Longitudinal Modulation of Loco-Regional Immunity in Ovarian Cancer Patients Receiving Intraperitoneal Chemotherapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort and Sample Collection
2.2. NanoString
2.3. ELISA
2.4. Immunohistochemistry (IHC)
2.5. Meso Scale Discovery (MSD)
2.6. Statistics and Bioinformatics
3. Results
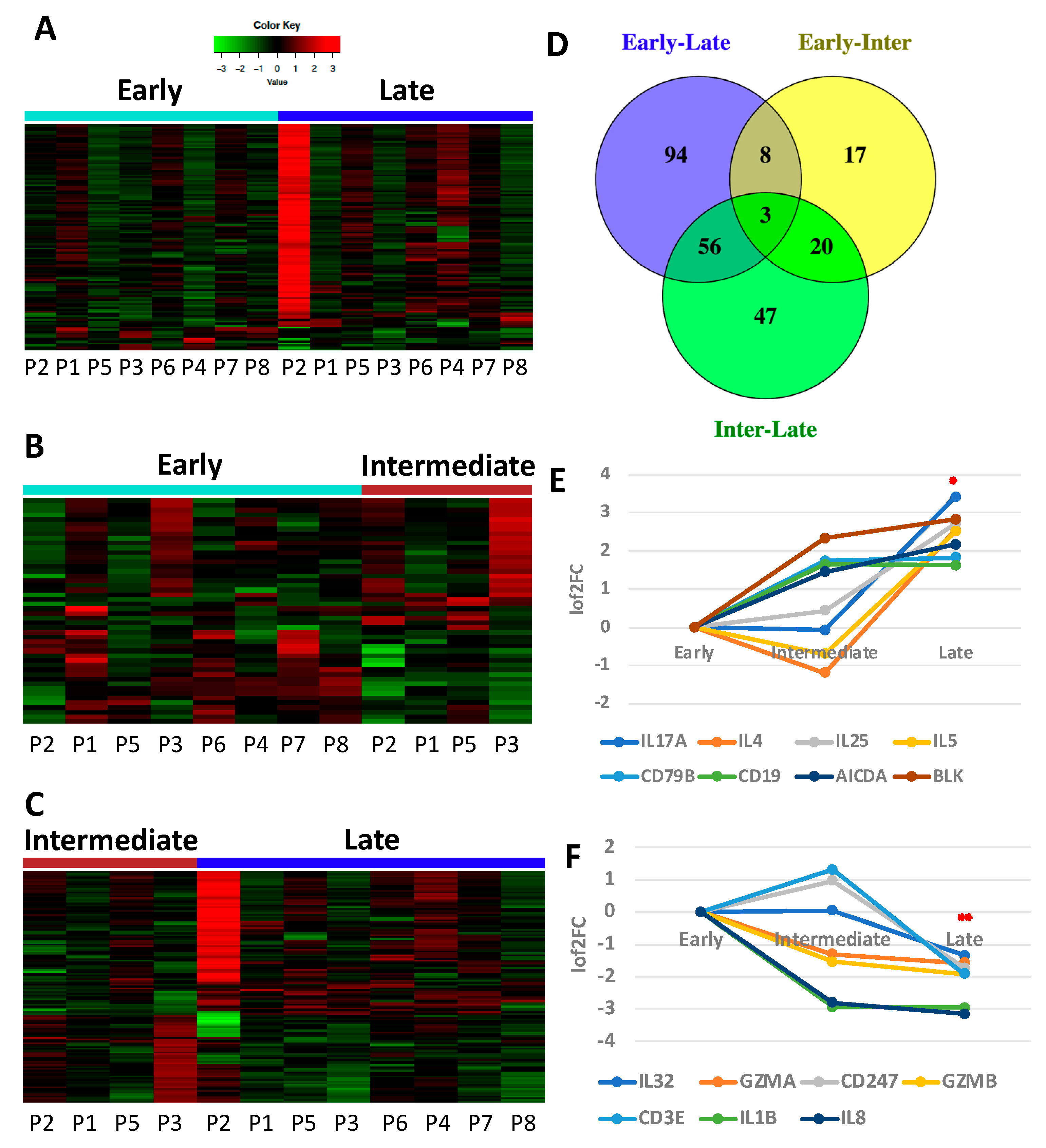
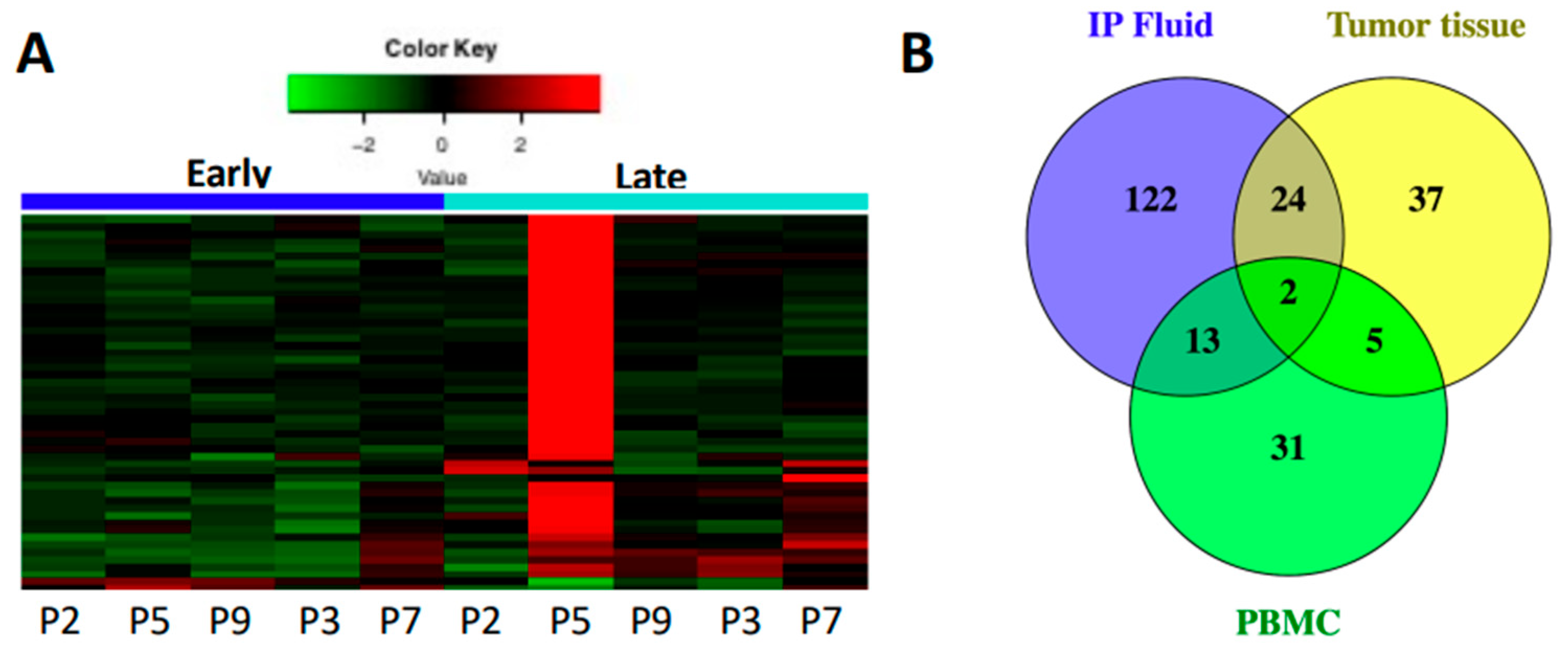
3.1. Immune Gene Expression Analyses Capture Chemotherapy-Induced Dynamic Changes in IP Fluid
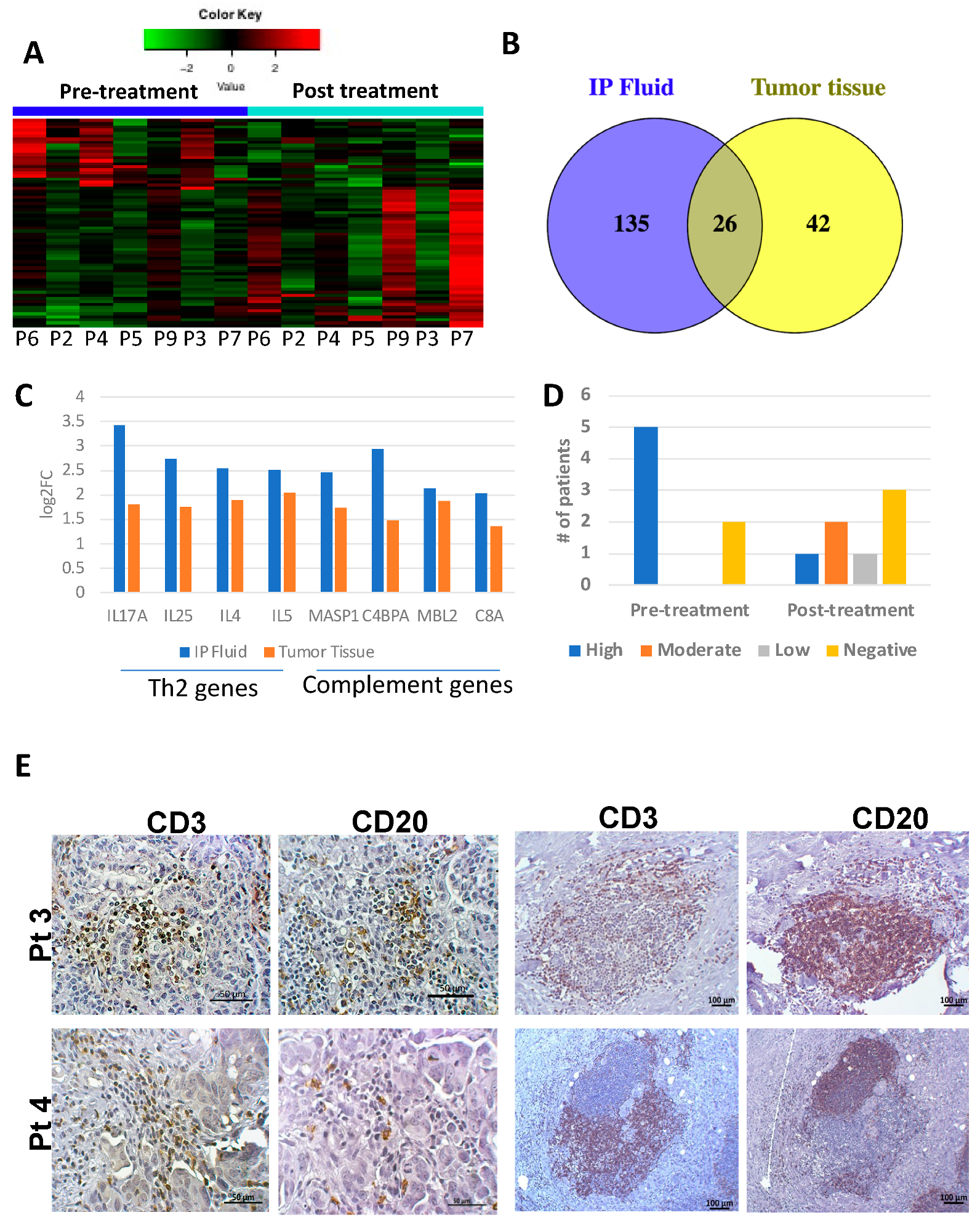
3.2. Tumor and IP Fluid Show Concordant Gene Expression Profiles of Th2 Genes
3.3. Tumor Infiltrating Immune Cell Populations Show Dampening of CD8+ T Cells with Chemotherapy Treatment
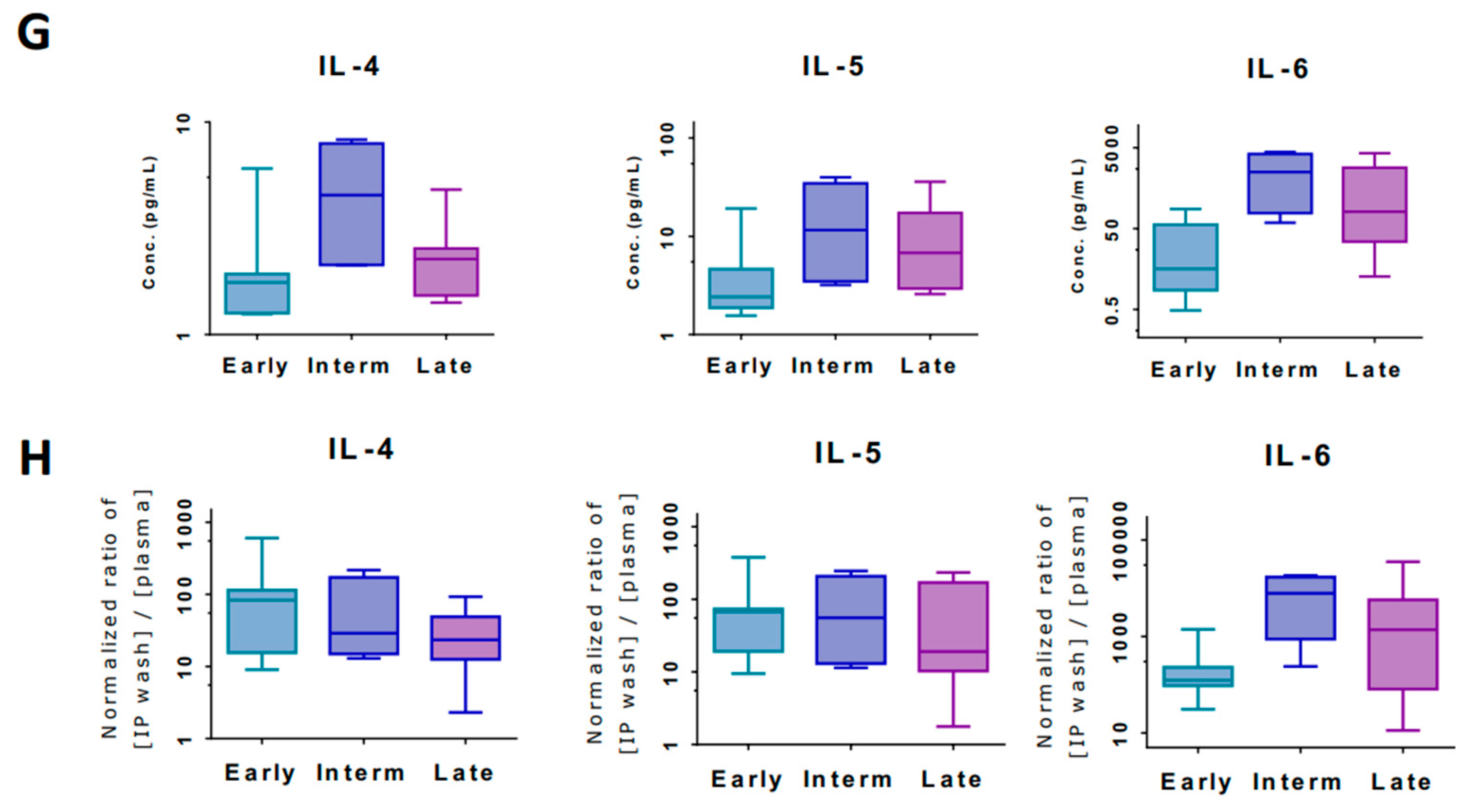
3.4. Markers of Inflammation Differ in Systemic Circulation versus Loco-Regional Environment
3.5. Secondary Patient Cohort Shows Similar Immune Gene Upregulation and B Cell Pathway Activation in Response to Chemotherapy
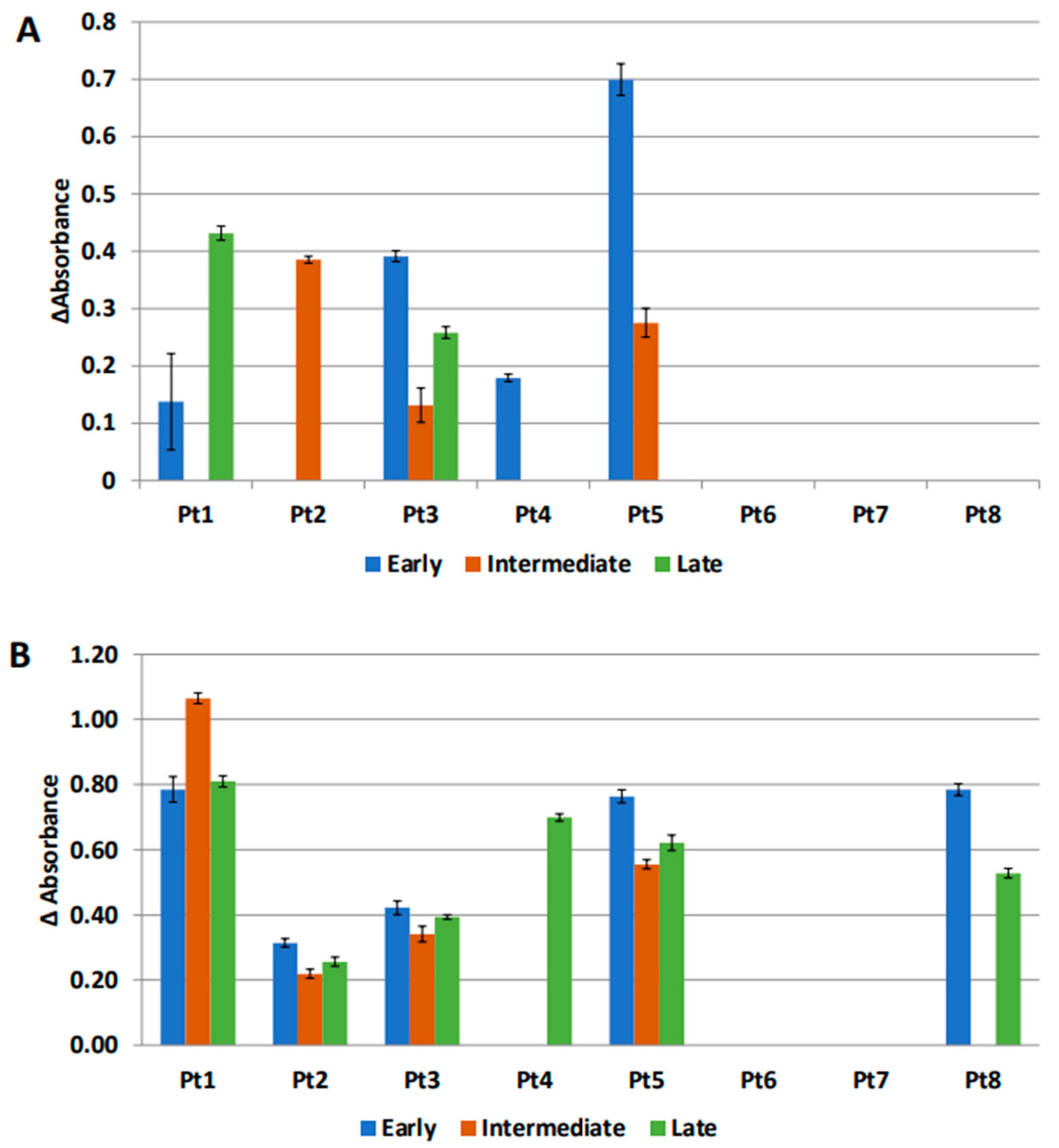
3.6. Anti-MUC1 Antibodies Are Present in the IP Fluid
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Cannistra, S.A. Cancer of the ovary. N. Engl. J. Med. 2004, 351, 2519–2529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Chen, R.; Bai, Y.; Lu, X. The prognostic value of tumor-infiltrating T lymphocytes in ovarian cancer. Oncotarget 2017, 8, 15621–15631. [Google Scholar] [CrossRef]
- Mariya, T.; Hirohashi, Y.; Torigoe, T.; Asano, T.; Kuroda, T.; Yasuda, K.; Mizuuchi, M.; Sonoda, T.; Saito, T.; Sato, N. Prognostic impact of human leukocyte antigen class I expression and association of platinum resistance with immunologic profiles in epithelial ovarian cancer. Cancer Immunol. Res. 2014, 2, 1220–1229. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef]
- Hamanishi, J.; Mandai, M.; Iwasaki, M.; Okazaki, T.; Tanaka, Y.; Yamaguchi, K.; Higuchi, T.; Yagi, H.; Takakura, K.; Minato, N.; et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 3360–3365. [Google Scholar] [CrossRef]
- Schachter, J.; Ribas, A.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus ipilimumab for advanced melanoma: Final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 2017, 390, 1853–1862. [Google Scholar] [CrossRef]
- Bellmunt, J.; de Wit, R.; Vaughn, D.J.; Fradet, Y.; Lee, J.L.; Fong, L.; Vogelzang, N.J.; Climent, M.A.; Petrylak, D.P.; Choueiri, T.K.; et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N. Engl. J. Med. 2017, 376, 1015–1026. [Google Scholar] [CrossRef]
- Reck, M.; Rodriguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csoszi, T.; Fulop, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef]
- Hamanishi, J.; Mandai, M.; Konishi, I. Immune checkpoint inhibition in ovarian cancer. Int. Immunol. 2016, 28, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Grabosch, S.; Tseng, G.; Edwards, R.P.; Lankes, H.A.; Moore, K.; Odunsi, K.; Vlad, A.; Ma, T.; Strange, M.; Brozick, J.; et al. Multiplex profiling identifies distinct local and systemic alterations during intraperitoneal chemotherapy for ovarian cancer: An NRG Oncology/Gynecologic Oncology Group Study. Gynecol. Oncol. 2017, 146, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Vlad, A.M.; Budiu, R.A.; Lenzner, D.E.; Wang, Y.; Thaller, J.A.; Colonello, K.; Crowley-Nowick, P.A.; Kelley, J.L.; Price, F.V.; Edwards, R.P. A phase II trial of intraperitoneal interleukin-2 in patients with platinum-resistant or platinum-refractory ovarian cancer. Cancer Immunol. Immunother. 2010, 59, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Lenzi, R.; Edwards, R.; June, C.; Seiden, M.V.; Garcia, M.E.; Rosenblum, M.; Freedman, R.S. Phase II study of intraperitoneal recombinant interleukin-12 (rhIL-12) in patients with peritoneal carcinomatosis (residual disease < 1 cm) associated with ovarian cancer or primary peritoneal carcinoma. J. Transl. Med. 2007, 5, 66. [Google Scholar] [CrossRef]
- Orr, B.; Mahdi, H.; Fang, Y.; Strange, M.; Uygun, I.; Rana, M.; Zhang, L.; Suarez Mora, A.; Pusateri, A.; Elishaev, E.; et al. Phase I trial combining chemokine-targeting with loco-regional chemo-immunotherapy for recurrent, platinum-sensitive ovarian cancer shows induction of CXCR3 ligands and markers of type 1 immunity. Clin. Cancer Res. 2022, 28, 2038–2049. [Google Scholar] [CrossRef]
- Budiu, R.A.; Mantia-Smaldone, G.; Elishaev, E.; Chu, T.; Thaller, J.; McCabe, K.; Lenzner, D.; Edwards, R.P.; Vlad, A.M. Soluble MUC1 and serum MUC1-specific antibodies are potential prognostic biomarkers for platinum-resistant ovarian cancer. Cancer Immunol. Immunother. 2011, 60, 975–984. [Google Scholar] [CrossRef]
- Ovarian Tumor Tissue Analysis (OTTA) Consortium; Goode, E.L.; Block, M.S.; Kalli, K.R.; Vierkant, R.A.; Chen, W.; Fogarty, Z.C.; Gentry-Maharaj, A.; Toloczko, A.; Hein, A.; et al. Dose-Response Association of CD8+ Tumor-Infiltrating Lymphocytes and Survival Time in High-Grade Serous Ovarian Cancer. JAMA Oncol. 2017, 3, e173290. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams (2007–2015). Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 4 October 2020).
- Goc, J.; Fridman, W.H.; Sautes-Fridman, C.; Dieu-Nosjean, M.C. Characteristics of tertiary lymphoid structures in primary cancers. Oncoimmunology 2013, 2, e26836. [Google Scholar] [CrossRef]
- Wang, L.; Ma, J.; Liu, F.; Yu, Q.; Chu, G.; Perkins, A.C.; Li, Y. Expression of MUC1 in primary and metastatic human epithelial ovarian cancer and its therapeutic significance. Gynecol. Oncol. 2007, 105, 695–702. [Google Scholar] [CrossRef]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Bamias, A.; Tsiatas, M.L.; Kafantari, E.; Liakou, C.; Rodolakis, A.; Voulgaris, Z.; Vlahos, G.; Papageorgiou, T.; Tsitsilonis, O.; Bamia, C.; et al. Significant differences of lymphocytes isolated from ascites of patients with ovarian cancer compared to blood and tumor lymphocytes. Association of CD3+CD56+ cells with platinum resistance. Gynecol. Oncol. 2007, 106, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Hato, S.V.; Khong, A.; de Vries, I.J.; Lesterhuis, W.J. Molecular pathways: The immunogenic effects of platinum-based chemotherapeutics. Clin. Cancer Res. 2014, 20, 2831–2837. [Google Scholar] [CrossRef] [PubMed]
- Grabosch, S.; Bulatovic, M.; Zeng, F.; Ma, T.; Zhang, L.; Ross, M.; Brozick, J.; Fang, Y.; Tseng, G.; Kim, E.; et al. Cisplatin-induced immune modulation in ovarian cancer mouse models with distinct inflammation profiles. Oncogene 2019, 38, 2380–2393. [Google Scholar] [CrossRef] [PubMed]
- Mesnage, S.J.L.; Auguste, A.; Genestie, C.; Dunant, A.; Pain, E.; Drusch, F.; Gouy, S.; Morice, P.; Bentivegna, E.; Lhomme, C.; et al. Neoadjuvant chemotherapy (NACT) increases immune infiltration and programmed death-ligand 1 (PD-L1) expression in epithelial ovarian cancer (EOC). Ann. Oncol. 2017, 28, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Polcher, M.; Braun, M.; Friedrichs, N.; Rudlowski, C.; Bercht, E.; Fimmers, R.; Sauerwald, A.; Keyver-Paik, M.D.; Kubler, K.; Buttner, R.; et al. Foxp3(+) cell infiltration and granzyme B(+)/Foxp3(+) cell ratio are associated with outcome in neoadjuvant chemotherapy-treated ovarian carcinoma. Cancer Immunol. Immunother. 2010, 59, 909–919. [Google Scholar] [CrossRef]
- Lo, C.S.; Sanii, S.; Kroeger, D.R.; Milne, K.; Talhouk, A.; Chiu, D.S.; Rahimi, K.; Shaw, P.A.; Clarke, B.A.; Nelson, B.H. Neoadjuvant Chemotherapy of Ovarian Cancer Results in Three Patterns of Tumor-Infiltrating Lymphocyte Response with Distinct Implications for Immunotherapy. Clin. Cancer Res. 2017, 23, 925–934. [Google Scholar] [CrossRef]
- Hiltbold, E.M.; Vlad, A.M.; Ciborowski, P.; Watkins, S.C.; Finn, O.J. The mechanism of unresponsiveness to circulating tumor antigen MUC1 is a block in intracellular sorting and processing by dendritic cells. J. Immunol. 2000, 165, 3730–3741. [Google Scholar] [CrossRef]
- Quartier, P.; Potter, P.K.; Ehrenstein, M.R.; Walport, M.J.; Botto, M. Predominant role of IgM-dependent activation of the classical pathway in the clearance of dying cells by murine bone marrow-derived macrophages in vitro. Eur. J. Immunol. 2005, 35, 252–260. [Google Scholar] [CrossRef]
- Suryawanshi, S.; Huang, X.; Elishaev, E.; Budiu, R.A.; Zhang, L.; Kim, S.; Donnellan, N.; Mantia-Smaldone, G.; Ma, T.; Tseng, G.; et al. Complement pathway is frequently altered in endometriosis and endometriosis-associated ovarian cancer. Clin. Cancer Res. 2014, 20, 6163–6174. [Google Scholar] [CrossRef]
- Senent, Y.; Ajona, D.; González-Martín, A.; Pio, R.; Tavira, B. The Complement System in Ovarian Cancer: An Underexplored Old Path. Cancers 2021, 13, 3806. [Google Scholar] [CrossRef] [PubMed]
- Afshar-Kharghan, V. The role of the complement system in cancer. J. Clin. Investig. 2017, 127, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Montfort, A.; Pearce, O.; Maniati, E.; Vincent, B.G.; Bixby, L.; Bohm, S.; Dowe, T.; Wilkes, E.H.; Chakravarty, P.; Thompson, R.; et al. A Strong B-cell Response Is Part of the Immune Landscape in Human High-Grade Serous Ovarian Metastases. Clin. Cancer Res. 2017, 23, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, R.; Lauss, M.; Sanna, A.; Donia, M.; Skaarup Larsen, M.; Mitra, S.; Johansson, I.; Phung, B.; Harbst, K.; Vallon-Christersson, J.; et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020, 577, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.S.; Sahota, R.A.; Milne, K.; Kost, S.E.; Nesslinger, N.J.; Watson, P.H.; Nelson, B.H. CD20+ tumor-infiltrating lymphocytes have an atypical CD27-memory phenotype and together with CD8+ T cells promote favorable prognosis in ovarian cancer. Clin. Cancer Res. 2012, 18, 3281–3292. [Google Scholar] [CrossRef] [PubMed]
- Milne, K.; Kobel, M.; Kalloger, S.E.; Barnes, R.O.; Gao, D.; Gilks, C.B.; Watson, P.H.; Nelson, B.H. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS ONE 2009, 4, e6412. [Google Scholar] [CrossRef]
- Helmink, B.A.; Reddy, S.M.; Gao, J.; Zhang, S.; Basar, R.; Thakur, R.; Yizhak, K.; Sade-Feldman, M.; Blando, J.; Han, G.; et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020, 577, 549–555. [Google Scholar] [CrossRef]
- Tsou, P.; Katayama, H.; Ostrin, E.J.; Hanash, S.M. The Emerging Role of B Cells in Tumor Immunity. Cancer Res. 2016, 76, 5597–5601. [Google Scholar] [CrossRef]
- Storkus, W.J.; Maurer, D.; Lin, Y.; Ding, F.; Bose, A.; Lowe, D.; Rose, A.; DeMark, M.; Karapetyan, L.; Taylor, J.L.; et al. Dendritic cell vaccines targeting tumor blood vessel antigens in combination with dasatinib induce therapeutic immune responses in patients with checkpoint-refractory advanced melanoma. J. Immunother. Cancer 2021, 9, e003675. [Google Scholar] [CrossRef]
- Waqar, S.N.; Boehmer, L.; Morgensztern, D.; Wang-Gillam, A.; Sorscher, S.; Lawrence, S.; Gao, F.; Guebert, K.; Williams, K.; Govindan, R. Immunogenicity of Influenza Vaccination in Patients With Cancer. Am. J. Clin. Oncol. 2018, 41, 248–253. [Google Scholar] [CrossRef]
- Haidar, G.; Agha, M.; Bilderback, A.; Lukanski, A.; Linstrum, K.; Troyan, R.; Rothenberger, S.; McMahon, D.K.; Crandall, M.D.; Sobolewksi, M.D.; et al. Prospective evaluation of COVID-19 vaccine responses across a broad spectrum of immunocompromising conditions: The COVICS study. Clin. Infect. Dis. 2022, 75, e630–e644. [Google Scholar] [CrossRef] [PubMed]
- Braly, P.; Nicodemus, C.F.; Chu, C.; Collins, Y.; Edwards, R.; Gordon, A.; McGuire, W.; Schoonmaker, C.; Whiteside, T.; Smith, L.M.; et al. The Immune adjuvant properties of front-line carboplatin-paclitaxel: A randomized phase 2 study of alternative schedules of intravenous oregovomab chemoimmunotherapy in advanced ovarian cancer. J. Immunother. 2009, 32, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Berek, J.; Taylor, P.; McGuire, W.; Smith, L.M.; Schultes, B.; Nicodemus, C.F. Oregovomab maintenance monoimmunotherapy does not improve outcomes in advanced ovarian cancer. J. Clin. Oncol. 2009, 27, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.M.; Butterfield, L.H. Dendritic Cell-Based Cancer Vaccines. J. Immunol. 2018, 200, 443–449. [Google Scholar] [CrossRef]
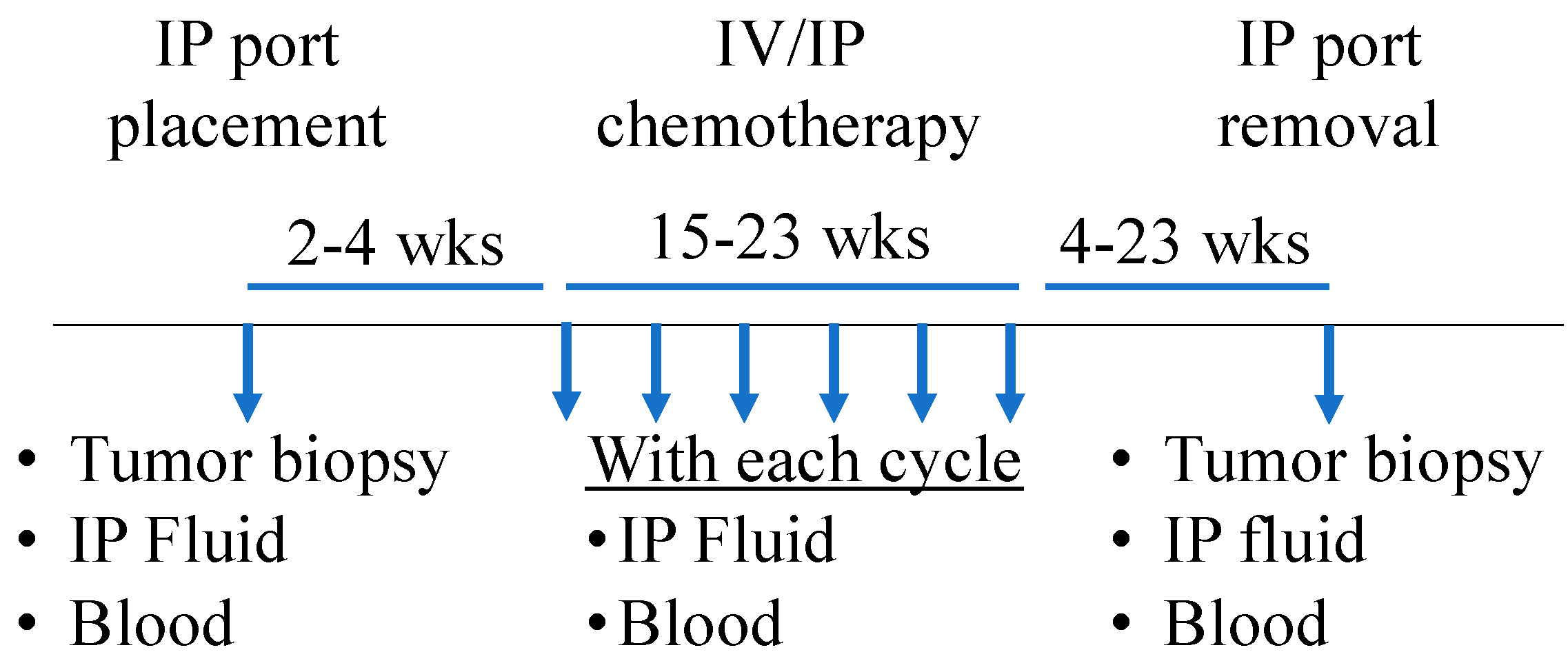

| Patients (N = 9) | |
|---|---|
| Median Age (range) | 52 (25–67) |
| Stage | |
| I | 1 (11%) |
| II | 1 (11%) |
| III | 7 (78%) |
| IV | 0 (0%) |
| Grade | |
| 1 | 2 (22%) |
| 2 | 0 (0%) |
| 3 | 7 (78%) |
| Histology | |
| Serous | 7 (78%) |
| Endometrioid | 1 (11%) |
| Clear Cell | 1 (11%) |
| Line of Therapy | |
| 1 | 5 (56%) |
| 2 | 2 (22%) |
| >2 | 2 (22%) |
| Platinum Sensitive | |
| Yes | 7 (78%) |
| No | 2 (22%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suarez Mora, A.; Strange, M.; Fang, Y.; Uygun, I.; Zhang, L.; Tseng, G.C.; Kalinski, P.; Edwards, R.P.; Vlad, A.M. Longitudinal Modulation of Loco-Regional Immunity in Ovarian Cancer Patients Receiving Intraperitoneal Chemotherapy. Cancers 2022, 14, 5647. https://doi.org/10.3390/cancers14225647
Suarez Mora A, Strange M, Fang Y, Uygun I, Zhang L, Tseng GC, Kalinski P, Edwards RP, Vlad AM. Longitudinal Modulation of Loco-Regional Immunity in Ovarian Cancer Patients Receiving Intraperitoneal Chemotherapy. Cancers. 2022; 14(22):5647. https://doi.org/10.3390/cancers14225647
Chicago/Turabian StyleSuarez Mora, Adria, Mary Strange, Yusi Fang, Ibrahim Uygun, Lixin Zhang, George C. Tseng, Pawel Kalinski, Robert P. Edwards, and Anda M. Vlad. 2022. "Longitudinal Modulation of Loco-Regional Immunity in Ovarian Cancer Patients Receiving Intraperitoneal Chemotherapy" Cancers 14, no. 22: 5647. https://doi.org/10.3390/cancers14225647
APA StyleSuarez Mora, A., Strange, M., Fang, Y., Uygun, I., Zhang, L., Tseng, G. C., Kalinski, P., Edwards, R. P., & Vlad, A. M. (2022). Longitudinal Modulation of Loco-Regional Immunity in Ovarian Cancer Patients Receiving Intraperitoneal Chemotherapy. Cancers, 14(22), 5647. https://doi.org/10.3390/cancers14225647







