HER2 mRNA Levels, Estrogen Receptor Activity and Susceptibility to Trastuzumab in Primary Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Breast Cancer Samples
2.2. Immunofluorescence Analyses
2.3. Estradiol Quantification
2.4. Breast Carcinoma Cell Lines
2.5. Fluorescence In Situ Hybridization
2.6. RNA Extraction and qRT-PCR
2.7. Flow Cytometry
2.8. Western Blotting and Immunoprecipitation
2.9. Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC) Assay
2.10. In Silico Analyses
2.11. Statistical Analyses
3. Results
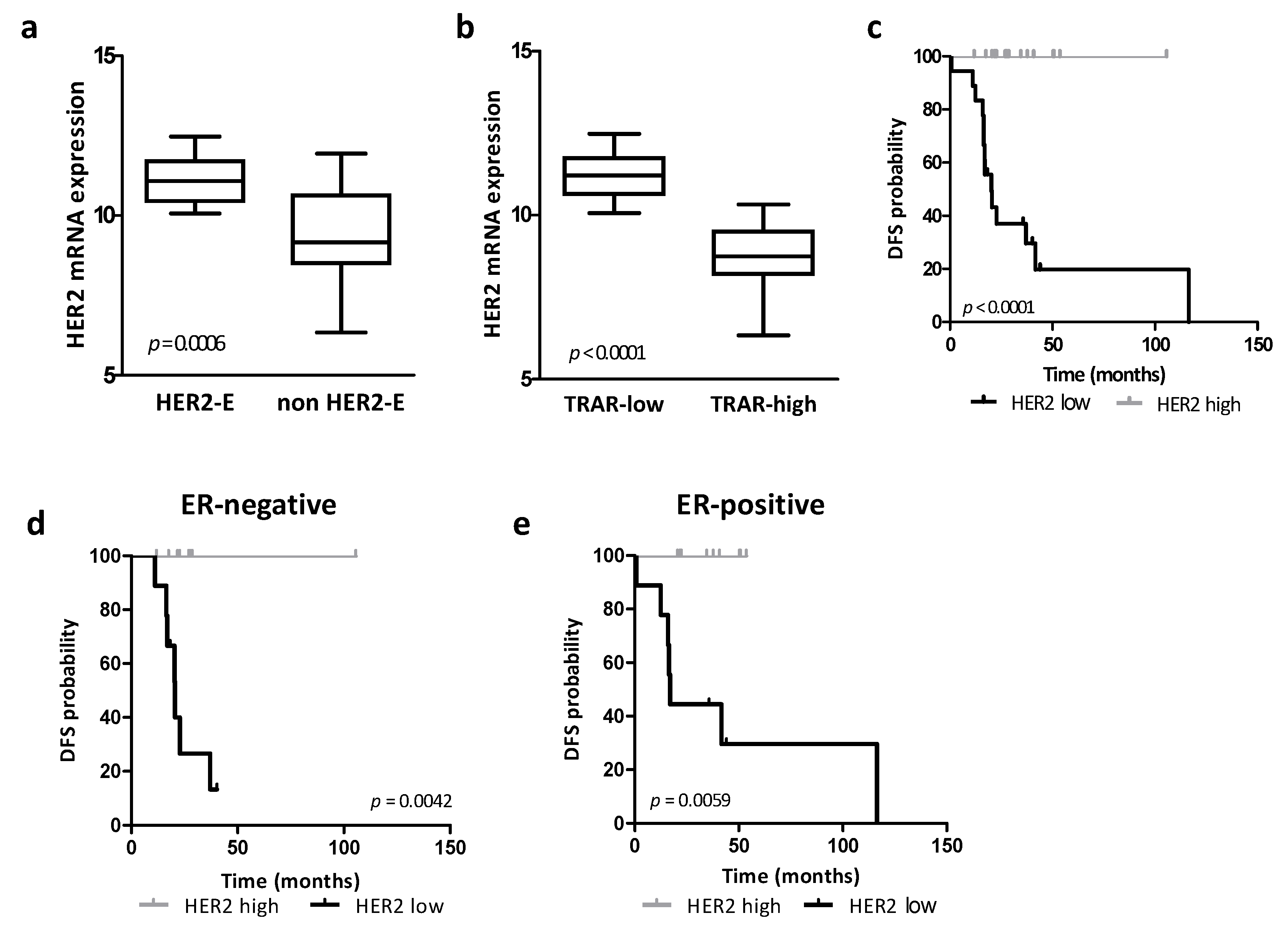
3.1. Association between HER2 Expression, HER2-Addiction and Patient Prognosis in Human Specimens
3.2. Association between HER2 Expression and HER2 Protein Levels
3.3. Dissection of the Tumor Molecular Features Associated with HER2 Levels
3.4. Regulation of HER2 Transcription by Ligand-Dependent ER Activity in BC Cell Lines
3.5. Regulation of HER2 Transcription by Ligand-Dependent ER Activity in HER2-Positive BC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ross, J.S.; Slodkowska, E.A.; Symmans, W.F.; Pusztai, L.; Ravdin, P.M.; Hortobagyi, G.N. The HER-2 Receptor and Breast Cancer: Ten Years of Targeted Anti-HER-2 Therapy and Personalized Medicine. Oncologist 2009, 14, 320–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Triulzi, T.; Bianchi, G.V.; Tagliabue, E. Predictive Biomarkers in the Treatment of HER2-Positive Breast Cancer: An Ongoing Challenge. Future Oncol. 2016, 12, 1413–1428. [Google Scholar] [CrossRef]
- di Modica, M.; Tagliabue, E.; Triulzi, T. Predicting the Efficacy of HER2-Targeted Therapies: A Look at the Host. Dis. Markers 2017, 2017, 7849108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schettini, F.; Prat, A. Dissecting the Biological Heterogeneity of HER2-Positive Breast Cancer. Breast 2021, 59, 339. [Google Scholar] [CrossRef] [PubMed]
- Schettini, F.; Pascual, T.; Chic, N.; Conte, B.; Martínez, O.; Adamo, B.; Vidal, M.; Muñoz, M.; Fernández-Martínez, A.; Griguolo, G.; et al. PAM50 HER2-Enriched Subtype and Pathological Complete Response in HER2-Positive Early Breast Cancer: A Meta-Analysis. Ann. Oncol. 2019, 30, 101965. [Google Scholar] [CrossRef]
- Spector, N.L.; Blackwell, K.L. Understanding the Mechanisms behind Trastuzumab Therapy for Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer. J. Clin. Oncol. 2009, 27, 5838–5847. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Loib, S.; Untch, M. What Is the Current Standard of Care for Anti-HER2 Neoadjuvant Therapy in Breast Cancer? Oncology 2012, 26, 20. [Google Scholar]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological Complete Response and Long-Term Clinical Benefit in Breast Cancer: The CTNeoBC Pooled Analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef] [Green Version]
- Bernard, P.S.; Parker, J.S.; Mullins, M.; Cheung, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Triulzi, T.; Cecco, L.D.; Sandri, M.; Prat, A.; Giussani, M.; Paolini, B.; Carcangiu, M.L.; Canevari, S.; Bottini, A.; Balsari, A.; et al. Whole-Transcriptome Analysis Links Trastuzumab Sensitivity of Breast Tumors to Both HER2 Dependence and Immune Cell Infiltration. Oncotarget 2015, 6, 28173. [Google Scholar] [CrossRef]
- Pogue-Geile, K.L.; Kim, C.; Jeong, J.H.; Tanaka, N.; Bandos, H.; Gavin, P.G.; Fumagalli, D.; Goldstein, L.C.; Sneige, N.; Burandt, E.; et al. Predicting Degree of Benefit From Adjuvant Trastuzumab in NSABP Trial B-31. J. Natl. Cancer Inst. 2013, 105, 1782. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Huober, J.; Loibl, S.; Prinzler, J.; Kronenwett, R.; Darb-Esfahani, S.; Brase, J.C.; Solbach, C.; Mehta, K.; Fasching, P.A.; et al. HER2 and ESR1 MRNA Expression Levels and Response to Neoadjuvant Trastuzumab plus Chemotherapy in Patients with Primary Breast Cancer. Breast Cancer Res. 2013, 15, R11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fumagalli, D.; Venet, D.; Ignatiadis, M.; Azim, H.A.; Maetens, M.; Rothé, F.; Salgado, R.; Bradbury, I.; Pusztai, L.; Harbeck, N.; et al. RNA Sequencing to Predict Response to Neoadjuvant Anti-HER2 Therapy: A Secondary Analysis of the NeoALTTO Randomized Clinical Trial. JAMA Oncol. 2017, 3, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Schneeweiss, A.; Chia, S.; Hegg, R.; Tausch, C.; Deb, R.; Ratnayake, J.; McNally, V.; Ross, G.; Kiermaier, A.; Cortés, J. Evaluating the Predictive Value of Biomarkers for Efficacy Outcomes in Response to Pertuzumab- and Trastuzumab-Based Therapy: An Exploratory Analysis of the TRYPHAENA Study. Breast Cancer Res. 2014, 16, 1–12. [Google Scholar] [CrossRef]
- Swain, S.M.; Tang, G.; Brauer, H.A.; Goerlitz, D.S.; Lucas, P.C.; Robidoux, A.; Harris, B.T.; Bandos, H.; Ren, Y.; Geyer, C.E.; et al. NSABP B-41, a Randomized Neoadjuvant Trial: Genes and Signatures Associated with Pathologic Complete Response. Clin. Cancer Res. 2020, 26, 4233. [Google Scholar] [CrossRef]
- Griguolo, G.; Brasó-Maristany, F.; González-Farré, B.; Pascual, T.; Chic, N.; Saurí, T.; Kates, R.; Gluz, O.; Martínez, D.; Paré, L.; et al. HER2 MRNA Expression and Response to Ado-Trastuzumab Emtansine (T-DM1) in HER2-Positive Breast Cancer. Cancers 2020, 12, 1902. [Google Scholar] [CrossRef]
- Carey, L.A.; Berry, D.A.; Cirrincione, C.T.; Barry, W.T.; Pitcher, B.N.; Harris, L.N.; Ollila, D.W.; Krop, I.E.; Henry, N.L.; Weckstein, D.J.; et al. Molecular Heterogeneity and Response to Neoadjuvant Human Epidermal Growth Factor Receptor 2 Targeting in CALGB 40601, a Randomized Phase III Trial of Paclitaxel Plus Trastuzumab With or Without Lapatinib. J. Clin. Oncol. 2016, 34, 542–549. [Google Scholar] [CrossRef] [Green Version]
- Bianchini, G.; Kiermaier, A.; Bianchi, G.V.; Im, Y.H.; Pienkowski, T.; Liu, M.C.; Tseng, L.M.; Dowsett, M.; Zabaglo, L.; Kirk, S.; et al. Biomarker Analysis of the NeoSphere Study: Pertuzumab, Trastuzumab, and Docetaxel versus Trastuzumab plus Docetaxel, Pertuzumab plus Trastuzumab, or Pertuzumab plus Docetaxel for the Neoadjuvant Treatment of HER2-Positive Breast Cancer. Breast Cancer Res. 2017, 19, 16. [Google Scholar] [CrossRef] [Green Version]
- Prat, A.; Pascual, T.; de Angelis, C.; Gutierrez, C.; Llombart-Cussac, A.; Wang, T.; Cortés, J.; Rexer, B.; Paré, L.; Forero, A.; et al. HER2-Enriched Subtype and HER2 Expression in HER2-Positive Breast Cancer Treated with Dual HER2 Blockade. J. Natl. Cancer Inst. 2020, 112, 46. [Google Scholar] [CrossRef]
- Baehner, F.L.; Achacoso, N.; Maddala, T.; Shak, S.; Quesenberry, C.P.; Goldstein, L.C.; Gown, A.M.; Habel, L.A. Human Epidermal Growth Factor Receptor 2 Assessment in a Case-Control Study: Comparison of Fluorescence in Situ Hybridization and Quantitative Reverse Transcription Polymerase Chain Reaction Performed by Central Laboratories. J. Clin. Oncol. 2010, 28, 4300–4306. [Google Scholar] [CrossRef]
- Viale, G.; Slaets, L.; Bogaerts, J.; Rutgers, E.; Van’t veer, L.; Piccart-gebhart, M.J.; de snoo, F.A.; Stork-sloots, L.; Russo, L.; Dell’orto, P.; et al. Editor’s Choice: High Concordance of Protein (by IHC), Gene (by FISH; HER2 Only), and Microarray Readout (by TargetPrint) of ER, PgR, and HER2: Results from the EORTC 10041/BIG 03-04 MINDACT Trial. Ann. Oncol. 2014, 25, 816. [Google Scholar] [CrossRef] [PubMed]
- Roepman, P.; Horlings, H.M.; Krijgsman, O.; Kok, M.; Bueno-de-Mesquita, J.M.; Bender, R.; Linn, S.C.; Glas, A.M.; van de Vijver, M.J. Microarray-Based Determination of Estrogen Receptor, Progesterone Receptor, and HER2 Receptor Status in Breast Cancer. Clin. Cancer Res. 2009, 15, 7003–7011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuliano, M.; Trivedi, M.V.; Schiff, R. Bidirectional Crosstalk between the Estrogen Receptor and Human Epidermal Growth Factor Receptor 2 Signaling Pathways in Breast Cancer: Molecular Basis and Clinical Implications. Breast Care 2013, 8, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venturelli, E.; Orenti, A.; Fabricio, A.S.C.; Garrone, G.; Agresti, R.; Paolini, B.; Bonini, C.; Gion, M.; Berrino, F.; Desmedt, C.; et al. Observational Study on the Prognostic Value of Testosterone and Adiposity in Postmenopausal Estrogen Receptor Positive Breast Cancer Patients. BMC Cancer 2018, 18, 651. [Google Scholar] [CrossRef] [Green Version]
- Triulzi, T.; Regondi, V.; de Cecco, L.; Cappelletti, M.R.; di Modica, M.; Paolini, B.; Lollini, P.L.; di Cosimo, S.; Sfondrini, L.; Generali, D.; et al. Early Immune Modulation by Single-Agent Trastuzumab as a Marker of Trastuzumab Benefit. Br. J. Cancer 2018, 119, 1487–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiu, K.K.; Wetterskog, D.; Mackay, A.; Natrajan, R.; Lambros, M.; Sims, D.; Bajrami, I.; Brough, R.; Frankum, J.; Sharpe, R.; et al. Integrative Molecular and Functional Profiling of HER2-Amplified Breast Cancers Identifies New Genetic Dependencies. Oncogene 2014, 33, 619–631. [Google Scholar] [CrossRef] [Green Version]
- Gasparini, P.; Bertolini, G.; Binda, M.; Magnifico, A.; Albano, L.; Tortoreto, M.; Pratesi, G.; Facchinetti, F.; Abolafio, G.; Roz, L.; et al. Molecular Cytogenetic Characterization of Stem-like Cancer Cells Isolated from Established Cell Lines. Cancer Lett. 2010, 296, 206–215. [Google Scholar] [CrossRef]
- Tagliabue, E.; Centis, F.; Campiglio, M.; Mastroianni, A.; Martignone, S.; Pellegrini, R.; Casalini, P.; Lanzi, C.; Ménard, S.; Colnaghi, M.I. Selection of Monoclonal Antibodies Which Induce Internalization and Phosphorylation of P185HER2 and Growth Inhibition of Cells with HER2/Neu Gene Amplification. Int. J. Cancer 1991, 47, 933–937. [Google Scholar] [CrossRef]
- di Modica, M.; Sfondrini, L.; Regondi, V.; Varchetta, S.; Oliviero, B.; Mariani, G.; Bianchi, G.V.; Generali, D.; Balsari, A.; Triulzi, T.; et al. Taxanes Enhance Trastuzumab-Mediated ADCC on Tumor Cells through NKG2D-Mediated NK Cell Recognition. Oncotarget 2016, 7, 255. [Google Scholar] [CrossRef] [Green Version]
- Bianchini, G.; Pusztai, L.; Karn, T.; Iwamoto, T.; Rody, A.; Kelly, C.M.; Müller, V.; Schmidt, M.; Qi, Y.; Holtrich, U.; et al. Proliferation and Estrogen Signaling Can Distinguish Patients at Risk for Early versus Late Relapse among Estrogen Receptor Positive Breast Cancers. Breast Cancer Res. 2013, 15, R86. [Google Scholar] [CrossRef] [Green Version]
- Lupien, M.; Meyer, C.A.; Bailey, S.T.; Eeckhoute, J.; Cook, J.; Westerling, T.; Zhang, X.; Carroll, J.S.; Rhodes, D.R.; Liu, X.S.; et al. Growth Factor Stimulation Induces a Distinct ER(Alpha) Cistrome Underlying Breast Cancer Endocrine Resistance. Genes Dev. 2010, 24, 2219–2227. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.J. Strategies for Subtypes--Dealing with the Diversity of Breast Cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Triulzi, T.; Bianchini, G.; di Cosimo, S.; Pienkowski, T.; Im, Y.-H.; Bianchi, G.V.; Galbardi, B.; Dugo, M.; de Cecco, L.; Tseng, L.-M.; et al. The TRAR Gene Classifier to Predict Response to Neoadjuvant Therapy in HER2-Positive and ER-Positive Breast Cancer Patients: An Explorative Analysis from the NeoSphere Trial. Mol. Oncol. 2022, 16, 2355–2366. [Google Scholar] [CrossRef] [PubMed]
- Dabbs, D.J.; Klein, M.E.; Mohsin, S.K.; Tubbs, R.R.; Shuai, Y.; Bhargava, R. High False-Negative Rate of HER2 Quantitative Reverse Transcription Polymerase Chain Reaction of the Oncotype DX Test: An Independent Quality Assurance Study. J. Clin. Oncol. 2011, 29, 4279–4285. [Google Scholar] [CrossRef]
- Joensuu, H.; Sperinde, J.; Leinonen, M.; Huang, W.; Weidler, J.; Bono, P.; Kataja, V.; Kokko, R.; Turpeenniemi-Hujanen, T.; Jyrkkiö, S.; et al. Very High Quantitative Tumor HER2 Content and Outcome in Early Breast Cancer. Ann. Oncol. 2011, 22, 2007–2013. [Google Scholar] [CrossRef]
- Filho, O.M.; Viale, G.; Stein, S.; Trippa, L.; Yardley, D.A.; Mayer, I.A.; Abramson, V.G.; Arteaga, C.L.; Spring, L.M.; Waks, A.G.; et al. Impact of HER2 Heterogeneity on Treatment Response of Early-Stage HER2-Positive Breast Cancer: Phase II Neoadjuvant Clinical Trial of T-DM1 Combined with Pertuzumab. Cancer Discov. 2021, 11, 2474. [Google Scholar] [CrossRef]
- Birnbaum, D.; Sircoulomb, F.; Imbert, J. A Reason Why the HER2 Gene Is Amplified and Not Mutated in Breast Cancer. Cancer Cell Int. 2009, 9, 5. [Google Scholar] [CrossRef] [Green Version]
- Awasthi, S.; Hamburger, A.W. Heregulin Negatively Regulates Transcription of HER2/3 Receptors via an AKT-Mediated Pathway. J. Cell. Physiol. 2014, 229, 1831. [Google Scholar] [CrossRef] [Green Version]
- Cuesta, R.; Gritsenko, M.A.; Petyuk, V.A.; Shukla, A.K.; Tsai, C.F.; Liu, T.; McDermott, J.E.; Holz, M.K. Phosphoproteome Analysis Reveals Estrogen-ER Pathway as a Modulator of MTOR Activity Via DEPTOR. Mol. Cell Proteom. 2019, 18, 1607. [Google Scholar] [CrossRef]
- Bosch, A.; Li, Z.; Bergamaschi, A.; Ellis, H.; Toska, E.; Prat, A.; Tao, J.J.; Spratt, D.E.; Viola-Villegas, N.T.; Castel, P.; et al. PI3K Inhibition Results in Enhanced Estrogen Receptor Function and Dependence in Hormone Receptor-Positive Breast Cancer. Sci. Transl. Med. 2015, 7, 283ra51. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Kusnadi, E.P.; Furic, L.; Selth, L.A. Regulation of MRNA Translation by Hormone Receptors in Breast and Prostate Cancer. Cancers 2021, 13, 3254. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.M.; Allen, P.; Gonzalez-Angulo, A.M.; Woodward, W.A.; Meric-Bernstam, F.; Buzdar, A.U.; Hunt, K.K.; Kuerer, H.M.; Litton, J.K.; Hortobagyi, G.N.; et al. Pathologic Complete Response to Neoadjuvant Chemotherapy with Trastuzumab Predicts for Improved Survival in Women with HER2-Overexpressing Breast Cancer. Ann. Oncol. 2013, 24, 1999. [Google Scholar] [CrossRef] [PubMed]
- Hamy-Petit, A.S.; Belin, L.; Bonsang-Kitzis, H.; Paquet, C.; Pierga, J.Y.; Lerebours, F.; Cottu, P.; Rouzier, R.; Savignoni, A.; Lae, M.; et al. Pathological Complete Response and Prognosis after Neoadjuvant Chemotherapy for HER2-Positive Breast Cancers before and after Trastuzumab Era: Results from a Real-Life Cohort. Br. J. Cancer 2015, 114, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yang, H.; Yu, X.; Qin, J.-J. Drug-Resistant HER2-Positive Breast Cancer: Molecular Mechanisms and Overcoming Strategies. Front. Pharm. 2022, 13, 3952. [Google Scholar] [CrossRef]
- Hurvitz, S.A.; Andre, F.; Jiang, Z.; Shao, Z.; Mano, M.S.; Neciosup, S.P.; Tseng, L.M.; Zhang, Q.; Shen, K.; Liu, D.; et al. Combination of Everolimus with Trastuzumab plus Paclitaxel as First-Line Treatment for Patients with HER2-Positive Advanced Breast Cancer (BOLERO-1): A Phase 3, Randomised, Double-Blind, Multicentre Trial. Lancet Oncol. 2015, 16, 816–829. [Google Scholar] [CrossRef] [Green Version]
- André, F.; O’Regan, R.; Ozguroglu, M.; Toi, M.; Xu, B.; Jerusalem, G.; Masuda, N.; Wilks, S.; Arena, F.; Isaacs, C.; et al. Everolimus for Women with Trastuzumab-Resistant, HER2-Positive, Advanced Breast Cancer (BOLERO-3): A Randomised, Double-Blind, Placebo-Controlled Phase 3 Trial. Lancet Oncol. 2014, 15, 580–591. [Google Scholar] [CrossRef]
- McAndrew, N.P.; Finn, R.S. Management of ER Positive Metastatic Breast Cancer. Semin. Oncol. 2020, 47, 270–277. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Triulzi, T.; Regondi, V.; Venturelli, E.; Gasparini, P.; Ghirelli, C.; Groppelli, J.; Di Modica, M.; Bianchi, F.; De Cecco, L.; Sfondrini, L.; et al. HER2 mRNA Levels, Estrogen Receptor Activity and Susceptibility to Trastuzumab in Primary Breast Cancer. Cancers 2022, 14, 5650. https://doi.org/10.3390/cancers14225650
Triulzi T, Regondi V, Venturelli E, Gasparini P, Ghirelli C, Groppelli J, Di Modica M, Bianchi F, De Cecco L, Sfondrini L, et al. HER2 mRNA Levels, Estrogen Receptor Activity and Susceptibility to Trastuzumab in Primary Breast Cancer. Cancers. 2022; 14(22):5650. https://doi.org/10.3390/cancers14225650
Chicago/Turabian StyleTriulzi, Tiziana, Viola Regondi, Elisabetta Venturelli, Patrizia Gasparini, Cristina Ghirelli, Jessica Groppelli, Martina Di Modica, Francesca Bianchi, Loris De Cecco, Lucia Sfondrini, and et al. 2022. "HER2 mRNA Levels, Estrogen Receptor Activity and Susceptibility to Trastuzumab in Primary Breast Cancer" Cancers 14, no. 22: 5650. https://doi.org/10.3390/cancers14225650
APA StyleTriulzi, T., Regondi, V., Venturelli, E., Gasparini, P., Ghirelli, C., Groppelli, J., Di Modica, M., Bianchi, F., De Cecco, L., Sfondrini, L., & Tagliabue, E. (2022). HER2 mRNA Levels, Estrogen Receptor Activity and Susceptibility to Trastuzumab in Primary Breast Cancer. Cancers, 14(22), 5650. https://doi.org/10.3390/cancers14225650









