Manta Ray Foraging Optimization Transfer Learning-Based Gastric Cancer Diagnosis and Classification on Endoscopic Images
Abstract
Simple Summary
Abstract
1. Introduction
2. Related Works
3. The Proposed Model
3.1. Stage I: Pre-Processing
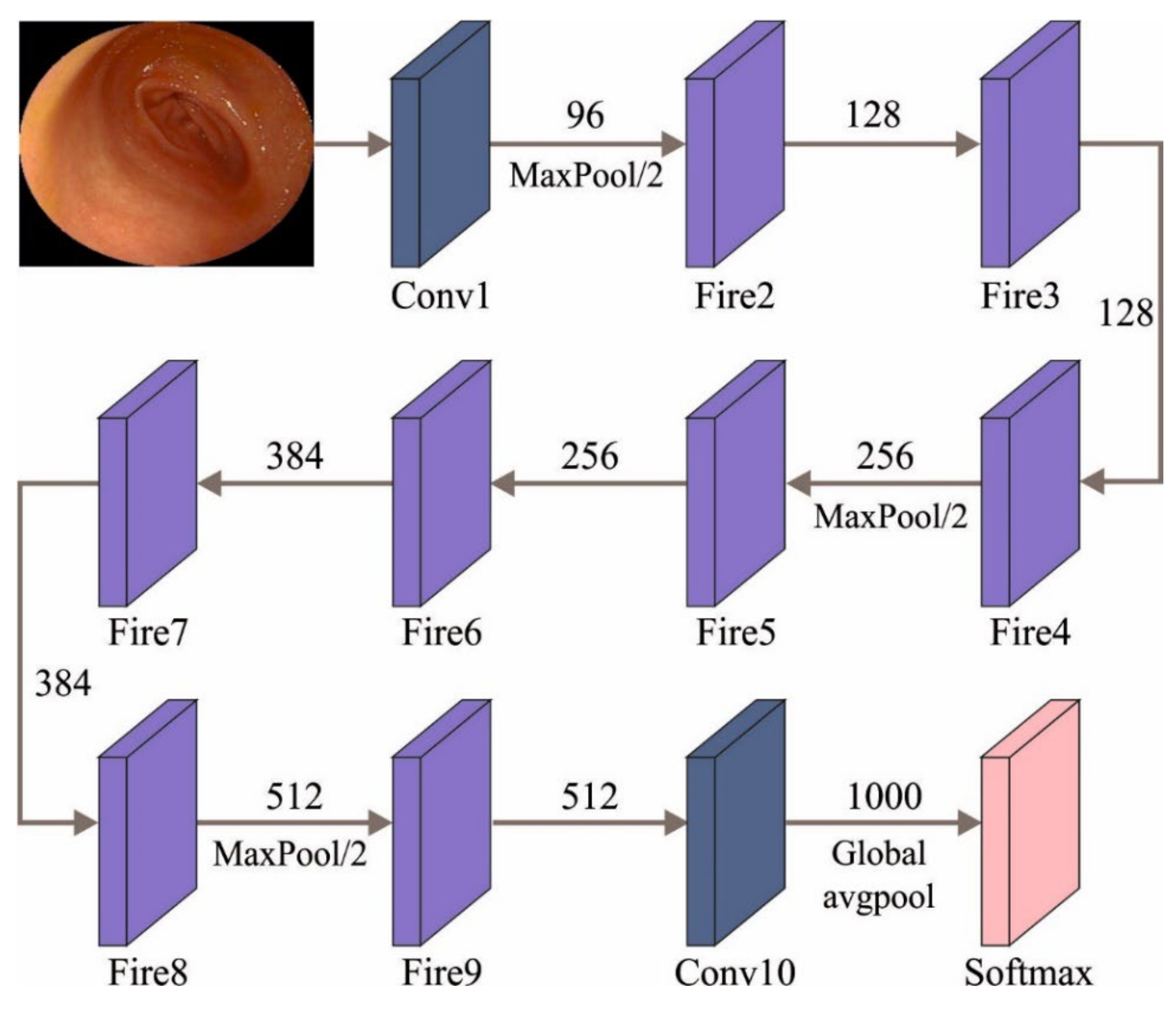
3.2. Stage II: Feature Extraction
3.2.1. Chain Foraging Phase
3.2.2. Cyclone Foraging
3.2.3. Somersault Foraging
3.3. Stage III: GC Classification
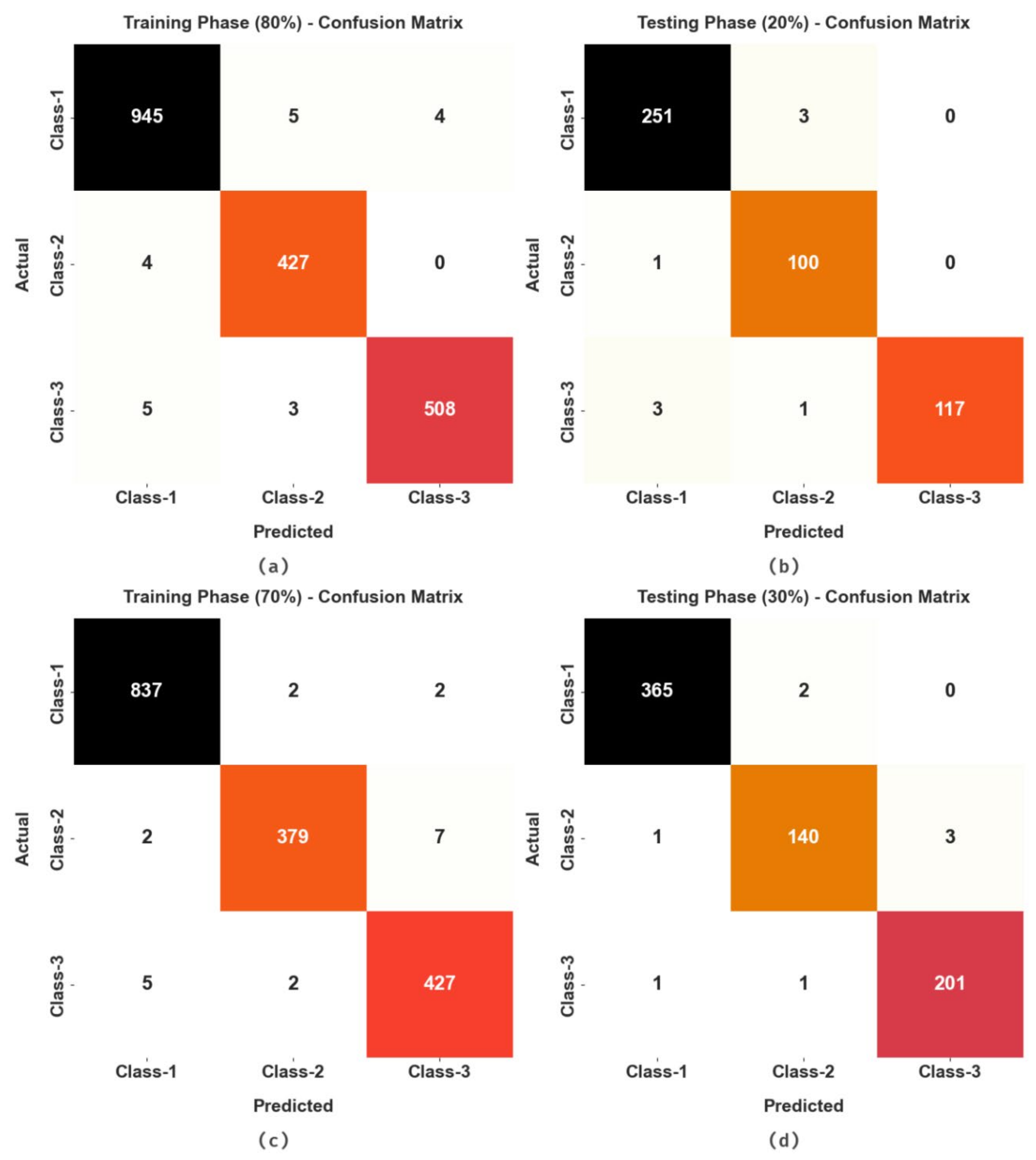
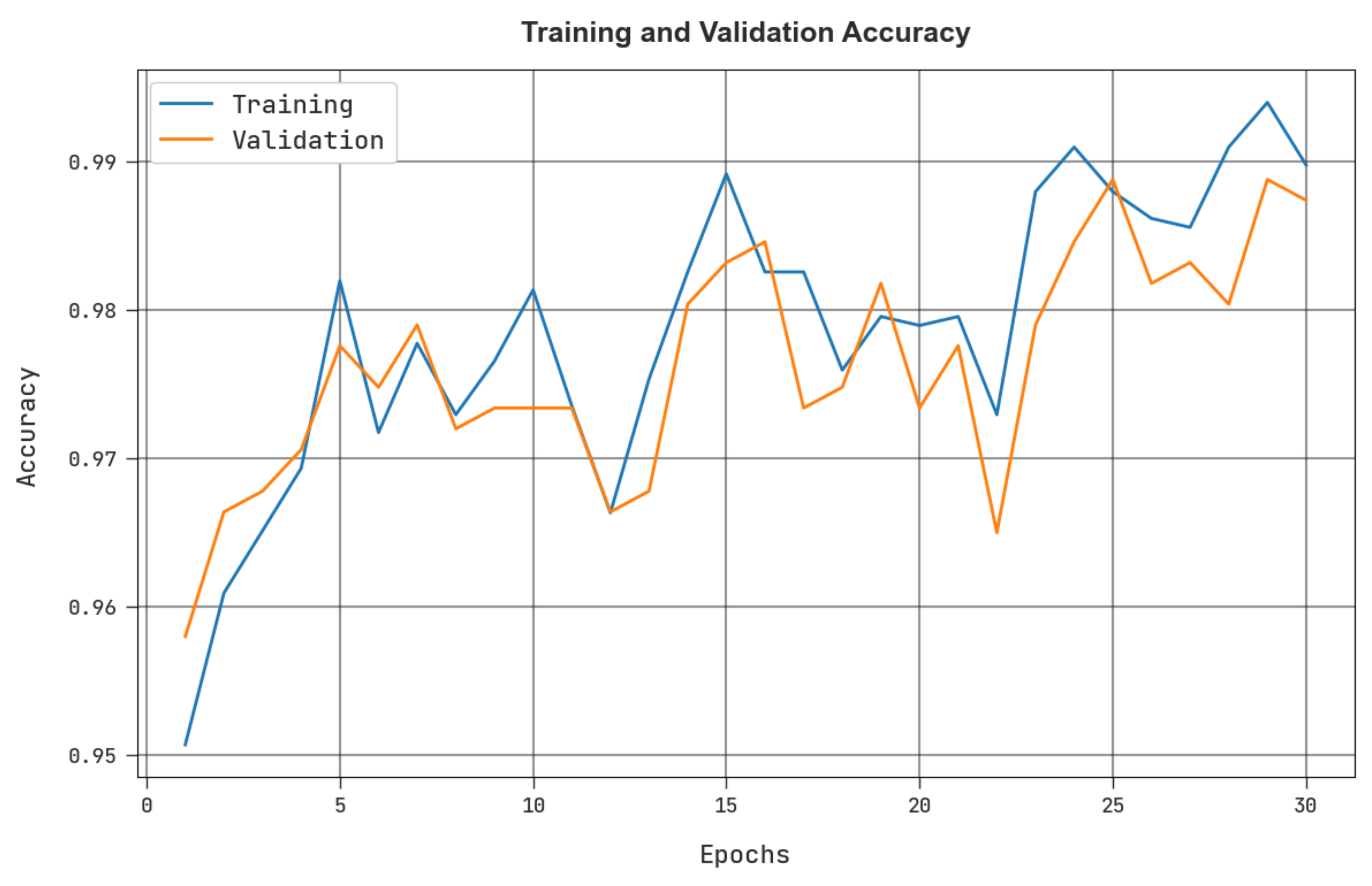
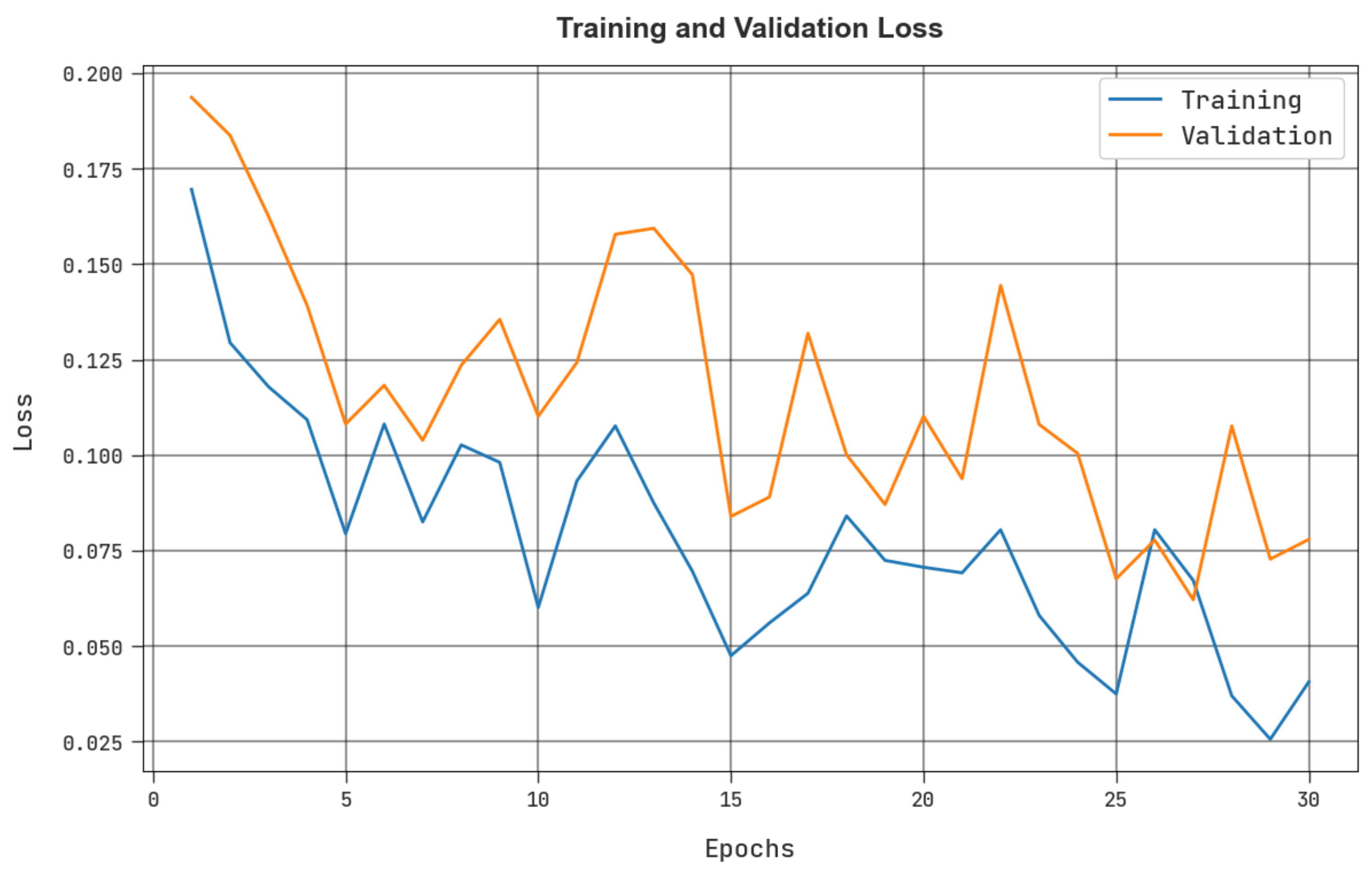
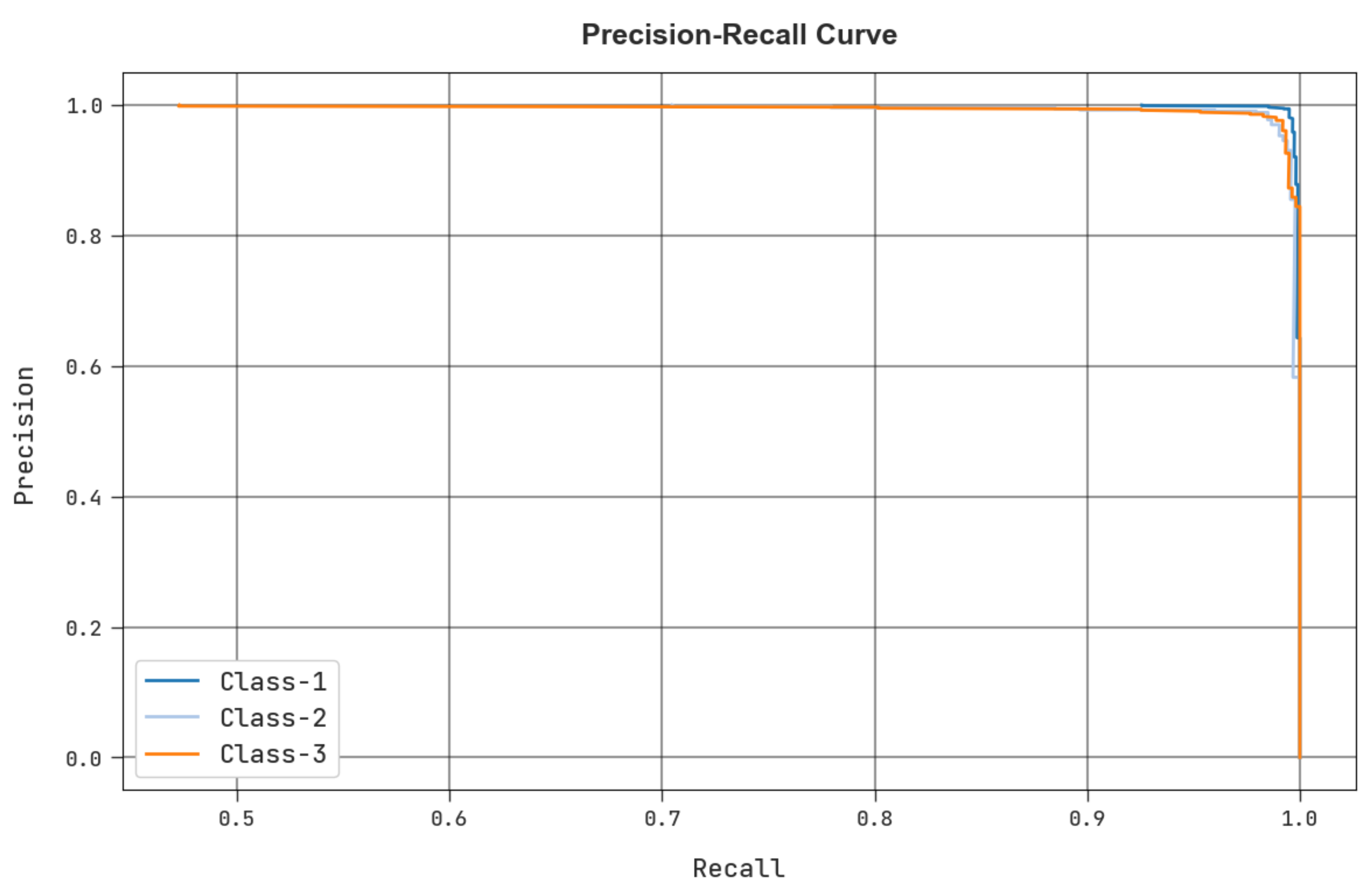
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, H.; Gong, L.; Dong, D.; Zhu, L.; Wang, M.; He, J.; Shu, L.; Cai, Y.; Cai, S.; Su, W.; et al. Identifying early gastric cancer under magnifying narrow-band images with deep learning: A multicenter study. Gastrointest. Endosc. 2021, 93, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qin, Y.; Zhang, W.H.; Jiang, H.; Song, B.; Bashir, M.R.; Xu, H.; Duan, T.; Fang, M.; Zhong, L.; et al. Deep learning-based AI model for signet-ring cell carcinoma diagnosis and chemotherapy response prediction in gastric cancer. Med. Phys. 2022, 49, 1535–1546. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Xie, X.; Shen, L. Deep Learning Based Gastric Cancer Identification. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 182–185. [Google Scholar]
- Wu, L.; Wang, J.; He, X.; Zhu, Y.; Jiang, X.; Chen, Y.; Wang, Y.; Huang, L.; Shang, R.; Dong, Z.; et al. Deep learning system compared with expert endoscopists in predicting early gastric cancer and its invasion depth and differentiation status (with videos). Gastrointest. Endosc. 2022, 95, 92–104. [Google Scholar] [CrossRef]
- Song, Z.; Zou, S.; Zhou, W.; Huang, Y.; Shao, L.; Yuan, J.; Gou, X.; Jin, W.; Wang, Z.; Chen, X.; et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat. Commun. 2020, 11, 4294. [Google Scholar] [CrossRef]
- Ba, W.; Wang, S.; Shang, M.; Zhang, Z.; Wu, H.; Yu, C.; Xing, R.; Wang, W.; Wang, L.; Liu, C.; et al. Assessment of deep learning assistance for the pathological diagnosis of gastric cancer. Mod. Pathol. 2022, 35, 1262–1268. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, W.; Wan, X.; Zhang, J.; Shen, L.; Hu, S.; Ding, Q.; Mu, G.; Yin, A.; Huang, X.; et al. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy 2019, 51, 522–531. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kim, J.H. Lesion-based convolutional neural network in diagnosis of early gastric cancer. Clin. Endosc. 2020, 53, 127–131. [Google Scholar] [CrossRef]
- Raihan, M.; Sarker, M.; Islam, M.M.; Fairoz, F.; Shams, A.B. Identification of the Resting Position Based on EGG, ECG, Respiration Rate and SPO2 Using Stacked Ensemble Learning. In Proceedings of the International Conference on Big Data, IoT, and Machine Learning; Springer: Singapore, 2022; pp. 789–798. [Google Scholar]
- Amri, M.F.; Yuliani, A.R.; Simbolon, A.I.; Ristiana, R.; Kusumandari, D.E. Toward Early Abnormalities Detection on Digestive System: Multi-Features Electrogastrogram (EGG) Signal Classification Based on Machine Learning. In Proceedings of the 2021 International Conference on Radar, Antenna, Microwave, Electronics, and Telecommunications (ICRAMET), Bandung, Indonesia, 23–24 November 2021; pp. 185–190. [Google Scholar]
- Hu, W.; Li, C.; Li, X.; Rahaman, M.M.; Ma, J.; Zhang, Y.; Chen, H.; Liu, W.; Sun, C.; Yao, Y.; et al. GasHisSDB: A new gastric histopathology image dataset for computer aided diagnosis of gastric cancer. Comput. Biol. Med. 2022, 142, 105207. [Google Scholar] [CrossRef] [PubMed]
- Sharanyaa, S.; Vijayalakshmi, S.; Therasa, M.; Kumaran, U.; Deepika, R. DCNET: A Novel Implementation of Gastric Cancer Detection System through Deep Learning Convolution Networks. In Proceedings of the 2022 International Conference on Advanced Computing Technologies and Applications (ICACTA), Virtual, 4–5 March 2022; pp. 1–5. [Google Scholar]
- Qiu, W.; Xie, J.; Shen, Y.; Xu, J.; Liang, J. Endoscopic image recognition method of gastric cancer based on deep learning model. Expert Syst. 2022, 39, e12758. [Google Scholar] [CrossRef]
- Jang, H.J.; Song, I.H.; Lee, S.H. Deep Learning for Automatic Subclassification of Gastric Carcinoma Using Whole-Slide Histopathology Images. Cancers 2021, 13, 3811. [Google Scholar] [CrossRef] [PubMed]
- Valieris, R.; Amaro, L.; Osório, C.A.B.D.T.; Bueno, A.P.; Rosales Mitrowsky, R.A.; Carraro, D.M.; Nunes, D.N.; Dias-Neto, E.; Silva, I.T.D. Deep learning predicts underlying features on pathology images with therapeutic relevance for breast and gastric cancer. Cancers 2020, 12, 3687. [Google Scholar] [CrossRef] [PubMed]
- Meier, A.; Nekolla, K.; Hewitt, L.C.; Earle, S.; Yoshikawa, T.; Oshima, T.; Miyagi, Y.; Huss, R.; Schmidt, G.; Grabsch, H.I. Hypothesis-free deep survival learning applied to the tumour microenvironment in gastric cancer. J. Pathol. Clin. Res. 2020, 6, 273–282. [Google Scholar] [CrossRef] [PubMed]
- An, P.; Yang, D.; Wang, J.; Wu, L.; Zhou, J.; Zeng, Z.; Huang, X.; Xiao, Y.; Hu, S.; Chen, Y.; et al. A deep learning method for delineating early gastric cancer resection margin under chromoendoscopy and white light endoscopy. Gastric Cancer 2020, 23, 884–892. [Google Scholar] [CrossRef] [PubMed]
- Alwazzan, M.J.; Ismael, M.A.; Ahmed, A.N. A hybrid algorithm to enhance colour retinal fundus images using a Wiener filter and CLAHE. J. Digit. Imaging 2021, 34, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Ragab, M.; Albukhari, A.; Alyami, J.; Mansour, R.F. Ensemble deep-learning-enabled clinical decision support system for breast cancer diagnosis and classification on ultrasound images. Biology 2022, 11, 439. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhang, Z.; Wang, L. Manta ray foraging optimization: An effective bio-inspired optimizer for engineering applications. Eng. Appl. Artif. Intell. 2020, 87, 103300. [Google Scholar] [CrossRef]
- Rizk-Allah, R.M.; Zineldin, M.I.; Mousa, A.A.A.; Abdel-Khalek, S.; Mohamed, M.S.; Snášel, V. On a Novel Hybrid Manta Ray Foraging Optimizer and Its Application on Parameters Estimation of Lithium-Ion Battery. Int. J. Comput. Intell. Syst. 2022, 15, 62. [Google Scholar] [CrossRef]
- Fan, Q.; Zhang, Z.; Huang, X. Parameter conjugate gradient with secant equation based Elman neural network and its convergence analysis. Adv. Theory Simul. 2022, 5, 2200047. [Google Scholar] [CrossRef]


| Label | Class | Count of Images |
|---|---|---|
| Class1 | Healthy | 1208 |
| Class2 | Early gastric cancer | 532 |
| Class3 | Advanced gastric cancer | 637 |
| Total No. of Images | 2377 | |
| Labels | AUC Score | ||||
|---|---|---|---|---|---|
| Training Phase (80%) | |||||
| Class-1 | 99.05 | 99.06 | 99.06 | 99.06 | 99.05 |
| Class-2 | 99.37 | 98.16 | 99.07 | 98.61 | 99.26 |
| Class-3 | 99.37 | 99.22 | 98.45 | 98.83 | 99.08 |
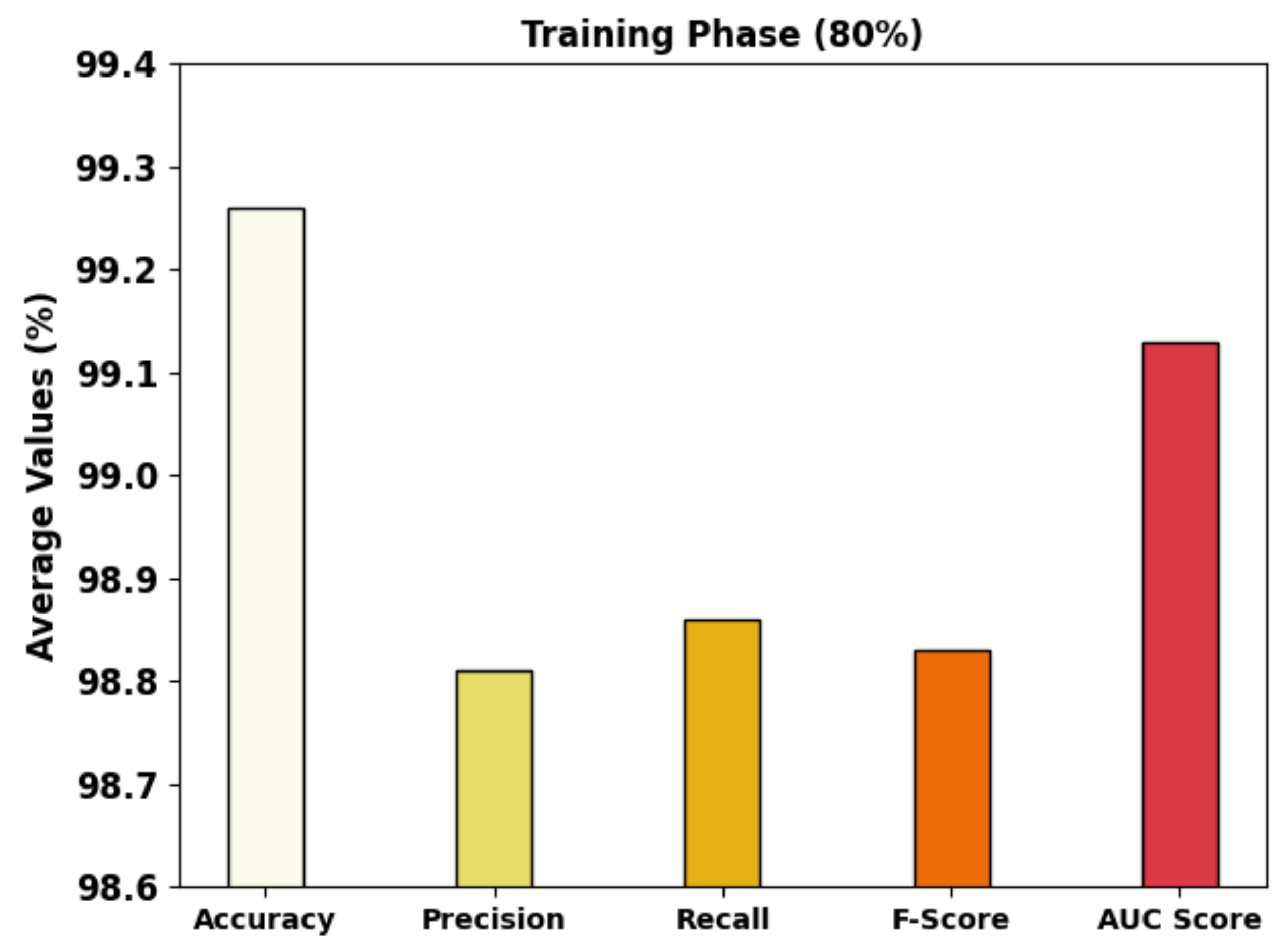
| Average | 99.26 | 98.81 | 98.86 | 98.83 | 99.13 |
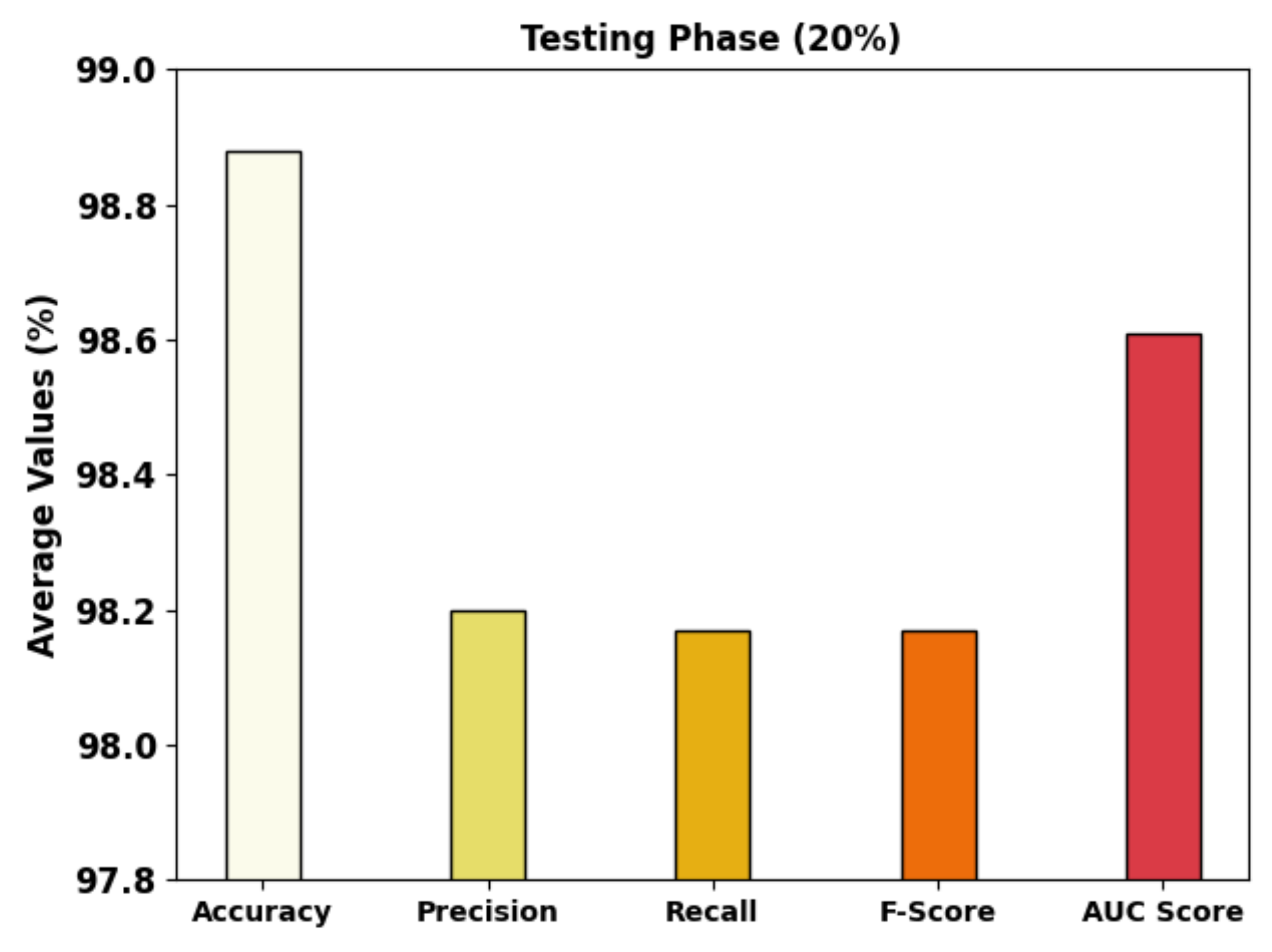
| Testing Phase (20%) | |||||
| Class-1 | 98.53 | 98.43 | 98.82 | 98.62 | 98.51 |
| Class-2 | 98.95 | 96.15 | 99.01 | 97.56 | 98.97 |
| Class-3 | 99.16 | 100.00 | 96.69 | 98.32 | 98.35 |
| Average | 98.88 | 98.20 | 98.17 | 98.17 | 98.61 |
| Labels | AUC Score | ||||
|---|---|---|---|---|---|
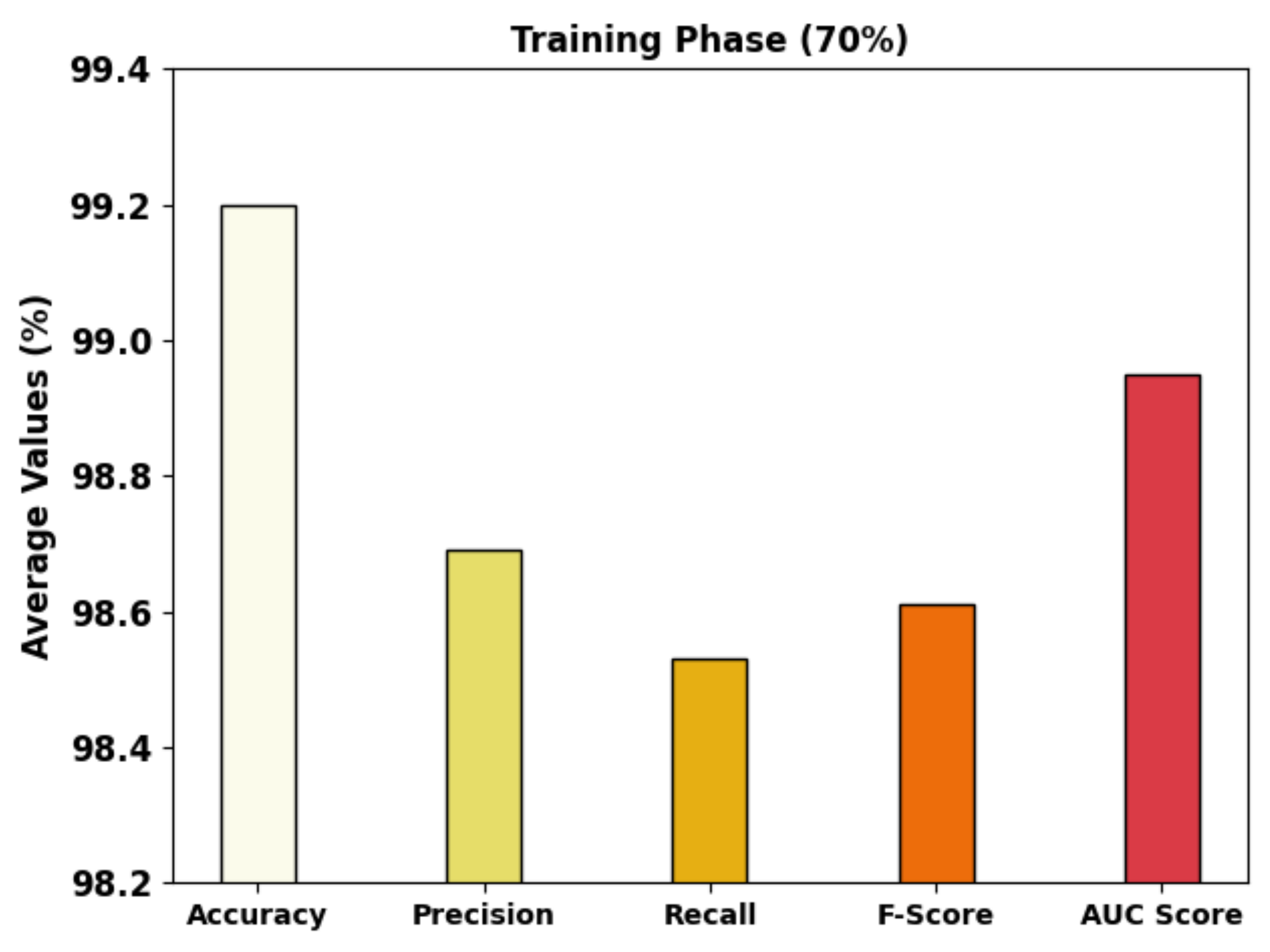
| Training Phase (70%) | |||||
| Class-1 | 99.34 | 99.17 | 99.52 | 99.35 | 99.34 |
| Class-2 | 99.22 | 98.96 | 97.68 | 98.31 | 98.68 |
| Class-3 | 99.04 | 97.94 | 98.39 | 98.16 | 98.83 |
| Average | 99.20 | 98.69 | 98.53 | 98.61 | 98.95 |
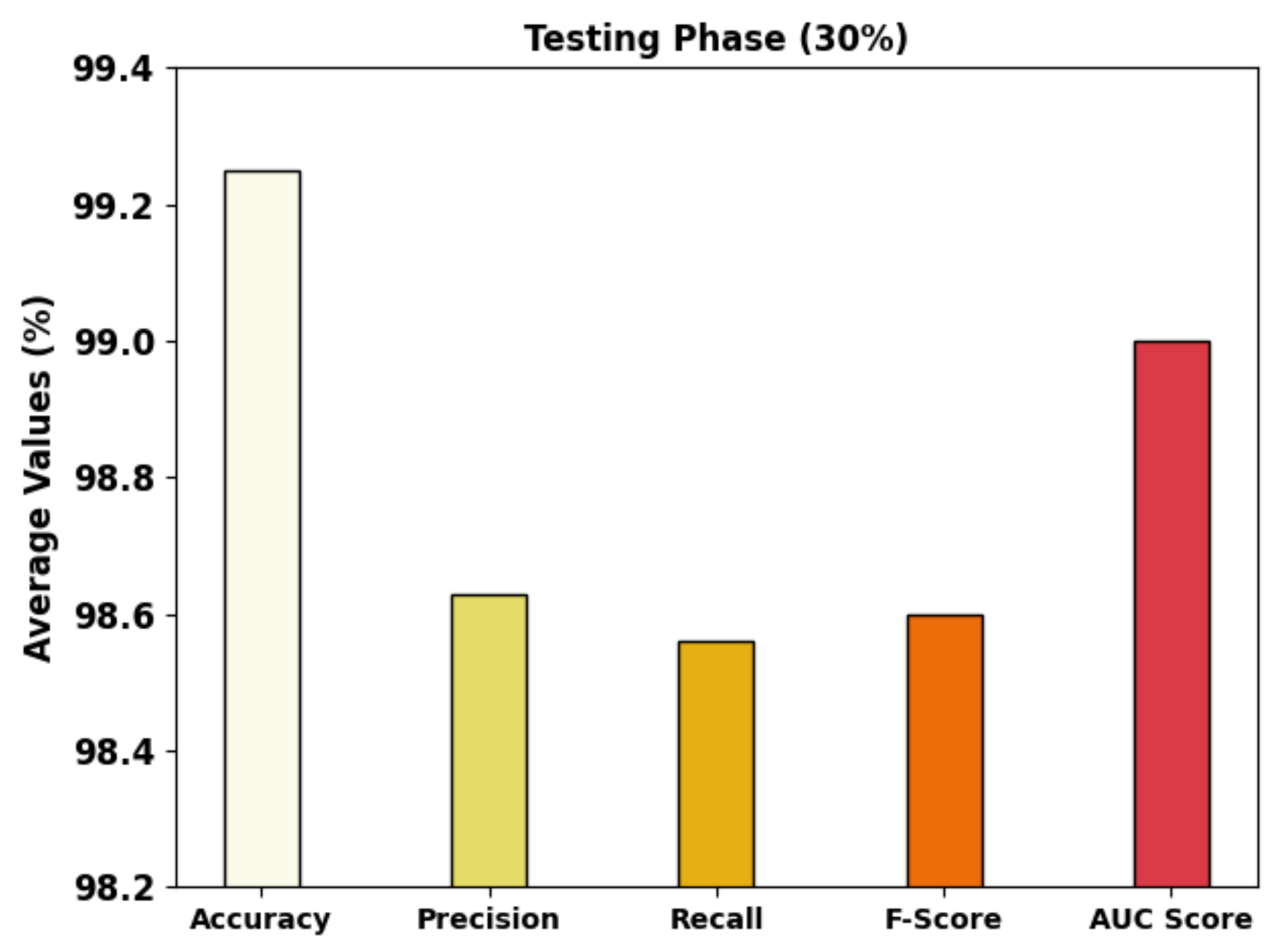
| Testing Phase (30%) | |||||
| Class-1 | 99.44 | 99.46 | 99.46 | 99.46 | 99.44 |
| Class-2 | 99.02 | 97.90 | 97.22 | 97.56 | 98.35 |
| Class-3 | 99.30 | 98.53 | 99.01 | 98.77 | 99.21 |
| Average | 99.25 | 98.63 | 98.56 | 98.60 | 99.00 |
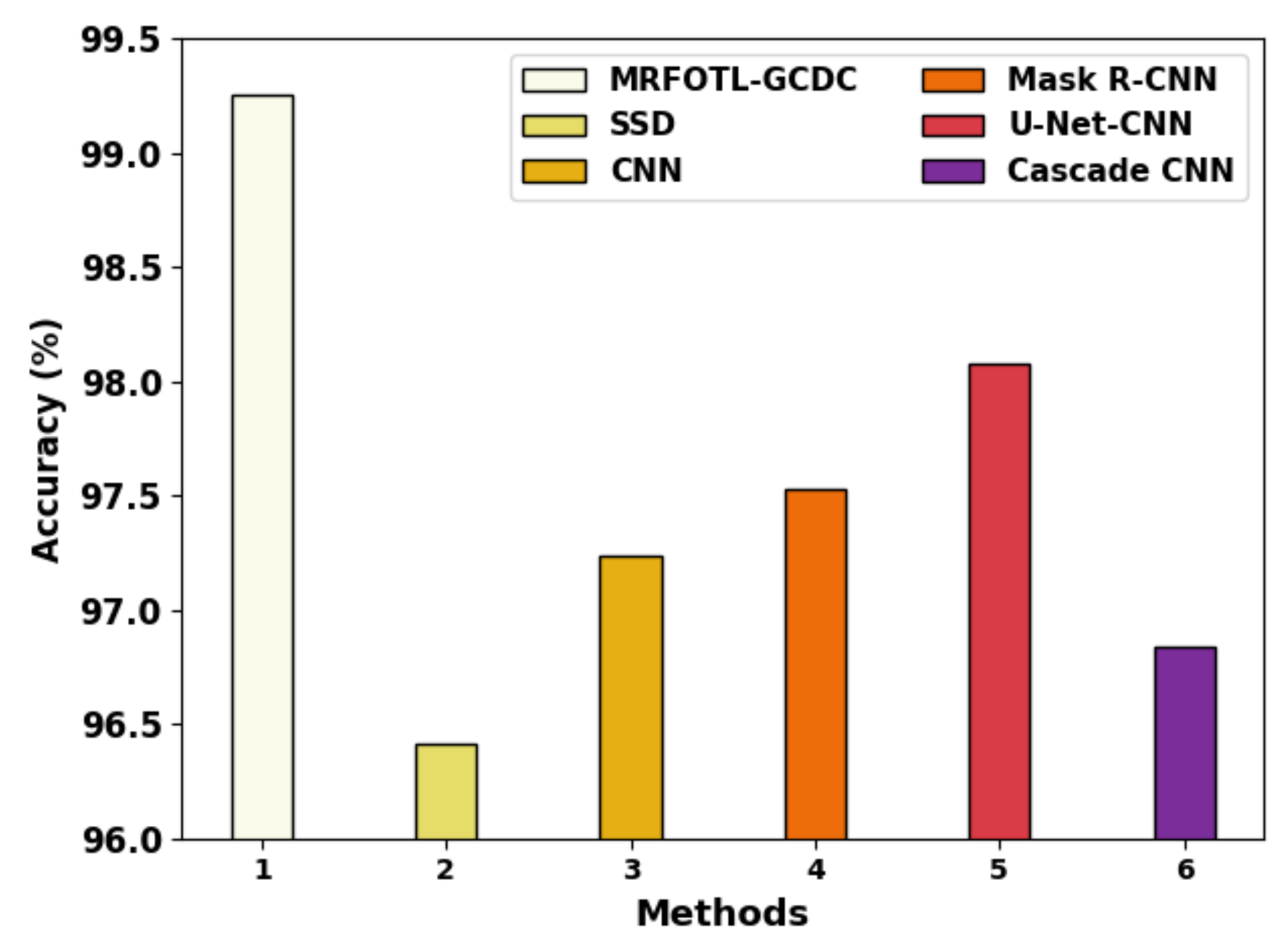
| Methods | ||||
|---|---|---|---|---|
| MRFOTL-GCDC | 99.25 | 98.63 | 98.56 | 98.60 |
| SSD | 96.41 | 96.16 | 95.61 | 96.26 |
| CNN | 97.24 | 95.38 | 98.00 | 97.91 |
| Mask R-CNN | 97.53 | 96.58 | 98.25 | 97.67 |
| U-Net-CNN | 98.08 | 97.54 | 96.99 | 95.00 |
| Cascade CNN | 96.84 | 95.95 | 98.00 | 97.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alrowais, F.; S. Alotaibi, S.; Marzouk, R.; S. Salama, A.; Rizwanullah, M.; Zamani, A.S.; Atta Abdelmageed, A.; I. Eldesouki, M. Manta Ray Foraging Optimization Transfer Learning-Based Gastric Cancer Diagnosis and Classification on Endoscopic Images. Cancers 2022, 14, 5661. https://doi.org/10.3390/cancers14225661
Alrowais F, S. Alotaibi S, Marzouk R, S. Salama A, Rizwanullah M, Zamani AS, Atta Abdelmageed A, I. Eldesouki M. Manta Ray Foraging Optimization Transfer Learning-Based Gastric Cancer Diagnosis and Classification on Endoscopic Images. Cancers. 2022; 14(22):5661. https://doi.org/10.3390/cancers14225661
Chicago/Turabian StyleAlrowais, Fadwa, Saud S. Alotaibi, Radwa Marzouk, Ahmed S. Salama, Mohammed Rizwanullah, Abu Sarwar Zamani, Amgad Atta Abdelmageed, and Mohamed I. Eldesouki. 2022. "Manta Ray Foraging Optimization Transfer Learning-Based Gastric Cancer Diagnosis and Classification on Endoscopic Images" Cancers 14, no. 22: 5661. https://doi.org/10.3390/cancers14225661
APA StyleAlrowais, F., S. Alotaibi, S., Marzouk, R., S. Salama, A., Rizwanullah, M., Zamani, A. S., Atta Abdelmageed, A., & I. Eldesouki, M. (2022). Manta Ray Foraging Optimization Transfer Learning-Based Gastric Cancer Diagnosis and Classification on Endoscopic Images. Cancers, 14(22), 5661. https://doi.org/10.3390/cancers14225661








