Quantitative Relaxometry Metrics for Brain Metastases Compared to Normal Tissues: A Pilot MR Fingerprinting Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Phantom
2.2. Patient Cohort
2.3. MRI Data Acquisition
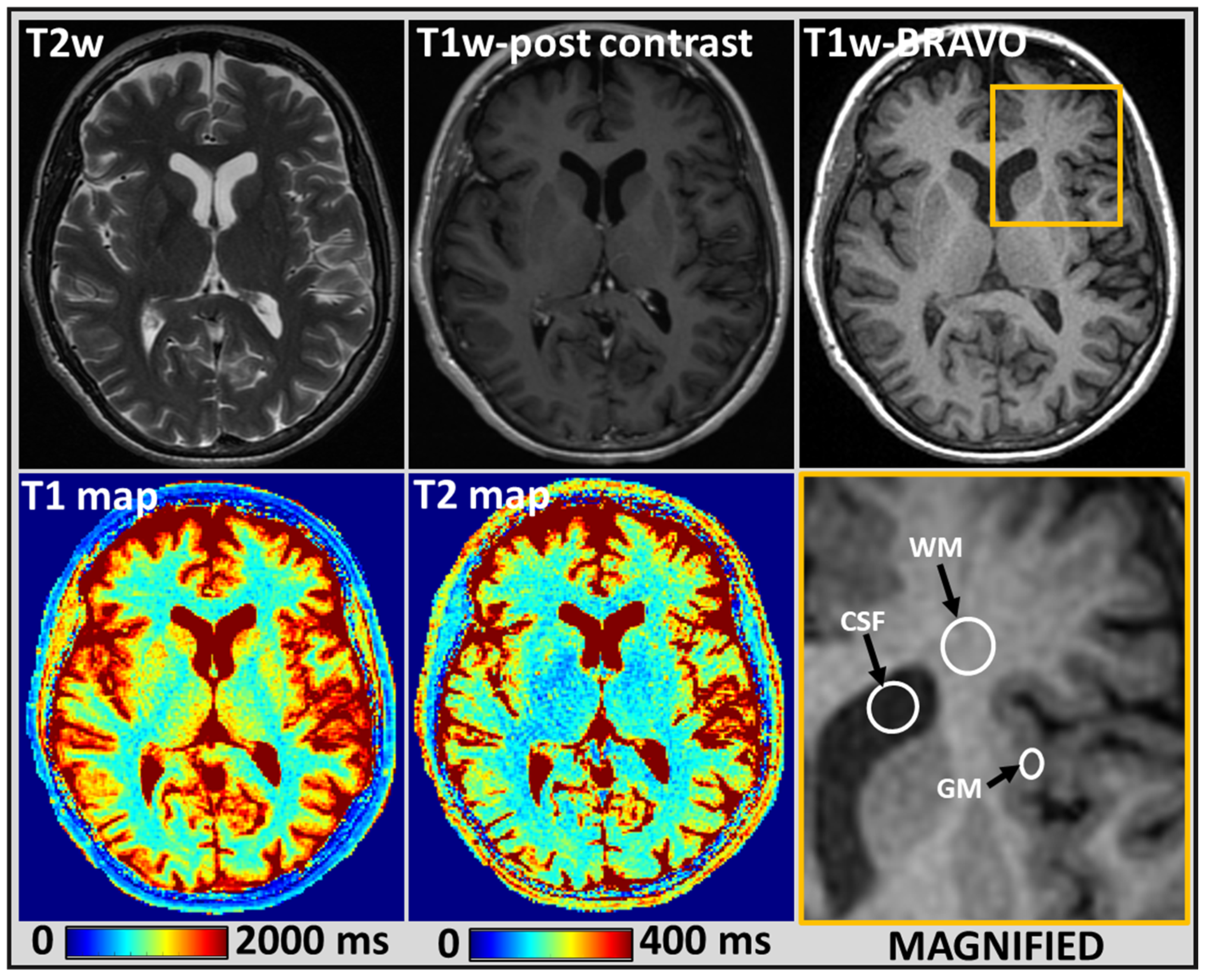
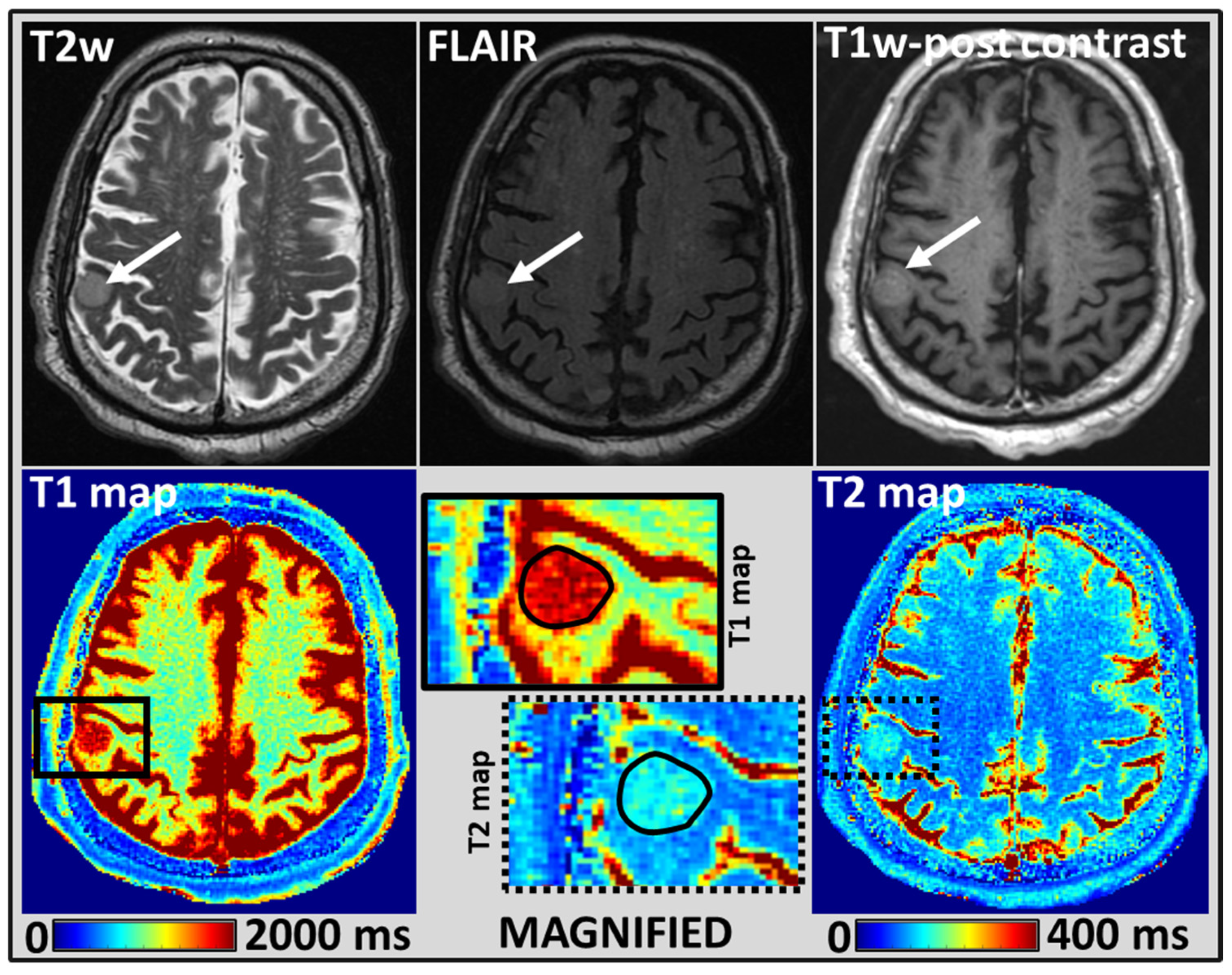
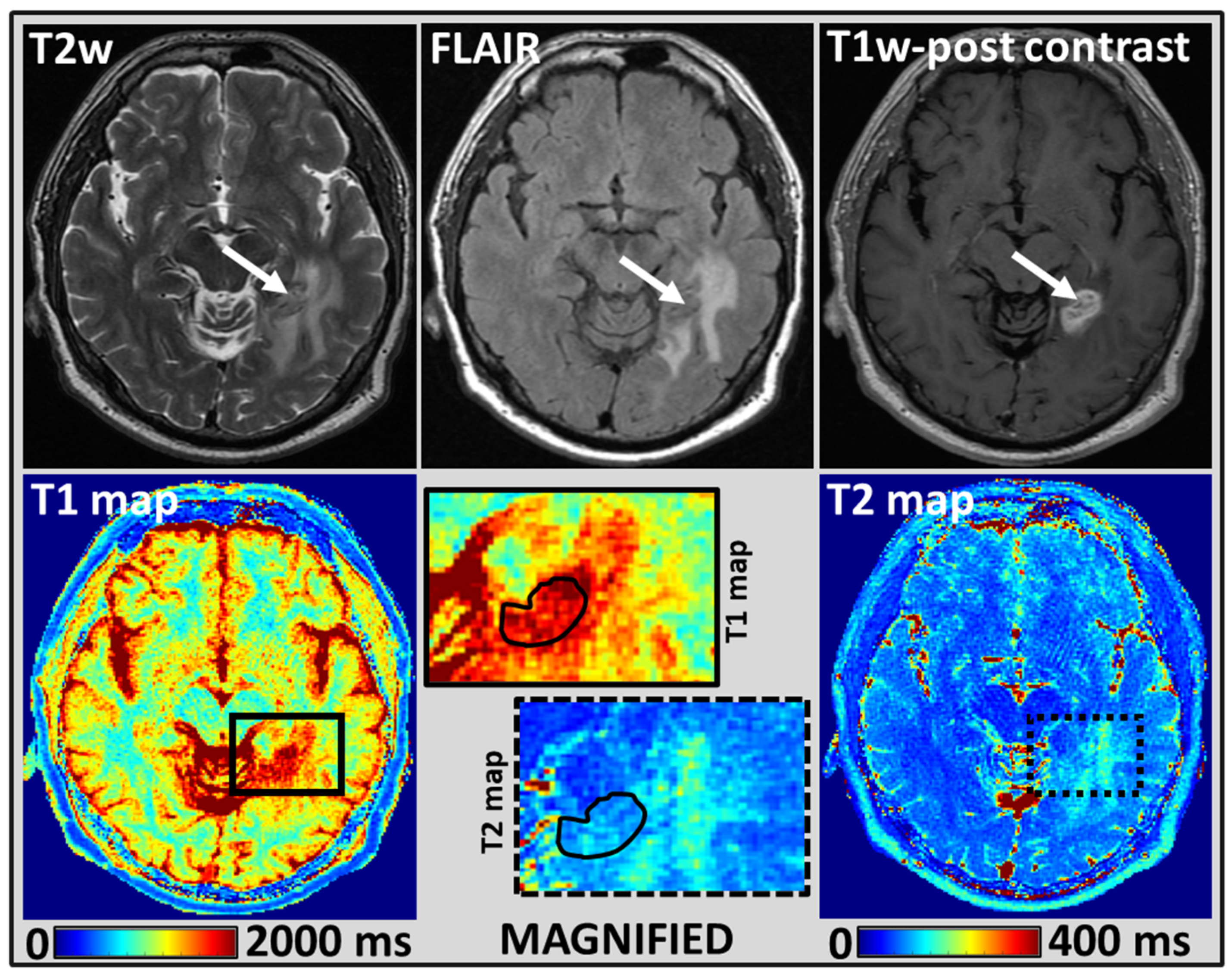
2.4. MRI Tumor Regions of Interest Analysis
2.5. Statistical Analysis
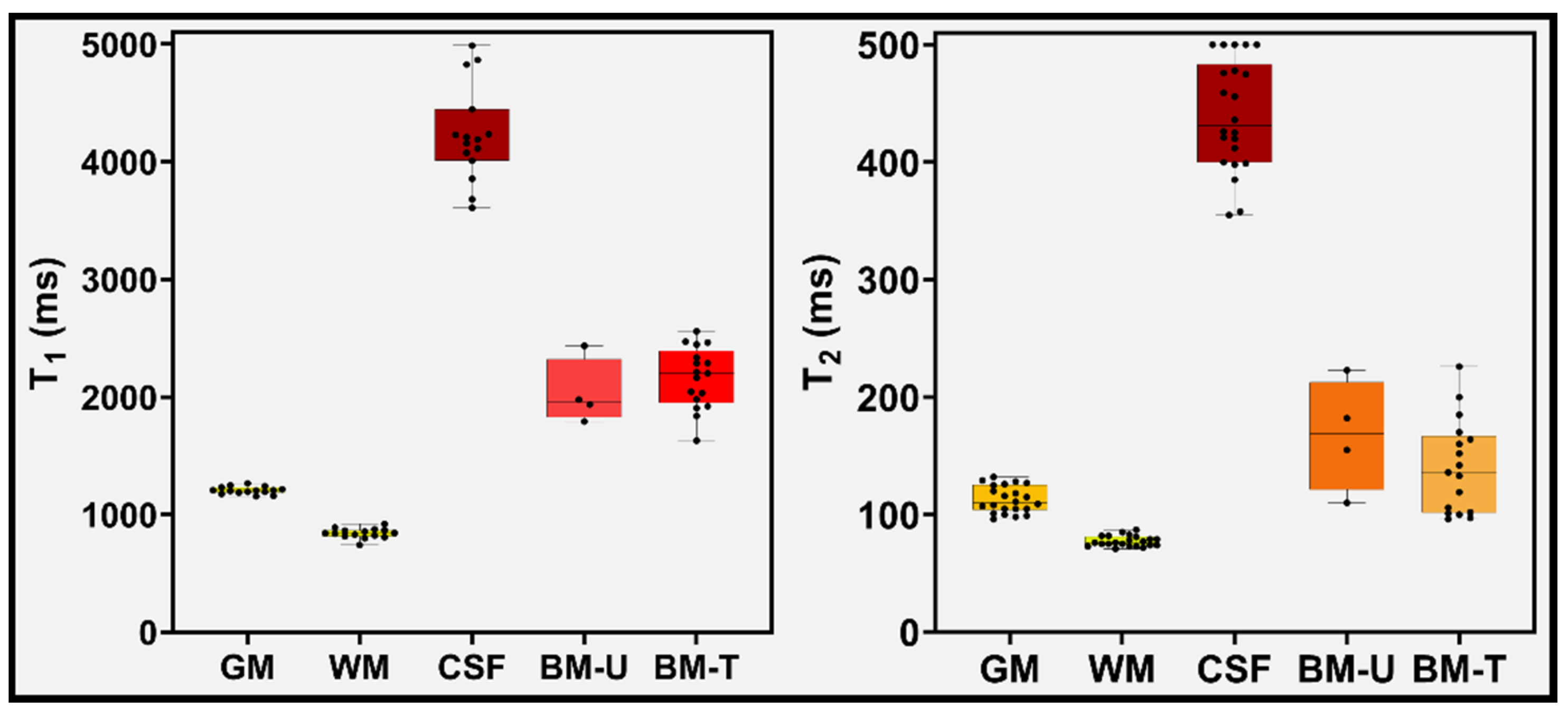
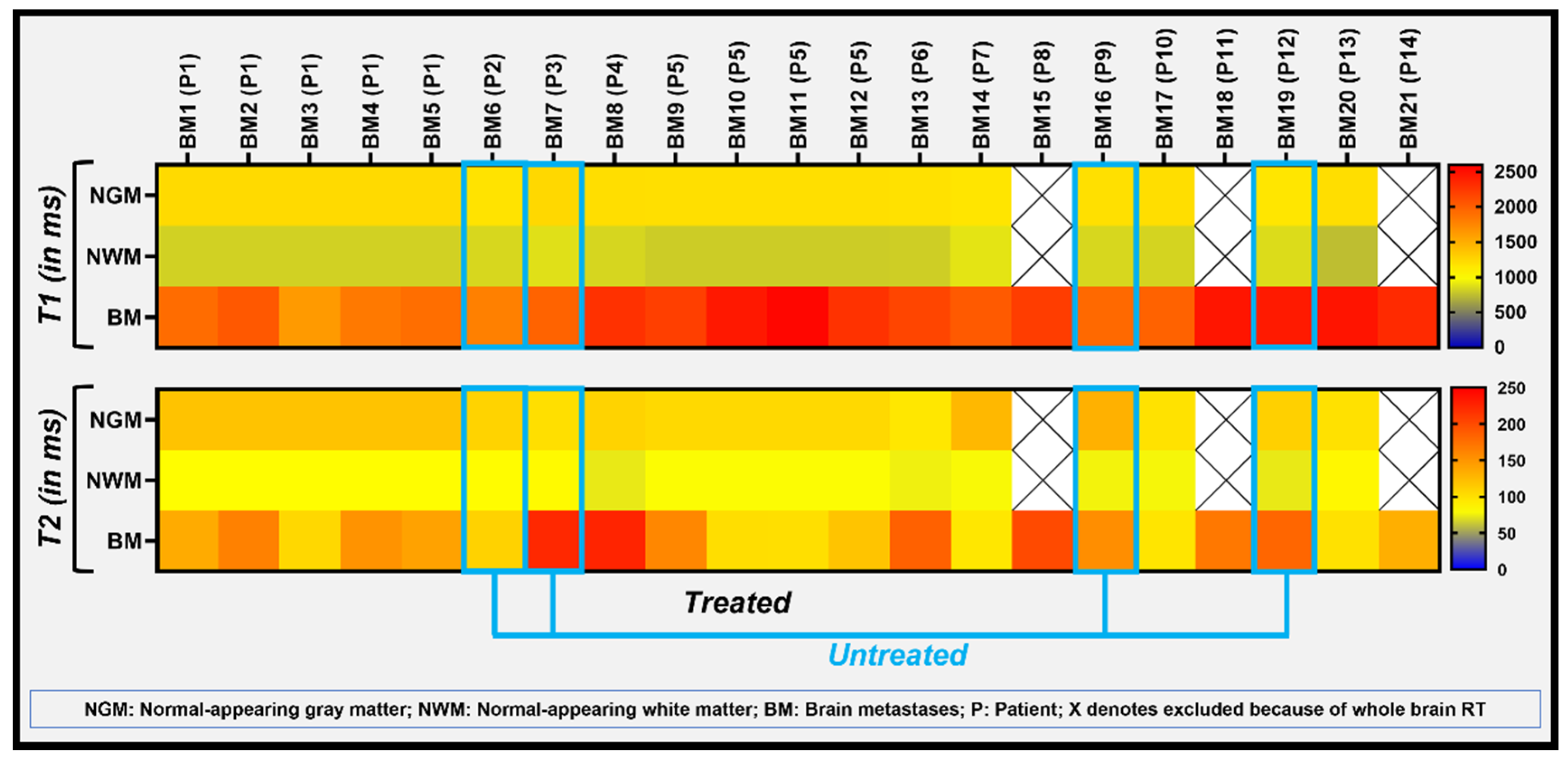
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nayak, L.; Lee, E.Q.; Wen, P.Y. Epidemiology of brain metastases. Curr. Oncol. Rep. 2012, 14, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Fecci, P.E.; Champion, C.D.; Hoj, J.; McKernan, C.M.; Goodwin, C.R.; Kirkpatrick, J.P.; Anders, C.K.; Pendergast, A.M.; Sampson, J.H. The Evolving Modern Management of Brain Metastasis. Clin. Cancer Res. 2019, 25, 6570–6580. [Google Scholar] [CrossRef] [PubMed]
- Richards, P.; Mc, K.W. Intracranial metastases. Br. Med. J. 1963, 1, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Sperduto, P.W.; Chao, S.T.; Sneed, P.K.; Luo, X.; Suh, J.; Roberge, D.; Bhatt, A.; Jensen, A.W.; Brown, P.D.; Shih, H.; et al. Diagnosis-specific prognostic factors, indexes, and treatment outcomes for patients with newly diagnosed brain metastases: A multi-institutional analysis of 4,259 patients. Int. J. Radiat. Oncol. Biol. Phys. 2010, 77, 655–661. [Google Scholar] [CrossRef]
- Di Lorenzo, R.; Ahluwalia, M.S. Targeted therapy of brain metastases: Latest evidence and clinical implications. Ther. Adv. Med. Oncol. 2017, 9, 781–796. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.V.; Kluger, H.M. Systemic Immunotherapy for the Treatment of Brain Metastases. Front. Oncol. 2016, 6, 49. [Google Scholar] [CrossRef][Green Version]
- Leung, D.; Han, X.; Mikkelsen, T.; Nabors, L.B. Role of MRI in primary brain tumor evaluation. J. Natl. Compr. Cancer Netw. 2014, 12, 1561–1568. [Google Scholar] [CrossRef]
- Cheng, H.L.; Stikov, N.; Ghugre, N.R.; Wright, G.A. Practical medical applications of quantitative MR relaxometry. J. Magn. Reson. Imaging 2012, 36, 805–824. [Google Scholar] [CrossRef]
- Lin, N.U.; Lee, E.Q.; Aoyama, H.; Barani, I.J.; Barboriak, D.P.; Baumert, B.G.; Bendszus, M.; Brown, P.D.; Camidge, D.R.; Chang, S.M.; et al. Response assessment criteria for brain metastases: Proposal from the RANO group. Lancet Oncol. 2015, 16, e270–e278. [Google Scholar] [CrossRef]
- Deoni, S.C. Magnetic resonance relaxation and quantitative measurement in the brain. Methods Mol. Biol. 2011, 711, 65–108. [Google Scholar] [CrossRef]
- Keenan, K.E.; Biller, J.R.; Delfino, J.G.; Boss, M.A.; Does, M.D.; Evelhoch, J.L.; Griswold, M.A.; Gunter, J.L.; Hinks, R.S.; Hoffman, S.W.; et al. Recommendations towards standards for quantitative MRI (qMRI) and outstanding needs. J. Magn. Reson. Imaging 2019, 49, e26–e39. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.P.B.; Aboagye, E.O.; Adams, J.E.; Aerts, H.J.W.L.; Barrington, S.F.; Beer, A.J.; Boellaard, R.; Bohndiek, S.E.; Brady, M.; Brown, G.; et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 2017, 14, 169–186. [Google Scholar] [CrossRef] [PubMed]
- Little, R.A.; Barjat, H.; Hare, J.I.; Jenner, M.; Watson, Y.; Cheung, S.; Holliday, K.; Zhang, W.; O’Connor, J.P.B.; Barry, S.T.; et al. Evaluation of dynamic contrast-enhanced MRI biomarkers for stratified cancer medicine: How do permeability and perfusion vary between human tumours? Magn. Reson. Imaging 2018, 46, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gulotta, B.; Thomas, A.; Kaley, T.; Karimi, S.; Gavrilovic, I.; Woo, K.M.; Zhang, Z.; Arevalo-Perez, J.; Holodny, A.I.; et al. Large-volume low apparent diffusion coefficient lesions predict poor survival in bevacizumab-treated glioblastoma patients. Neuro. Oncol. 2016, 18, 735–743. [Google Scholar] [CrossRef]
- Shah, A.D.; Shridhar Konar, A.; Paudyal, R.; Oh, J.H.; LoCastro, E.; Nuñez, D.A.; Swinburne, N.; Vachha, B.; Ulaner, G.A.; Young, R.J.; et al. Diffusion and Perfusion MRI Predicts Response Preceding and Shortly After Radiosurgery to Brain Metastases: A Pilot Study. J. Neuroimaging 2021, 31, 317–323. [Google Scholar] [CrossRef]
- Gharzeddine, K.; Hatzoglou, V.; Holodny, A.I.; Young, R.J. MR Perfusion and MR Spectroscopy of Brain Neoplasms. Radiol. Clin. N. Am. 2019, 57, 1177–1188. [Google Scholar] [CrossRef]
- Stanisz, G.J.; Odrobina, E.E.; Pun, J.; Escaravage, M.; Graham, S.J.; Bronskill, M.J.; Henkelman, R.M. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn. Reson. Med. 2005, 54, 507–512. [Google Scholar] [CrossRef]
- Shiguetomi-Medina, J.M.; Ramirez-Gl, J.L.; Stødkilde-Jørgensen, H.; Møller-Madsen, B. Systematized water content calculation in cartilage using T1-mapping MR estimations: Design and validation of a mathematical model. J. Orthop. Traumatol. 2017, 18, 217–220. [Google Scholar] [CrossRef]
- Chen, Q.; Zuo, Q.; Hu, Q.; Feng, Y.; Cui, W.; Fan, W.; Zou, Y. Morphological MRI and T2 mapping of cartilage repair tissue after mosaicplasty with tissue-engineered cartilage in a pig model. J. Biomed. Res. 2014, 28, 309–319. [Google Scholar] [CrossRef]
- Lescher, S.; Jurcoane, A.; Veit, A.; Bähr, O.; Deichmann, R.; Hattingen, E. Quantitative T1 and T2 mapping in recurrent glioblastomas under bevacizumab: Earlier detection of tumor progression compared to conventional MRI. Neuroradiology 2015, 57, 11–20. [Google Scholar] [CrossRef]
- Barral, J.K.; Gudmundson, E.; Stikov, N.; Etezadi-Amoli, M.; Stoica, P.; Nishimura, D.G. A robust methodology for in vivo T1 mapping. Magn. Reson. Med. 2010, 64, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Gelman, N.; Ewing, J.R.; Gorell, J.M.; Spickler, E.M.; Solomon, E.G. Interregional variation of longitudinal relaxation rates in human brain at 3.0 T: Relation to estimated iron and water contents. Magn. Reson. Med. 2001, 45, 71–79. [Google Scholar] [CrossRef]
- Carr, M.E.; Keenan, K.E.; Rai, R.; Metcalfe, P.; Walker, A.; Holloway, L. Determining the longitudinal accuracy and reproducibility of T(1) and T(2) in a 3T MRI scanner. J. Appl. Clin. Med. Phys. 2021, 22, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Deoni, S.C.; Peters, T.M.; Rutt, B.K. High-resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn. Reson. Med. 2005, 53, 237–241. [Google Scholar] [CrossRef]
- Warntjes, J.; Leinhard, O.D.; West, J.; Lundberg, P. Rapid magnetic resonance quantification on the brain: Optimization for clinical usage. Magn. Reson. Med. 2008, 60, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Gulani, V.; Seiberlich, N.; Liu, K.; Sunshine, J.L.; Duerk, J.L.; Griswold, M.A. Magnetic resonance fingerprinting. Nature 2013, 495, 187–192. [Google Scholar] [CrossRef]
- Vardhanabhuti, V.; Au, H.; Ding, J.; Lee, E.; Cao, P.; Hui, S. Repeatability of Magnetic Resonance Fingerprinting using ISMRM/NIST MRI Phantom in Philips 3T MRI Scanner. In Proceedings of the International Society of Magnetic Resonance Imaging (ISMRM), Virtual Conference & Exhibition, 8–14 August 2020. [Google Scholar]
- Jiang, Y.; Ma, D.; Keenan, K.E.; Stupic, K.F.; Gulani, V.; Griswold, M.A. Repeatability of magnetic resonance fingerprinting T1 and T2 estimates assessed using the ISMRM/NIST MRI system phantom. Magn. Reson. Med. 2017, 78, 1452–1457. [Google Scholar] [CrossRef]
- Shridhar Konar, A.; Qian, E.; Geethanath, S.; Buonincontri, G.; Obuchowski, N.A.; Fung, M.; Gomez, P.; Schulte, R.; Cencini, M.; Tosetti, M. Quantitative imaging metrics derived from magnetic resonance fingerprinting using ISMRM/NIST MRI system phantom: An international multicenter repeatability and reproducibility study. Med. Phys. 2021, 48, 2438–2447. [Google Scholar] [CrossRef]
- Buonincontri, G.; Biagi, L.; Retico, A.; Cecchi, P.; Cosottini, M.; Gallagher, F.A.; Gómez, P.A.; Graves, M.J.; McLean, M.A.; Riemer, F. Multi-site repeatability and reproducibility of MR fingerprinting of the healthy brain at 1.5 and 3.0 T. NeuroImage 2019, 195, 362–372. [Google Scholar] [CrossRef]
- Ma, D.; Jiang, Y.; Chen, Y.; McGivney, D.; Mehta, B.; Gulani, V.; Griswold, M. Fast 3D magnetic resonance fingerprinting for a whole-brain coverage. Magn. Reson. Med. 2018, 79, 2190–2197. [Google Scholar] [CrossRef]
- Korzdorfer, G.; Kirsch, R.; Liu, K.; Pfeuffer, J.; Hensel, B.; Jiang, Y.; Ma, D.; Gratz, M.; Bar, P.; Bogner, W.; et al. Reproducibility and Repeatability of MR Fingerprinting Relaxometry in the Human Brain. Radiology 2019, 292, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Velasco, C.; Ye, H.; Lindner, T.; Grech-Sollars, M.; O’Callaghan, J.; Hiley, C.; Chouhan, M.D.; Niendorf, T.; Koh, D.M.; et al. Current Applications and Future Development of Magnetic Resonance Fingerprinting in Diagnosis, Characterization, and Response Monitoring in Cancer. Cancers 2021, 13, 4742. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, V.G.; Körzdörfer, G.; Gall, P. Toward Quantification: Microstructure and Magnetic Resonance Fingerprinting. Investig. Radiol. 2021, 56, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tippareddy, C.; Zhao, W.; Sunshine, J.L.; Griswold, M.; Ma, D.; Badve, C. Magnetic resonance fingerprinting: An overview. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4189–4200. [Google Scholar] [CrossRef] [PubMed]
- Badve, C.; Yu, A.; Dastmalchian, S.; Rogers, M.; Ma, D.; Jiang, Y.; Margevicius, S.; Pahwa, S.; Lu, Z.; Schluchter, M.; et al. MR Fingerprinting of Adult Brain Tumors: Initial Experience. AJNR Am. J. Neuroradiol. 2017, 38, 492–499. [Google Scholar] [CrossRef]
- Dastmalchian, S.; Kilinc, O.; Onyewadume, L.; Tippareddy, C.; McGivney, D.; Ma, D.; Griswold, M.; Sunshine, J.; Gulani, V.; Barnholtz-Sloan, J.S.; et al. Radiomic analysis of magnetic resonance fingerprinting in adult brain tumors. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 683–693. [Google Scholar] [CrossRef]
- Ma, D.; Jones, S.E.; Deshmane, A.; Sakaie, K.; Pierre, E.Y.; Larvie, M.; McGivney, D.; Blümcke, I.; Krishnan, B.; Lowe, M. Development of high-resolution 3D MR fingerprinting for detection and characterization of epileptic lesions. J. Magn. Reson. Imaging 2019, 49, 1333–1346. [Google Scholar] [CrossRef]
- Russek, S.; Boss, M.; Jackson, E.; Jennings, D.; Evelhoch, J.; Gunter, J.; Sorensen, A. Characterization of NIST/ISMRM MRI system phantom. In Proceedings of the 20th Annual Meeting of ISMRM, Melbourne, VIC, Australia, 5–11 May 2012; p. 2456. [Google Scholar]
- Keenan, K.E.; Stupic, K.F.; Boss, M.A.; Russek, S.E.; Chenevert, T.L.; Prasad, P.V.; Reddick, W.E.; Zheng, J.; Hu, P.; Jackson, E.F. Comparison of T1 measurement using ISMRM/NIST system phantom. In Proceedings of the 24th Annual Meeting of ISMRM, Singapore, 7–13 May 2016; p. 3290. [Google Scholar]
- Jiang, Y.; Ma, D.; Seiberlich, N.; Gulani, V.; Griswold, M.A. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn. Reson. Med. 2015, 74, 1621–1631. [Google Scholar] [CrossRef]
- Mazor, G.; Weizman, L.; Tal, A.; Eldar, Y.C. Low rank magnetic resonance fingerprinting. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2016, 2016, 439–442. [Google Scholar] [CrossRef]
- Jackson, J.I.; Meyer, C.H.; Nishimura, D.G.; Macovski, A. Selection of a convolution function for Fourier inversion using gridding (computerised tomography application). IEEE Trans. Med. Imaging 1991, 10, 473–478. [Google Scholar] [CrossRef]
- McGivney, D.F.; Pierre, E.; Ma, D.; Jiang, Y.; Saybasili, H.; Gulani, V.; Griswold, M.A. SVD compression for magnetic resonance fingerprinting in the time domain. IEEE Trans. Med. Imaging 2014, 33, 2311–2322. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.O.; Gmitro, A.F.; Marcellin, M.W. Adaptive reconstruction of phased array MR imagery. Magn. Reson. Med. 2000, 43, 682–690. [Google Scholar] [CrossRef]
- Thrower, S.L.; Al Feghali, K.A.; Luo, D.; Paddick, I.; Hou, P.; Briere, T.; Li, J.; McAleer, M.F.; McGovern, S.L.; Woodhouse, K.D. The effect of slice thickness on contours of brain metastases for stereotactic radiosurgery. Adv. Radiat. Oncol. 2021, 6, 100708. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Hao, X.; Liu, F.; Xu, D.; Liu, J.; Peterson, B.S. The effects of changing water content, relaxation times, and tissue contrast on tissue segmentation and measures of cortical anatomy in MR images. Magn. Reson. Imaging 2013, 31, 1709–1730. [Google Scholar] [CrossRef]
- de Blank, P.; Badve, C.; Gold, D.R.; Stearns, D.; Sunshine, J.; Dastmalchian, S.; Tomei, K.; Sloan, A.E.; Barnholtz-Sloan, J.S.; Lane, A.; et al. Magnetic Resonance Fingerprinting to Characterize Childhood and Young Adult Brain Tumors. Pediatr. Neurosurg. 2019, 54, 310–318. [Google Scholar] [CrossRef]
- Rieger, B.; Akçakaya, M.; Pariente, J.C.; Llufriu, S.; Martinez-Heras, E.; Weingärtner, S.; Schad, L.R. Time efficient whole-brain coverage with MR Fingerprinting using slice-interleaved echo-planar-imaging. Sci. Rep. 2018, 8, 6667. [Google Scholar] [CrossRef]
- Steen, R.G.; Koury, B.S.M.; Granja, C.I.; Xiong, X.; Wu, S.; Glass, J.O.; Mulhern, R.K.; Kun, L.E.; Merchant, T.E. Effect of ionizing radiation on the human brain: White matter and gray matter T1 in pediatric brain tumor patients treated with conformal radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2001, 49, 79–91. [Google Scholar] [CrossRef]
- Wagner, M.; Helfrich, M.; Volz, S.; Magerkurth, J.; Blasel, S.; Porto, L.; Singer, O.C.; Deichmann, R.; Jurcoane, A.; Hattingen, E. Quantitative T2, T2*, and T2′ MR imaging in patients with ischemic leukoaraiosis might detect microstructural changes and cortical hypoxia. Neuroradiology 2015, 57, 1023–1030. [Google Scholar] [CrossRef]
- Vymazal, J.; Hajek, M.; Patronas, N.; Giedd, J.N.; Bulte, J.W.; Baumgarner, C.; Tran, V.; Brooks, R.A. The quantitative relation between T1-weighted and T2-weighted MRI of normal gray matter and iron concentration. J. Magn. Reson. Imaging 1995, 5, 554–560. [Google Scholar] [CrossRef]
- Shah, R.; Vattoth, S.; Jacob, R.; Manzil, F.F.; O’Malley, J.P.; Borghei, P.; Patel, B.N.; Curé, J.K. Radiation necrosis in the brain: Imaging features and differentiation from tumor recurrence. Radiographics 2012, 32, 1343–1359. [Google Scholar] [CrossRef] [PubMed]
- Stockham, A.L.; Tievsky, A.L.; Koyfman, S.A.; Reddy, C.A.; Suh, J.H.; Vogelbaum, M.A.; Barnett, G.H.; Chao, S.T. Conventional MRI does not reliably distinguish radiation necrosis from tumor recurrence after stereotactic radiosurgery. J. Neurooncol. 2012, 109, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Badve, C.; Yu, A.; Rogers, M.; Ma, D.; Liu, Y.; Schluchter, M.; Sunshine, J.; Griswold, M.; Gulani, V. Simultaneous T(1) and T(2) Brain Relaxometry in Asymptomatic Volunteers using Magnetic Resonance Fingerprinting. Tomography 2015, 1, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Wansapura, J.P.; Holland, S.K.; Dunn, R.S.; Ball Jr, W.S. NMR relaxation times in the human brain at 3.0 tesla. J. Magn. Reson. Imaging 1999, 9, 531–538. [Google Scholar] [CrossRef]
- Pirkl, C.M.; Nunez-Gonzalez, L.; Kofler, F.; Endt, S.; Grundl, L.; Golbabaee, M.; Gómez, P.A.; Cencini, M.; Buonincontri, G.; Schulte, R.F.; et al. Accelerated 3D whole-brain T1, T2, and proton density mapping: Feasibility for clinical glioma MR imaging. Neuroradiology 2021, 63, 1831–1851. [Google Scholar] [CrossRef]
- Konar, A.S.; Shah, A.D.; Paudyal, R.; Fung, M.; Banerjee, S.; Dave, A.; Hatzoglou, V.; Shukla-Dave, A. Quantitative Synthetic Magnetic Resonance Imaging for Brain Metastases: A Feasibility Study. Cancers 2022, 14, 2651. [Google Scholar] [CrossRef]
- Konar, A.S.; Paudyal, R.; Shah, A.D.; Fung, M.; Banerjee, S.; Dave, A.; Lee, N.; Hatzoglou, V.; Shukla-Dave, A. Qualitative and Quantitative Performance of Magnetic Resonance Image Compilation (MAGiC) Method: An Exploratory Analysis for Head and Neck Imaging. Cancers 2022, 14, 3624. [Google Scholar] [CrossRef]
- Poon, C.S.; Henkelman, R.M. Practical T2 quantitation for clinical applications. J. Magn. Reson. Imaging 1992, 2, 541–553. [Google Scholar] [CrossRef]
- Vymazal, J.; Righini, A.; Brooks, R.A.; Canesi, M.; Mariani, C.; Leonardi, M.; Pezzoli, G. T1 and T2 in the brain of healthy subjects, patients with Parkinson disease, and patients with multiple system atrophy: Relation to iron content. Radiology 1999, 211, 489–495. [Google Scholar] [CrossRef]
- Whittall, K.P.; Mackay, A.L.; Graeb, D.A.; Nugent, R.A.; Li, D.K.; Paty, D.W. In vivo measurement of T2 distributions and water contents in normal human brain. Magn. Reson. Med. 1997, 37, 34–43. [Google Scholar] [CrossRef]
- Magalhaes, A.; Godfrey, W.; Shen, Y.; Hu, J.; Smith, W. Proton magnetic resonance spectroscopy of brain tumors correlated with pathology. Acad. Radiol. 2005, 12, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Tong, E.; McCullagh, K.L.; Iv, M. Advanced imaging of brain metastases: From augmenting visualization and improving diagnosis to evaluating treatment response. Front. Neurol. 2020, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Mehrabian, H.; Detsky, J.; Soliman, H.; Sahgal, A.; Stanisz, G.J. Advanced magnetic resonance imaging techniques in management of brain metastases. Front. Oncol. 2019, 9, 440. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.L.; Chang, S. Pseudoprogression and pseudoresponse: Challenges in brain tumor imaging. Curr. Neurol. Neurosci. Rep. 2009, 9, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Shukla-Dave, A.; Obuchowski, N.A.; Chenevert, T.L.; Jambawalikar, S.; Schwartz, L.H.; Malyarenko, D.; Huang, W.; Noworolski, S.M.; Young, R.J.; Shiroishi, M.S.; et al. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials. J. Magn. Reson. Imaging 2019, 49, e101–e121. [Google Scholar] [CrossRef]
| Characteristics | Value |
|---|---|
| Total Patients | 14 |
| Total number of brain metastases lesions | 21 |
| Demographics Median age (Y) Age range (Y) Male/Female | 53 25–72 6/8 |
| Location of primary tumor | |
| Lung Colon Melanoma Other | 6 2 3 3 |
| Untreated/Treated | 4/10 |
| Vial | T1 (ms) | Relative Difference (%) | ||||
| VP | GS | MRF | MRF and VP | MRF and GS | ||
| 1 | 1838 | 1780 | 1881 | 2.3 | 5.7 | |
| 2 | 1398 | 1351 | 1301 | 6.9 | 3.7 | |
| 3 | 998.3 | 958 | 927 | 7.1 | 3.2 | |
| 4 | 725.8 | 678 | 671 | 7.6 | 1 | |
| 5 | 509 | 483 | 461 | 9.4 | 4.6 | |
| 6 | 367 | 346 | 352 | 4.1 | 1.7 | |
| 7 | 258.7 | 242 | 237 | 8.4 | 2.1 | |
| Vial | T2 (ms) | Relative Difference (%) | ||||
| VP | GS | MRF | MRF and VP | MRF and GS | ||
| 1 | 645.8 | 537 | 637 | 1.4 | 18.6 | |
| 2 | 423.6 | 357 | 440 | 3.9 | 23.2 | |
| 3 | 286 | 246 | 288 | 0.7 | 17.1 | |
| 4 | 184.8 | 163 | 206 | 11.5 | 26.4 | |
| 5 | 134.1 | 118 | 155 | 15.6 | 31.4 | |
| 6 | 94.4 | 82 | 115 | 21.8 | 40.2 | |
| 7 | 62.5 | 57 | 84 | 34.4 | 47.4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konar, A.S.; Shah, A.D.; Paudyal, R.; Fung, M.; Banerjee, S.; Dave, A.; Hatzoglou, V.; Shukla-Dave, A. Quantitative Relaxometry Metrics for Brain Metastases Compared to Normal Tissues: A Pilot MR Fingerprinting Study. Cancers 2022, 14, 5606. https://doi.org/10.3390/cancers14225606
Konar AS, Shah AD, Paudyal R, Fung M, Banerjee S, Dave A, Hatzoglou V, Shukla-Dave A. Quantitative Relaxometry Metrics for Brain Metastases Compared to Normal Tissues: A Pilot MR Fingerprinting Study. Cancers. 2022; 14(22):5606. https://doi.org/10.3390/cancers14225606
Chicago/Turabian StyleKonar, Amaresha Shridhar, Akash Deelip Shah, Ramesh Paudyal, Maggie Fung, Suchandrima Banerjee, Abhay Dave, Vaios Hatzoglou, and Amita Shukla-Dave. 2022. "Quantitative Relaxometry Metrics for Brain Metastases Compared to Normal Tissues: A Pilot MR Fingerprinting Study" Cancers 14, no. 22: 5606. https://doi.org/10.3390/cancers14225606
APA StyleKonar, A. S., Shah, A. D., Paudyal, R., Fung, M., Banerjee, S., Dave, A., Hatzoglou, V., & Shukla-Dave, A. (2022). Quantitative Relaxometry Metrics for Brain Metastases Compared to Normal Tissues: A Pilot MR Fingerprinting Study. Cancers, 14(22), 5606. https://doi.org/10.3390/cancers14225606





