. Clin. Med. 2020, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Bredemeier, M.; Edimiris, P.; Mach, P.; Kubista, M.; Sjöback, R.; Rohlova, E.; Kolostova, K.; Hauch, S.; Aktas, B.; Tewes, M.; et al. Gene Expression Signatures in Circulating Tumor Cells Correlate with Response to Therapy in Metastatic Breast Cancer. Clin Chem. 2017, 63, 1585–1593. [Google Scholar] [CrossRef]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell. 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during, E.M.T. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352, Erratum in: Nat. Rev. Mol. Cell Biol. 2021, 22, 834. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.H.; Donaher, J.L.; Murphy, D.A.; Chau, S.; Yang, J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 2012, 22, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Mege, R.M.; Matsuzaki, F.; Gallin, W.J.; Goldberg, J.I.; Cunningham, B.A.; Edelman, G.M. Construction of epithelioid sheets by transfection of mouse sarcoma cells with cDNAs for chicken cell adhesion molecules. Proc. Natl. Acad. Sci. USA 1988, 85, 7274–7278. [Google Scholar] [CrossRef]
- Chao, Y.L.; Shepard, C.R.; Wells, A. Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Mol. Cancer 2010, 9, 179. [Google Scholar] [CrossRef]
- Padmanaban, V.; Krol, I.; Suhail, Y.; Szczerba, B.M.; Aceto, N.; Bader, J.S.; Ewald, A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019, 573, 439–444. [Google Scholar] [CrossRef]
- Spizzo, G.; Went, P.; Dirnhofer, S.; Obrist, P.; Simon, R.; Spichtin, H.; Maurer, R.; Metzger, U.; von Castelberg, B.; Bart, R.; et al. High Ep-CAM expression is associated with poor prognosis in node-positive breast cancer. Breast Cancer Res. Treat. 2004, 86, 207–213. [Google Scholar] [CrossRef]
- Went, P.T.; Lugli, A.; Meier, S.; Bundi, M.; Mirlacher, M.; Sauter, G.; Dirnhofer, S. Frequent EpCam protein expression in human carcinomas. Hum. Pathol. 2004, 35, 122–128. [Google Scholar] [CrossRef]
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J. Clin. Oncol. 2014, 32, 3483–3489. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584, Erratum in: Science 2019, 363, eaaw7579. [Google Scholar] [CrossRef]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009, 11, R46. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Agelaki, S.; Kalykaki, A.; Stournaras, C.; Mavroudis, D.; Georgoulias, V. Phosphorylated EGFR and PI3K/Akt signaling kinases are expressed in circulating tumor cells of breast cancer patients. Breast Cancer Res. 2008, 10, R80. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Markomanolaki, H.; Giannoukaraki, V.; Papadaki, M.A.; Strati, A.; Lianidou, E.S.; Georgoulias, V.; Mavroudis, D.; Agelaki, S. Hypoxia-inducible factor-1alpha and vascular endothelial growth factor expression in circulating tumor cells of breast cancer patients. Breast Cancer Res. 2009, 11, R84. [Google Scholar] [CrossRef] [PubMed]
- Markou, A.; Strati, A.; Malamos, N.; Georgoulias, V.; Lianidou, E.S. Molecular characterization of circulating tumor cells in breast cancer by a liquid bead array hybridization assay. Clin. Chem. 2011, 57, 421–430. [Google Scholar] [CrossRef]
- Strati, A.; Markou, A.; Parisi, C.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Lianidou, E. Gene expression profile of circulating tumor cells in breast cancer by RT-qPCR. BMC Cancer 2011, 11, 422. [Google Scholar] [CrossRef]
- Strati, A.; Zavridou, M.; Kallergi, G.; Politaki, E.; Kuske, A.; Gorges, T.M.; Riethdorf, S.; Joosse, S.A.; Koch, C.; Bohnen, A.L.; et al. A Comprehensive Molecular Analysis of in Vivo Isolated EpCAM-Positive Circulating Tumor Cells in Breast Cancer. Clin. Chem. 2021, 67, 1395–1405. [Google Scholar] [CrossRef]
- Kasimir-Bauer, S.; Hoffmann, O.; Wallwiener, D.; Kimmig, R.; Fehm, T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012, 14, R15. [Google Scholar] [CrossRef]
- Fina, E. Biological and Clinical Significance of Circulating Tumor Cells in Breast Cancer. Ph.D. Thesis, The Open University, Milton Keynes, UK, 2017. [Google Scholar] [CrossRef]
- Kallergi, G.; Papadaki, M.A.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011, 13, R59. [Google Scholar] [CrossRef]
- Mego, M.; Mani, S.A.; Lee, B.N.; Li, C.; Evans, K.W.; Cohen, E.N.; Gao, H.; Jackson, S.A.; Giordano, A.; Hortobagyi, G.N.; et al. Expression of epithelial-mesenchymal transition-inducing transcription factors in primary breast cancer: The effect of neoadjuvant therapy. Int. J. Cancer 2012, 130, 808–816. [Google Scholar] [CrossRef]
- Mego, M.; Karaba, M.; Minarik, G.; Benca, J.; Silvia, J.; Sedlackova, T.; Manasova, D.; Kalavska, K.; Pindak, D.; Cristofanilli, M.; et al. Circulating Tumor Cells with Epithelial-to-mesenchymal Transition Phenotypes Associated with Inferior Outcomes in Primary Breast Cancer. Anticancer Res. 2019, 39, 1829–1837. [Google Scholar] [CrossRef]
- Strati, A.; Nikolaou, M.; Georgoulias, V.; Lianidou, E.S. Prognostic Significance of TWIST1, CD24, CD44, and ALDH1 Transcript Quantification in EpCAM-Positive Circulating Tumor Cells from Early Stage Breast Cancer Patients. Cells 2019, 8, 652. [Google Scholar] [CrossRef] [PubMed]
- Jakabova, A.; Bielcikova, Z.; Pospisilova, E.; Matkowski, R.; Szynglarewicz, B.; Staszek-Szewczyk, U.; Zemanova, M.; Petruzelka, L.; Eliasova, P.; Kolostova, K.; et al. Molecular characterization and heterogeneity of circulating tumor cells in breast cancer. Breast Cancer Res. Treat. 2017, 166, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Onstenk, W.; Sieuwerts, A.M.; Weekhout, M.; Mostert, B.; Reijm, E.A.; van Deurzen, C.H.; Bolt-de Vries, J.B.; Peeters, D.J.; Hamberg, P.; Seynaeve, C.; et al. Gene expression profiles of circulating tumor cells versus primary tumors in metastatic breast cancer. Cancer Lett. 2015, 362, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Reijm, E.A.; Sieuwerts, A.M.; Smid, M.; Vries, J.B.; Mostert, B.; Onstenk, W.; Peeters, D.; Dirix, L.Y.; Seynaeve, C.M.; Jager, A.; et al. An 8-gene mRNA expression profile in circulating tumor cells predicts response to aromatase inhibitors in metastatic breast cancer patients. BMC Cancer 2016, 16, 123. [Google Scholar] [CrossRef] [PubMed]
- Mostert, B.; Sieuwerts, A.M.; Kraan, J.; Bolt-de Vries, J.; van der Spoel, P.; van Galen, A.; Peeters, D.J.; Dirix, L.Y.; Seynaeve, C.M.; Jager, A.; et al. Gene expression profiles in circulating tumor cells to predict prognosis in metastatic breast cancer patients. Ann Oncol. 2015, 26, 510–516. [Google Scholar] [CrossRef]
- Keup, C.; Suryaprakash, V.; Storbeck, M.; Hoffmann, O.; Kimmig, R.; Kasimir-Bauer, S. Longitudinal Multi-Parametric Liquid Biopsy Approach Identifies Unique Features of Circulating Tumor Cell, Extracellular Vesicle, and Cell-Free DNA Characterization for Disease Monitoring in Metastatic Breast Cancer Patients. Cells 2021, 10, 212. [Google Scholar] [CrossRef]
- Pereira-Veiga, T.; Martínez-Fernández, M.; Abuin, C.; Piñeiro, R.; Cebey, V.; Cueva, J.; Palacios, P.; Blanco, C.; Muinelo-Romay, L.; Abalo, A.; et al. CTCs Expression Profiling for Advanced Breast Cancer Monitoring. Cancers 2019, 11, 1941. [Google Scholar] [CrossRef]
- Strati, A.; Nikolaou, M.; Georgoulias, V.; Lianidou, E.S. RNA-Based CTC Analysis Provides Prognostic Information in Metastatic Breast Cancer. Diagnostics 2021, 11, 513. [Google Scholar] [CrossRef]
- Yu, T.; Wang, C.; Xie, M.; Zhu, C.; Shu, Y.; Tang, J.; Guan, X. Heterogeneity of CTC contributes to the organotropism of breast cancer. Biomed. Pharmacother. 2021, 137, 111314. [Google Scholar] [CrossRef]
- Boral, D.; Vishnoi, M.; Liu, H.N.; Yin, W.; Sprouse, M.L.; Scamardo, A.; Hong, D.S.; Tan, T.Z.; Thiery, J.P.; Chang, J.C.; et al. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 2017, 8, 196. [Google Scholar] [CrossRef]
- Klotz, R.; Thomas, A.; Teng, T.; Han, S.M.; Iriondo, O.; Li, L.; Restrepo-Vassalli, S.; Wang, A.; Izadian, N.; MacKay, M.; et al. Circulating Tumor Cells Exhibit Metastatic Tropism and Reveal Brain Metastasis Drivers. Cancer Discov. 2020, 10, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Vishnoi, M.; Liu, N.H.; Yin, W.; Boral, D.; Scamardo, A.; Hong, D.; Marchetti, D. The identification of a TNBC liver metastasis gene signature by sequential CTC-xenograft modeling. Mol. Oncol. 2019, 13, 1913–1926. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Kuske, A.; Joosse, S.A.; Yigit, G.; Sflomos, G.; Thaler, S.; Smit, D.J.; Werner, S.; Borgmann, K.; Gärtner, S.; et al. Characterization of circulating breast cancer cells with tumorigenic and metastatic capacity. EMBO Mol. Med. 2020, 12, e11908. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ule, N.; González-Conde, M.; Abuín, C.; Cueva, J.F.; Palacios, P.; López-López, R.; Costa, C.; Dávila-Ibáñez, A.B. Short-Term Ex Vivo Culture of CTCs from Advance Breast Cancer Patients: Clinical Implications. Cancers 2021, 13, 2668. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.C.; Kingston, B.; Kilburn, L.S.; Kernaghan, S.; Wardley, A.M.; Macpherson, I.R.; Baird, R.D.; Roylance, R.; Stephens, P.; Oikonomidou, O.; et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): A multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020, 21, 1296–1308. [Google Scholar] [CrossRef]
- Keup, C.; Storbeck, M.; Hauch, S.; Hahn, P.; Sprenger-Haussels, M.; Hoffmann, O.; Kimmig, R.; Kasimir-Bauer, S. Multimodal Targeted Deep Sequencing of Circulating Tumor Cells and Matched Cell-Free DNA Provides a More Comprehensive Tool to Identify Therapeutic Targets in Metastatic Breast Cancer Patients. Cancers 2020, 12, 1084. [Google Scholar] [CrossRef]
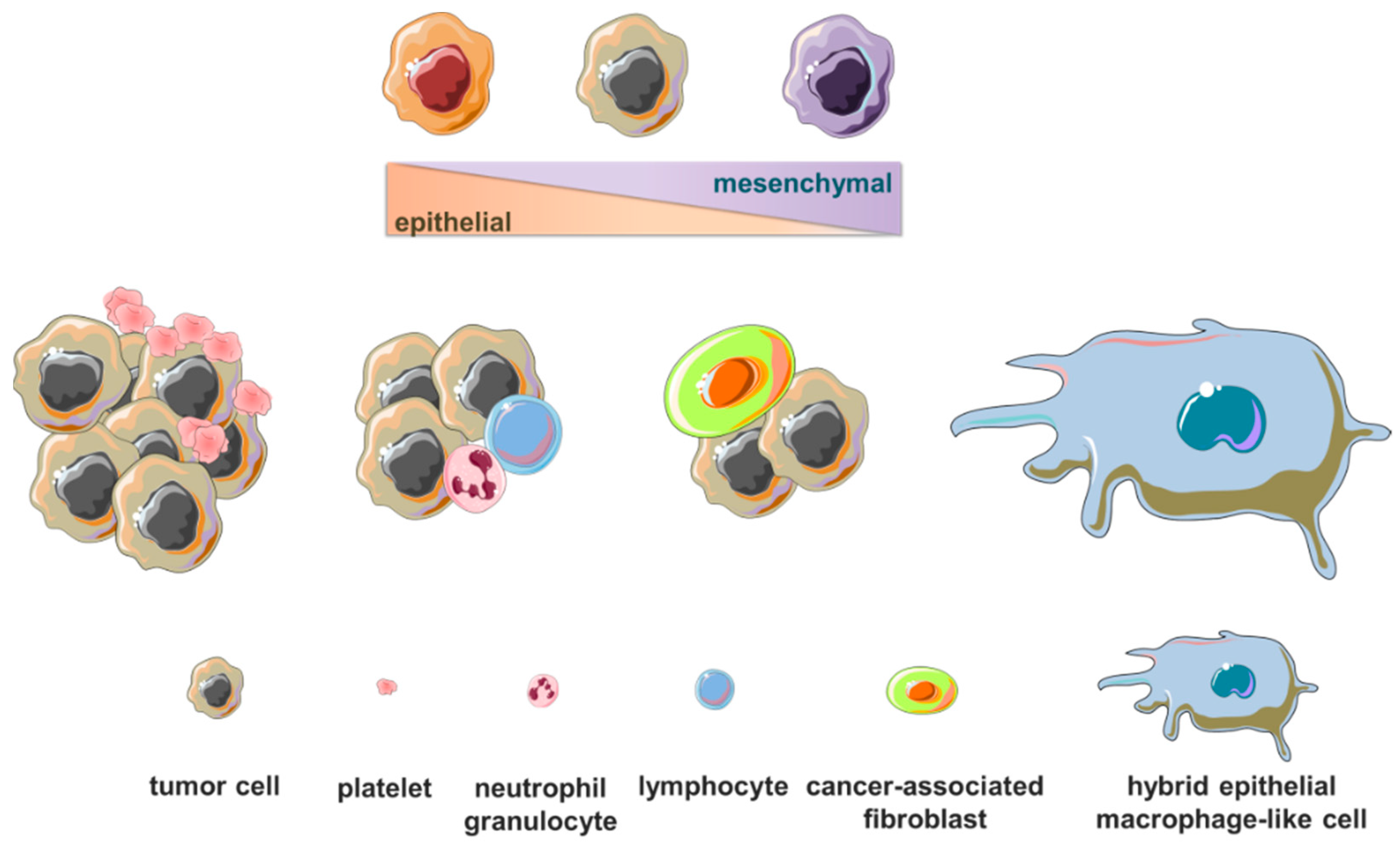
 ) or modulated in circulating tumor cells in patients with early-stage or metastatic breast cancer are related to normal and tumor breast tissue development, epithelial-to-mesenchymal transition, cell motility, migration, invasion and metastatic ability, drug resistance and immune system evasion, tumor growth and progression. (Cartoon depicting breast cancer cell hematogenous dissemination and transcription process was adapted from templates available in smart.servier.com, accessed on 26 October 2022).
) or modulated in circulating tumor cells in patients with early-stage or metastatic breast cancer are related to normal and tumor breast tissue development, epithelial-to-mesenchymal transition, cell motility, migration, invasion and metastatic ability, drug resistance and immune system evasion, tumor growth and progression. (Cartoon depicting breast cancer cell hematogenous dissemination and transcription process was adapted from templates available in smart.servier.com, accessed on 26 October 2022).
 ) or modulated in circulating tumor cells in patients with early-stage or metastatic breast cancer are related to normal and tumor breast tissue development, epithelial-to-mesenchymal transition, cell motility, migration, invasion and metastatic ability, drug resistance and immune system evasion, tumor growth and progression. (Cartoon depicting breast cancer cell hematogenous dissemination and transcription process was adapted from templates available in smart.servier.com, accessed on 26 October 2022).
) or modulated in circulating tumor cells in patients with early-stage or metastatic breast cancer are related to normal and tumor breast tissue development, epithelial-to-mesenchymal transition, cell motility, migration, invasion and metastatic ability, drug resistance and immune system evasion, tumor growth and progression. (Cartoon depicting breast cancer cell hematogenous dissemination and transcription process was adapted from templates available in smart.servier.com, accessed on 26 October 2022).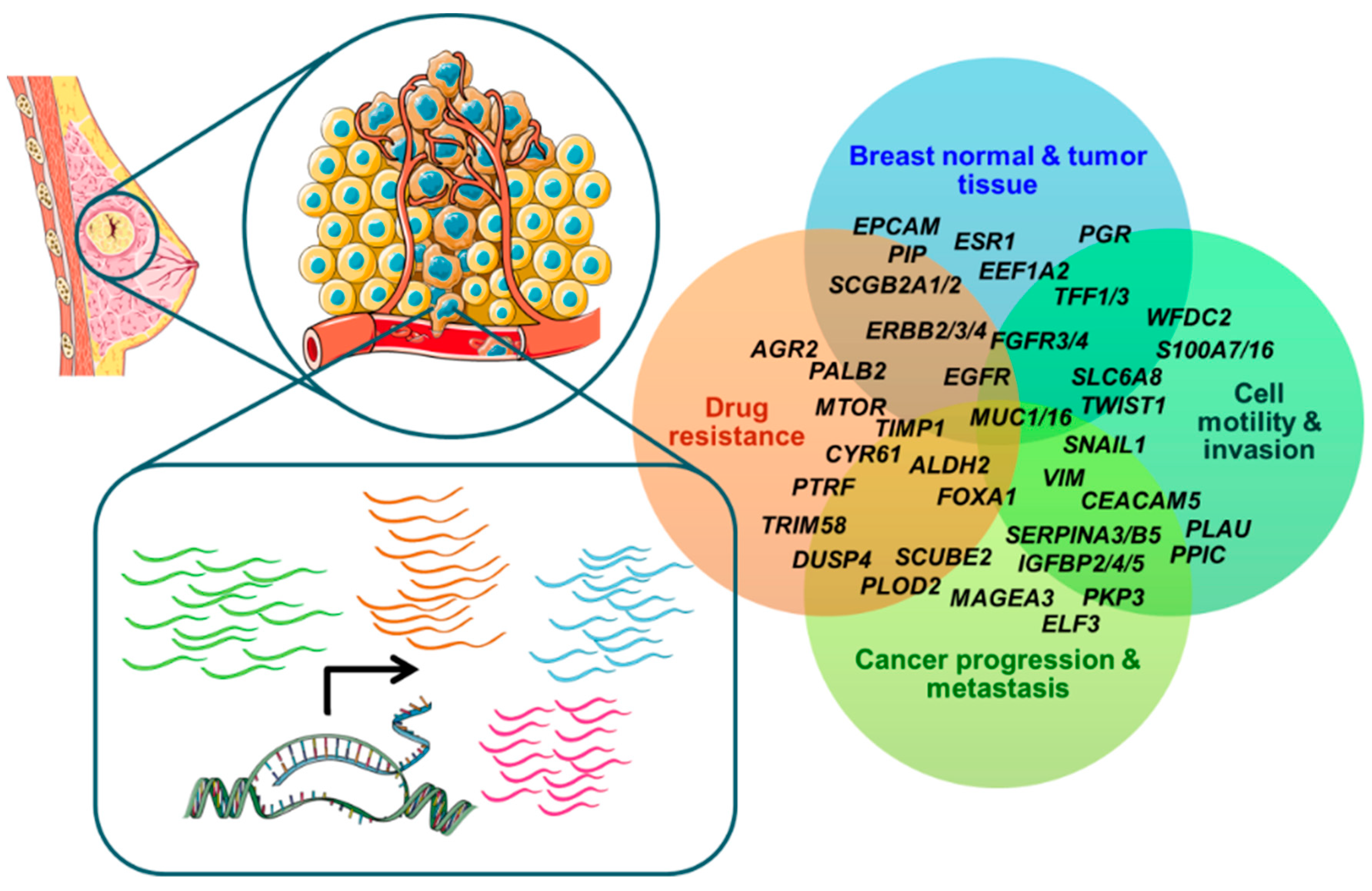
| Gene Abbreviation | Gene Function or Role in Cancer | Reference to CTC Studies |
|---|---|---|
| ACTA2 | invasion | [134] |
| ADPRHL1 | ADP-ribosylhydrolase | [135] |
| AGR2 | cancer progression, chemoresistance | [129,131,134] |
| ALDH2 | drug resistance, cancer stemness | [136] |
| AR | breast cancer development | [136] |
| BCL11A | tumorigenesis, metastasis | [136] |
| CAVIN2 | tumor suppressor | [133] |
| CCND1 | cell cycle | [131,134] |
| CCNE2 | cell cycle | [127] |
| CD24 | migration, invasion, anti-phagocytic | [134,137] |
| CD44 | cell adhesion, migration, cancer stemness | [136] |
| CEA | cancer progression | [132] |
| CEACAM5 | cell polarity, differentiation | [134] |
| CEP55 | mitotic cytokinesis, development, cancer progression | [134] |
| CLDN3 | cell adhesion | [134] |
| CRABP2 | retinoic acid shuttling | [134] |
| CRIPTO | embryonic development, tumor growth | [136] |
| CTTN | cytoskeleton, cell adhesion structure | [134] |
| CXCL13 | epithelial-derived cytokine | [129] |
| CXCL14 | epithelial-derived cytokine | [129,134] |
| CYR61 | chemoresistance | [131] |
| DKFZp762E1312 | cell cycle | [127] |
| DPP4 | metastasis | [128] |
| DTL | cell motility | [134] |
| DTX3 | cell proliferation | [134] |
| DUSP4 | chemoresistance | [134] |
| EEF1A2 | breast cancer development | [134] |
| EFHD1 | highly expressed in breast cancer | [129] |
| EGFR | tumor progression | [132] |
| ELF3 | breast cancer progression | [135] |
| EMP2 | cancer stemness, metastasis | [127] |
| EPCAM | cell adhesion, migration | [128,131,134,137] |
| ERBB2 | cancer survival and progression | [131] |
| ERBB3 | metastasis, therapy resistance | [134] |
| ERBB4 | cancer progression, therapy resistance | [132,134] |
| ESR1 | breast carcinogenesis | [128,131,134] |
| FAT1 | cell adhesion, signaling | [129] |
| FAT2 | cell adhesion, signaling | [129] |
| FCF1 | pre-rRNA processing | [135] |
| FEN1 | invasion, metastasis | [134] |
| FKBP10 | migration, invasion | [134] |
| FGFR3 | breast cancer development, endocrine resistance | [134] |
| FGFR4 | luminal differentiation, metastasis | [134] |
| FOXA1 | migration, invasion, endocrine resistance | [134] |
| GRB7 | EGFR/ERBB2 signaling | [131] |
| IGFBP2 | tumor growth | [134] |
| IGFBP4 | tumor growth | [134] |
| IGFBP5 | cell adhesion, survival | [132,134] |
| IL17BR3 | endocrine resistance | [134] |
| ITGA6 | cancer stemness, invasion | [134,136] |
| ITGB3 | cell adhesion, signaling | [133] |
| KIF11 | cell proliferation | [134] |
| KPNA2 | cell proliferation | [134] |
| KRT7 | invasion, metastasis | [134] |
| KRT8 | regulation of invasion | [128] |
| KRT16 | invasion, metastasis | [128] |
| KRT17 | proliferation, invasion | [134] |
| KRT18 | regulation of invasion | [134] |
| KRT19 | cell proliferation | [128,131,134,137] |
| LAD1 | breast cancer progression | [134] |
| LTBP1 | metastasis | [133] |
| LY6G6F | breast cancer progression | [133] |
| MAGEA3 | breast cancer progression | [132] |
| MAL2 | breast cancer immune evasion | [127] |
| MELK | breast carcinogenesis | [134] |
| MGP | highly expressed in breast cancer | [129] |
| MKI67 | cell proliferation | [131,134] |
| MUC1 | cell survival, therapy resistance | [131,134] |
| MUC16 | cancer growth and metastasis | [128,129] |
| MUCL1 | cell proliferation | [134] |
| PGR | breast lineage specific | [128,129,132] |
| PIP | breast lineage specific | [129,132,134] |
| PKP3 | breast cancer growth and progression | [134] |
| PLAU | motility, invasion | [134] |
| PLOD2 | tumor progression, metastasis | [134] |
| PPIC | cell migration, invasion | [127] |
| PRAME | oncofetal antigens | [129] |
| PTRF | multidrug resistance | [134] |
| S100A7 | cell survival, chemotaxis | [134] |
| S100A16 | invasion | [134] |
| SCGB1D2 | breast lineage specific, cell growth | [132,134] |
| SCGB2A1 | breast lineage specific, cell growth | [129] |
| SCGB2A2 | breast lineage specific, cell growth | [132,134] |
| SCUBE2 | motility and invasion regulation | [131] |
| SEPP1 | oxidative stress reduction | [134] |
| SERPINA3 | EMT, invasion | [129] |
| SERPINB5 | tumor growth and metastasis regulation | [132] |
| SFRP1 | cellular signaling | [129] |
| SFRP2 | cellular signaling | [129] |
| SLC6A8 | invasion | [127] |
| SNAIL1 | EMT inducer | [136] |
| SPDEF | pro- and anti-oncogenic among breast cancer subtypes | [134] |
| TERT | cancer development | [128] |
| TFF1 | proliferation, migration | [131,132,134,135] |
| TFF3 | migration, angiogenesis | [131,134,135] |
| TIMP1 | cell survival, angiogenesis | [136] |
| TIMP3 | cell growth, invasion, angiogenesis | [134] |
| TM4SF13 | breast cancer growth regulation | [134] |
| TMPRSS4 | cancer growth and metastasis | [129] |
| TNRC9 | breast cancer progression | [132,134] |
| TRIM58 | chemoresistance | [133] |
| TUBB1 | β-tubulin | [133] |
| TWIST1 | EMT inducer | [128] |
| VIM | cell motility | [136] |
| WFDC2 | EMT inducer | [129] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fina, E. Signatures of Breast Cancer Progression in the Blood: What Could Be Learned from Circulating Tumor Cell Transcriptomes. Cancers 2022, 14, 5668. https://doi.org/10.3390/cancers14225668
Fina E. Signatures of Breast Cancer Progression in the Blood: What Could Be Learned from Circulating Tumor Cell Transcriptomes. Cancers. 2022; 14(22):5668. https://doi.org/10.3390/cancers14225668
Chicago/Turabian StyleFina, Emanuela. 2022. "Signatures of Breast Cancer Progression in the Blood: What Could Be Learned from Circulating Tumor Cell Transcriptomes" Cancers 14, no. 22: 5668. https://doi.org/10.3390/cancers14225668
APA StyleFina, E. (2022). Signatures of Breast Cancer Progression in the Blood: What Could Be Learned from Circulating Tumor Cell Transcriptomes. Cancers, 14(22), 5668. https://doi.org/10.3390/cancers14225668






