Genomic and Transcriptomic Predictors of Response to Immune Checkpoint Inhibitors in Melanoma Patients: A Machine Learning Approach
Abstract
Simple Summary
Abstract
1. Introduction
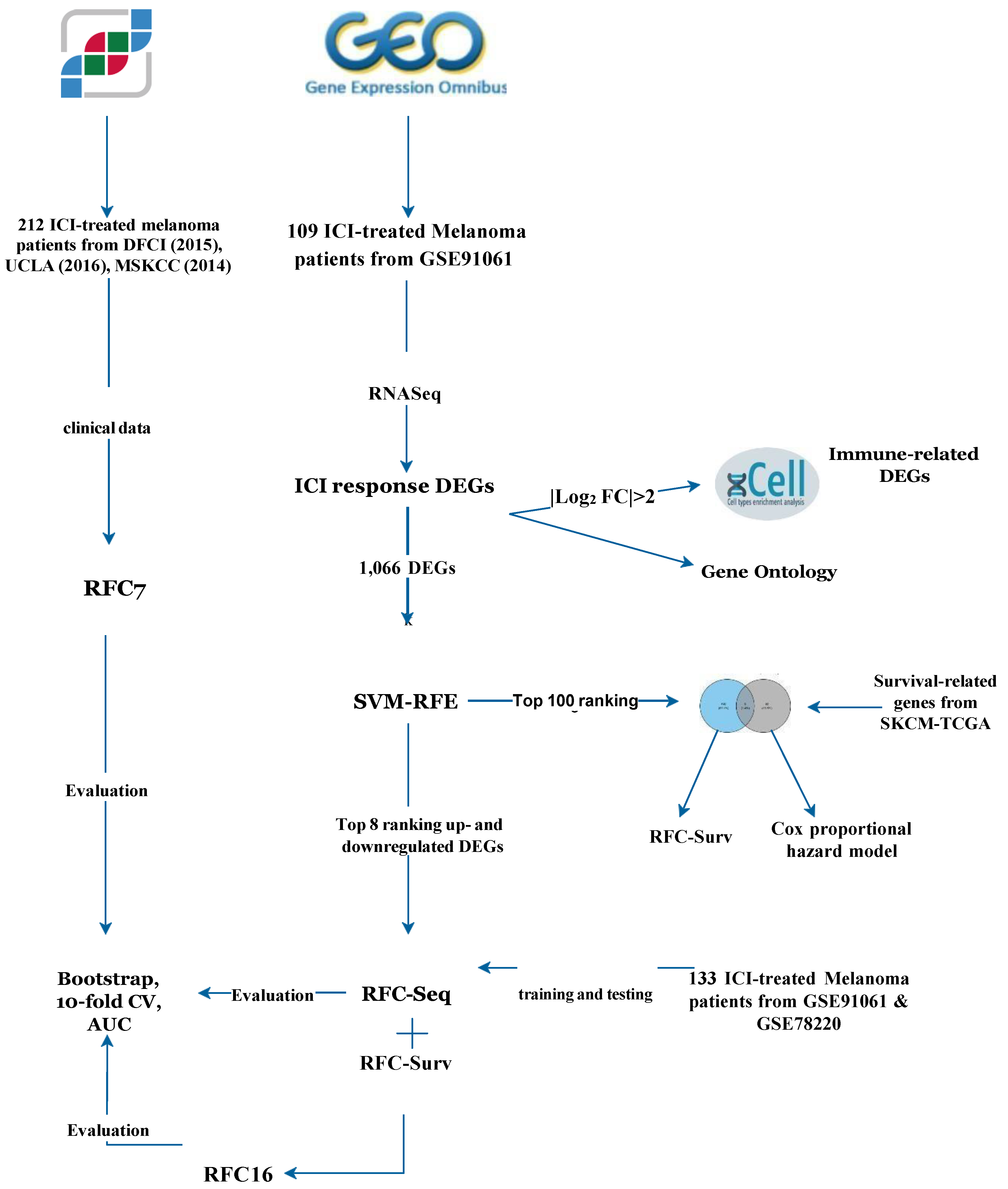
2. Materials and Methods
2.1. Data Acquisition
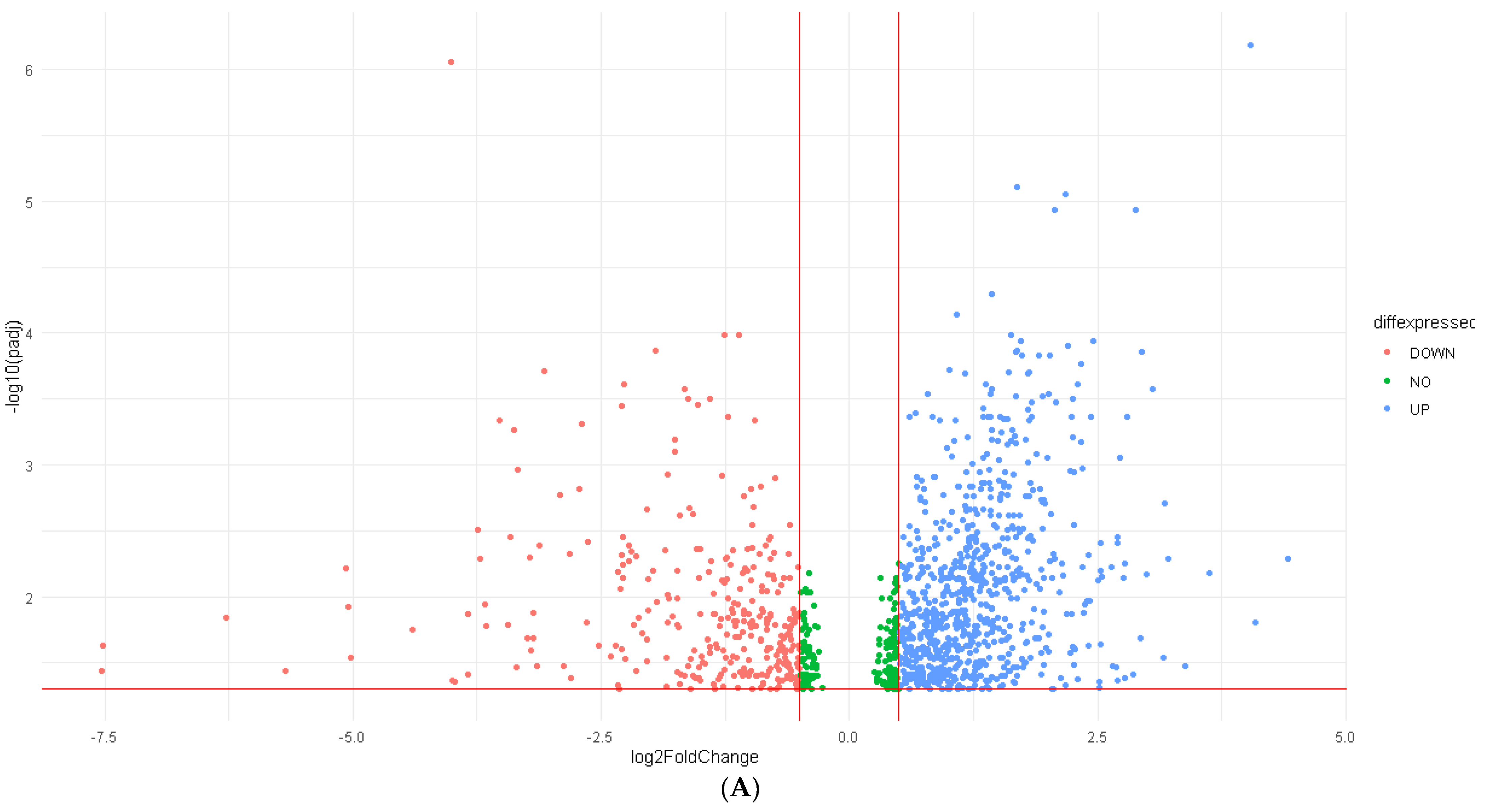
2.2. Differential Expression Analysis (DEA)
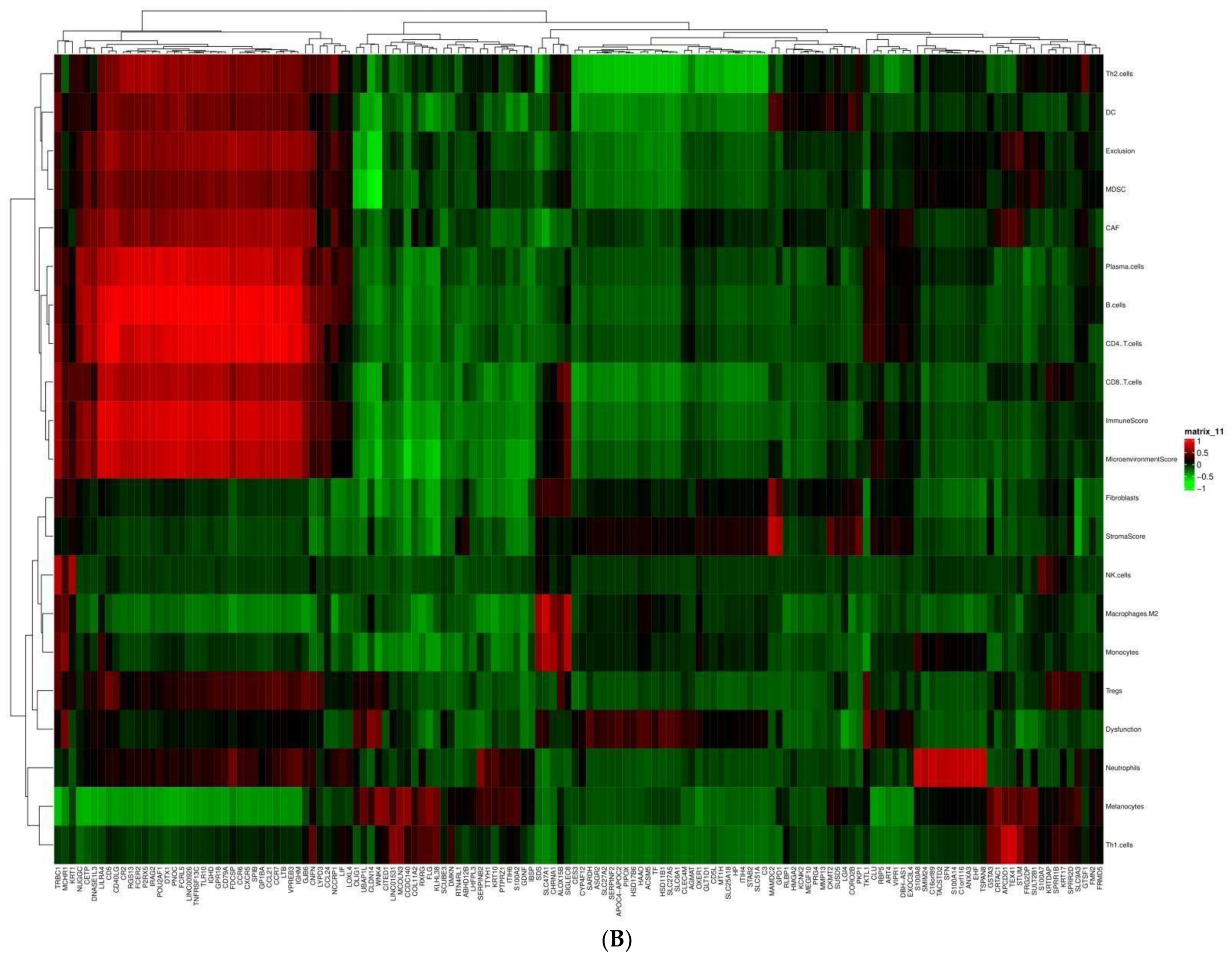
2.3. Immune Infiltration Analysis
2.4. Gene Ontology (GO) Enrichment Analysis
2.5. Support Vector Machine—Recursive Feature Elimination (SVM-RFE)
2.6. Survival-Associated DEGs
2.7. Random Forest Classifier
2.8. Data Availability
3. Results
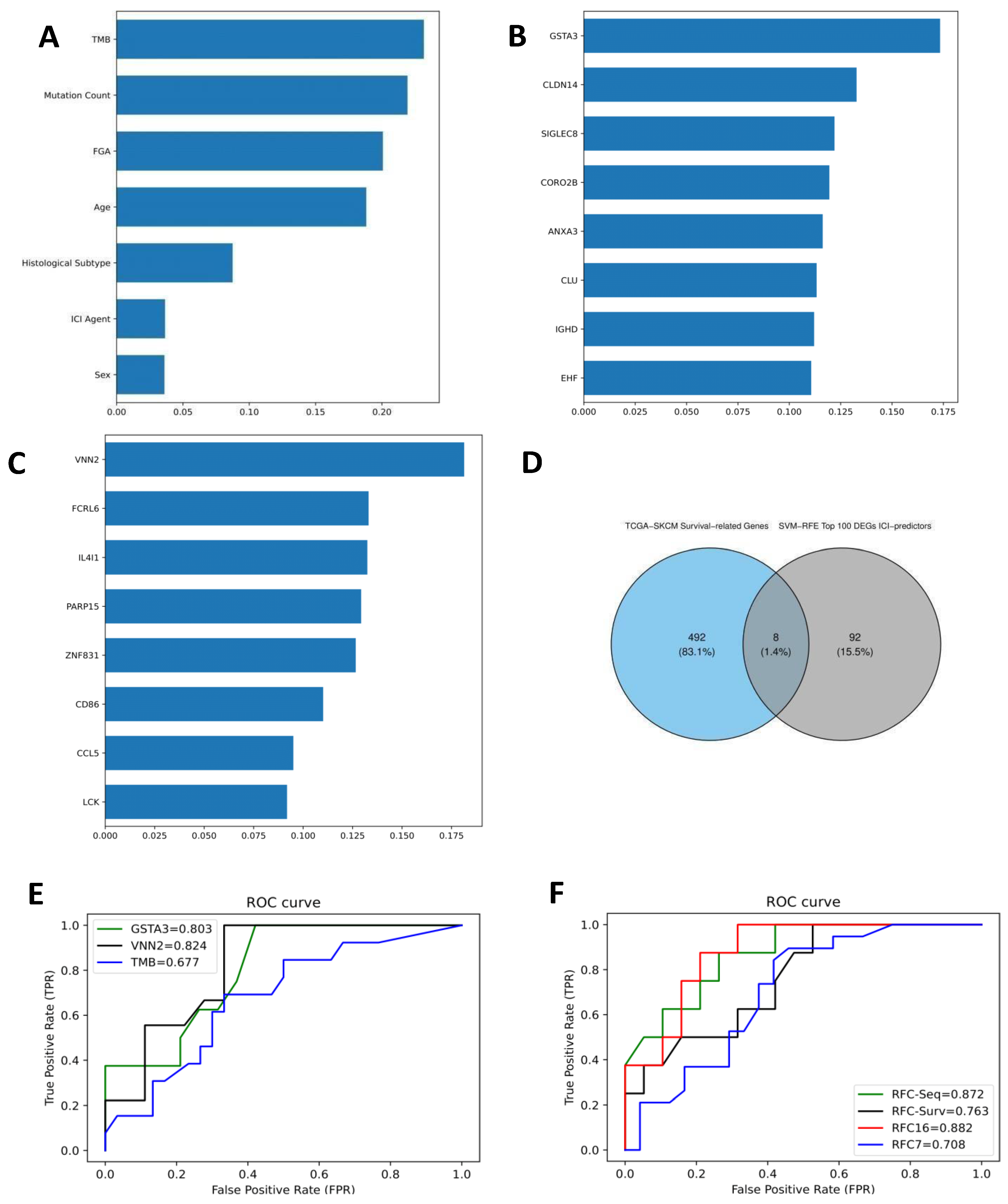
3.1. Clinical Data and ICI Response Model
3.2. Differentially Expressed Genes (DEGs) and RFC-Seq Model
3.3. Tumor Immune Microenvironment and DEGs
3.4. Gene Ontology (GO) Enrichment Analysis
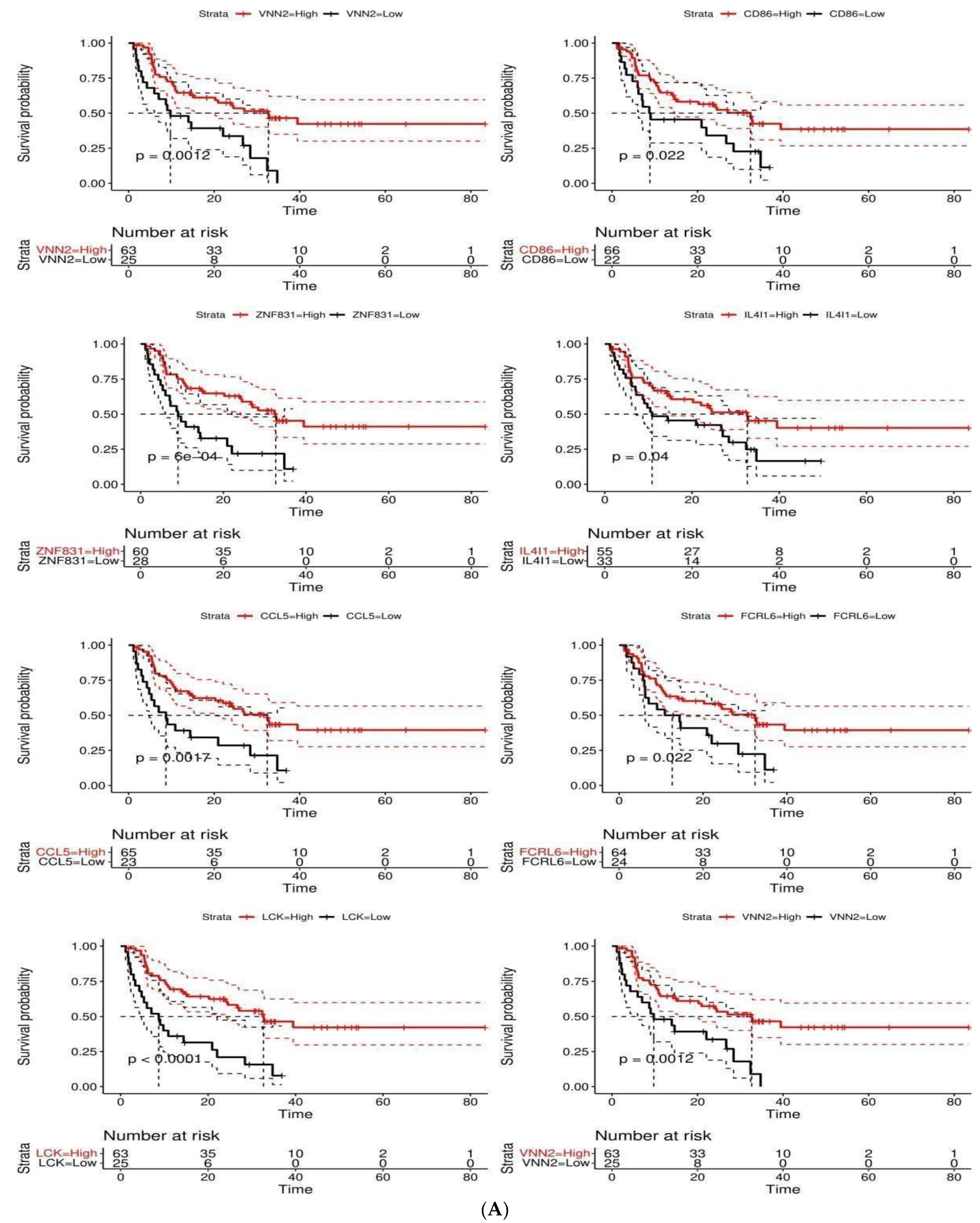
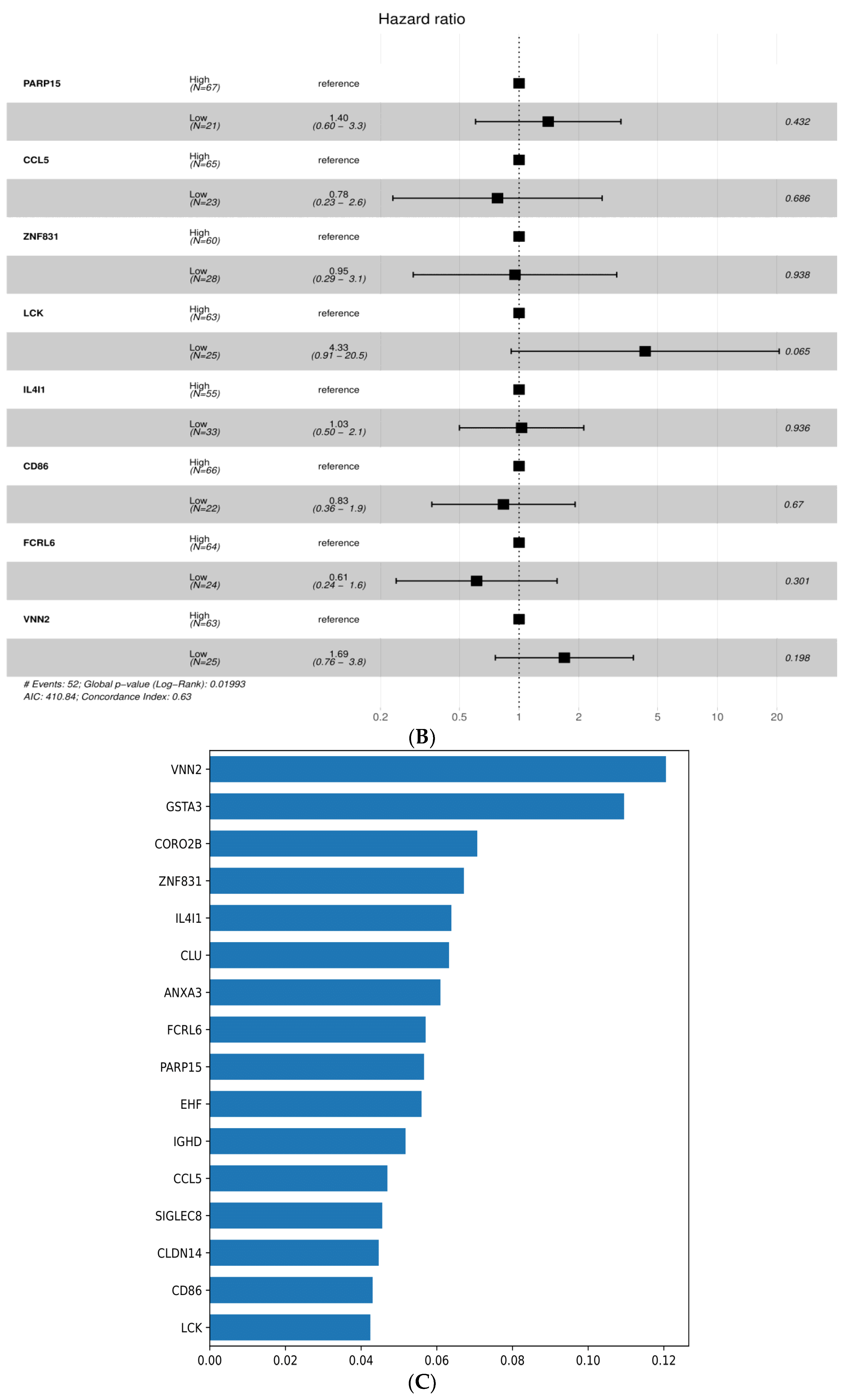
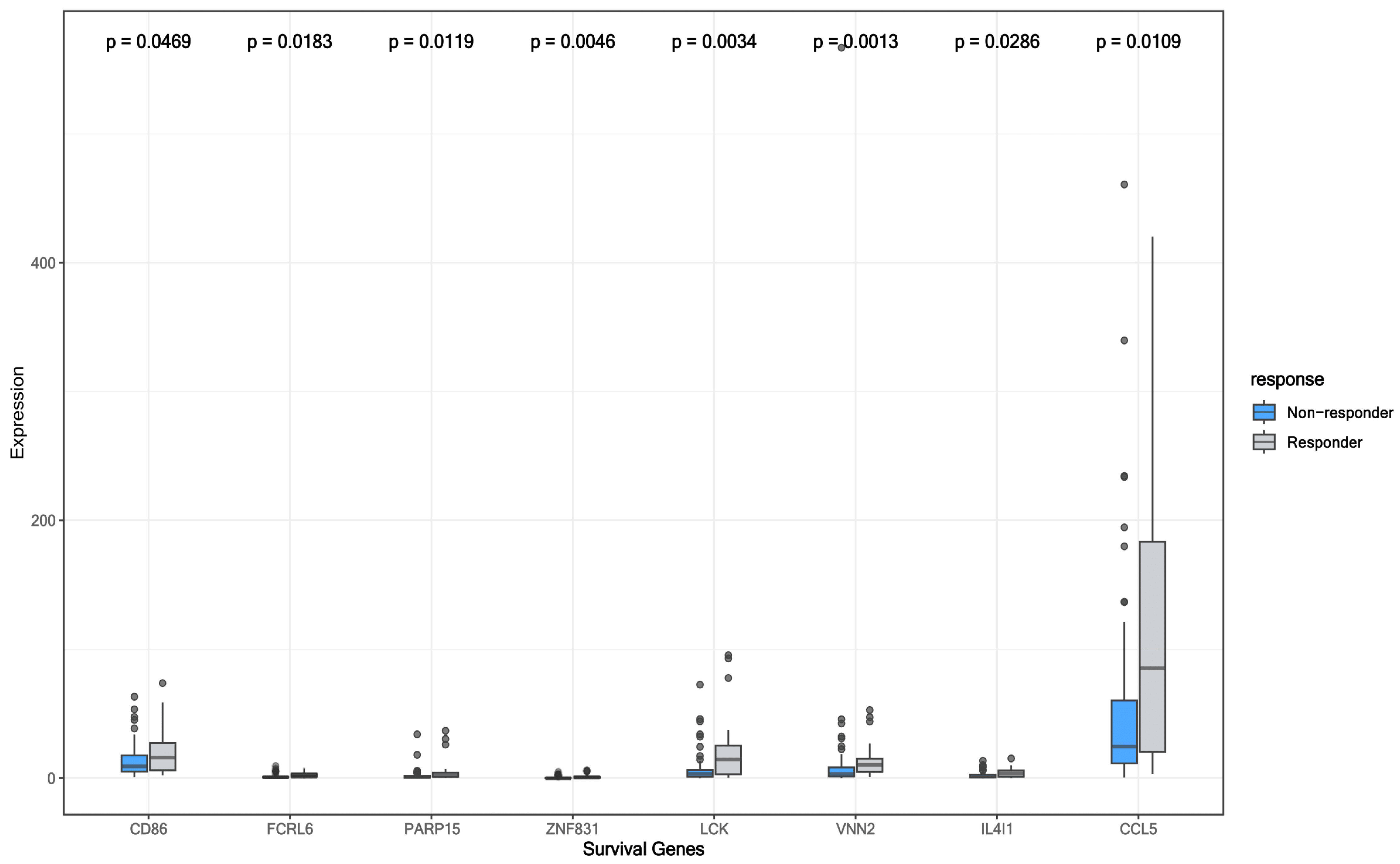
3.5. Survival-Associated DEGs and RFC-Surv Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Madden, K.; Kasler, M.K. Immune Checkpoint Inhibitors in Lung Cancer and Melanoma. Semin. Oncol. Nurs. 2019, 35, 683432. [Google Scholar] [CrossRef] [PubMed]
- Olbryt, M.; Rajczykowski, M.; Widłak, W. Biological Factors behind Melanoma Response to Immune Checkpoint Inhibitors. Int. J. Mol. Sci. 2020, 21, 4071. [Google Scholar] [CrossRef] [PubMed]
- Gajic, Z.Z.; Deshpande, A.; Legut, M.; Imieliński, M.; Sanjana, N.E. Recurrent somatic mutations as predictors of immunotherapy response. Nat. Commun. 2022, 13, 3938. [Google Scholar] [CrossRef]
- Taube, J.M.; Klein, A.; Brahmer, J.R.; Xu, H.; Pan, X.; Kim, J.H.; Chen, L.; Pardoll, D.M.; Topalian, S.L.; Anders, R.A. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 2014, 20, 5064–5074. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Liu, D.; Yang, X.; Wu, X. Tumor Immune Microenvironment Characterization Identifies Prognosis and Immunotherapy-Related Gene Signatures in Melanoma. Front. Immunol. 2021, 12, 663495. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.-Y.; Cheong, J.-H. Machine Learning Predictor of Immune Checkpoint Blockade Response in Gastric Cancer. Cancers 2022, 14, 3191. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Goodison, S.; Li, J.; Liu, L.; Farmerie, W. Improved breast cancer prognosis through the combination of clinical and genetic markers. Bioinformatics 2007, 23, 30–37. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.L.; Hung, Y.H.; Lee, W.M.; Li, R.K.; Jiang, B.R. SVM-RFE based feature selection and taguchi parameters optimization for multiclass SVM Classifier. Sci. World J. 2014, 2014, 795624. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. Available online: http://scikit-learn.sourceforge.net (accessed on 11 June 2022).
- 3.1. Cross-Validation: Evaluating Estimator Performance—Scikit-Learn 1.1.1 Documentation. Available online: https://scikit-learn.org/stable/modules/cross_validation.html (accessed on 11 June 2022).
- Chowell, D.; Yoo, S.-K.; Valero, C.; Pastore, A.; Krishna, C.; Lee, M.; Hoen, D.; Shi, H.; Kelly, D.W.; Patel, N.; et al. Improved prediction of immune checkpoint blockade efficacy across multiple cancer types. Nat. Biotechnol. 2022, 40, 499–506. [Google Scholar] [CrossRef]
- Patel, S.P.; Kurzrock, R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015, 14, 847–856. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Jacquelot, N.; Roberti, M.P.; Enot, D.P.; Rusakiewicz, S.; Ternès, N.; Jegou, S.; Woods, D.M.; Sodré, A.L.; Hansen, M.; Meirow, Y.; et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat. Commun. 2017, 8, 592. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Solit, D.B.; Chan, T.A.; Kurzrock, R. The FDA approval of pembrolizumab for adult and pediatric patients with tumor mutational burden (TMB) ≥ 10: A decision centered on empowering patients and their physicians. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020, 31, 1115–1118. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef]
- Tran, E.; Ahmadzadeh, M.; Lu, Y.-C.; Gros, A.; Turcotte, S.; Robbins, P.F.; Gartner, J.J.; Zheng, Z.; Li, Y.F.; Ray, S.; et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015, 350, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Merino, D.M.; McShane, L.M.; Fabrizio, D.; Funari, V.; Chen, S.J.; White, J.R.; Wenz, P.; Baden, J.; Barrett, J.C.; Chaudhary, R.; et al. Establishing guidelines to harmonize tumor mutational burden (TMB): In silico assessment of variation in TMB quantification across diagnostic platforms: Phase I of the Friends of Cancer Research TMB Harmonization Project. J. Immunother. Cancer 2020, 8, e000147. [Google Scholar] [CrossRef]
- Campbell, B.B.; Light, N.; Fabrizio, D.; Zatzman, M.; Fuligni, F.; De Borja, R.; Davidson, S.; Edwards, M.; Elvin, J.A.; Hodel, K.P.; et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017, 171, 1042–1056.e10. [Google Scholar] [CrossRef]
- Liu, D.; Schilling, B.; Liu, D.; Sucker, A.; Livingstone, E.; Jerby-Arnon, L.; Zimmer, L.; Gutzmer, R.; Satzger, I.; Loquai, C.; et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019, 25, 1916–1927. [Google Scholar] [CrossRef]
- Pfeifer, G.P. Environmental exposures and mutational patterns of cancer genomes. Genome Med. 2010, 2, 54. [Google Scholar] [CrossRef]
- Chakraborty, G.; Ghosh, A.; Nandakumar, S.; Armenia, J.; Mazzu, Y.Z.; Atiq, M.O.; Lee, G.-S.M.; Mucci, L.A.; Merghoub, T.; Wolchok, J.D.; et al. Fraction genome altered (FGA) to regulate both cell autonomous and non-cell autonomous functions in prostate cancer and its effect on prostate cancer aggressiveness. J. Clin. Oncol. 2020, 38 (Suppl. 6), 347. [Google Scholar] [CrossRef]
- Kong, J.; Ha, D.; Lee, J.; Kim, I.; Park, M.; Im, S.-H.; Shin, K.; Kim, S. Network-based machine learning approach to predict immunotherapy response in cancer patients. Nat. Commun. 2022, 13, 3703. [Google Scholar] [CrossRef]
- Lindström, H.; Peer, S.M.; Ing, N.H.; Mannervik, B. Characterization of equine GST A3-3 as a steroid isomerase. J. Steroid Biochem. Mol. Biol. 2018, 178, 117–126. [Google Scholar] [CrossRef]
- Chen, X.; Chen, X.Z.; Liu, M.; Lang, N.; Tang, Q.L.; Chen, J.; Zhao, X.F.; Bi, F. Analysis of gene expression patterns in an irinotecan-resistance colon cancer cell by cDNA microarray. Sichuan Da Xue Xue Bao. Yi Xue Ban = J. Sichuan Univ. Med. Sci. Ed. 2011, 42, 15–18, 36. [Google Scholar]
- Duan, S.; Gong, B.; Wang, P.; Huang, H.; Luo, L.; Liu, F. Novel prognostic biomarkers of gastric cancer based on gene expression microarray: COL12A1, GSTA3, FGA and FGG. Mol. Med. Rep. 2018, 18, 3727–3736. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qiu, C.; Wang, S.; Wu, L.; Zhao, T. Inhibitory effect of glutathione S-transferase A3 in the progression of cutaneous squamous cell carcinoma. J. Cosmet. Dermatol. 2021, 20, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Kluger, H.M.; Chiang, V.; Mahajan, A.; Zito, C.R.; Sznol, M.; Tran, T.; Weiss, S.A.; Cohen, J.V.; Yu, J.; Hegde, U.; et al. Long-Term Survival of Patients With Melanoma With Active Brain Metastases Treated With Pembrolizumab on a Phase II Trial. J. Clin. Oncol. 2019, 37, 52–60. [Google Scholar] [CrossRef]
- Cabrita, R.; Lauss, M.; Sanna, A.; Donia, M.; Larsen, M.S.; Mitra, S.; Johansson, I.; Phung, B.; Harbst, K.; Vallon-Christersson, J.; et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020, 577, 561–565. [Google Scholar] [CrossRef]
- De Cicco, P.; Ercolano, G.; Ianaro, A. The New Era of Cancer Immunotherapy: Targeting Myeloid-Derived Suppressor Cells to Overcome Immune Evasion. Front. Immunol. 2020, 11, 1680. [Google Scholar] [CrossRef]
- Nefedova, Y.; Fishman, M.; Sherman, S.; Wang, X.; Beg, A.A.; Gabrilovich, D.I. Mechanism of all-trans retinoic acid effect on tumor-associated myeloid-derived suppressor cells. Cancer Res. 2007, 67, 11021–11028. [Google Scholar] [CrossRef]
- Tobin, R.P.; Davis, D.; Jordan, K.R.; McCarter, M.D. The clinical evidence for targeting human myeloid-derived suppressor cells in cancer patients. J. Leukoc. Biol. 2017, 102, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Bønnelykke-Behrndtz, M.L.; Steiniche, T.; Damsgaard, T.E.; Georgsen, J.B.; Danielsen, A.; Bastholt, L.; Møller, H.J.; Nørgaard, P.H.; Schmidt, H. MelanA-negative spindle-cell associated melanoma, a distinct inflammatory phenotype correlated with dense infiltration of CD163 macrophages and loss of E-cadherin. Melanoma Res. 2015, 25, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Ock, C.-Y.; Kim, S.; Keam, B.; Kim, M.; Kim, T.M.; Kim, J.-H.; Jeon, Y.K.; Lee, J.-S.; Kwon, S.K.; Hah, J.H.; et al. PD-L1 expression is associated with epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Oncotarget 2016, 7, 15901–15914. [Google Scholar] [CrossRef] [PubMed]
- Yoshitake, H.; Takeda, Y.; Nitto, T.; Sendo, F.; Araki, Y. GPI-80, a beta2 integrin associated glycosylphosphatidylinositol-anchored protein, concentrates on pseudopodia without association with beta2 integrin during neutrophil migration. Immunobiology 2003, 208, 391–399. [Google Scholar] [CrossRef]
- Li, W.; Lu, J.; Ma, Z.; Zhao, J.; Liu, J. An Integrated Model Based on a Six-Gene Signature Predicts Overall Survival in Patients With Hepatocellular Carcinoma. Front. Genet. 2020, 10, 1323. [Google Scholar] [CrossRef]
- Bornhauser, B.; Cario, G.; Rinaldi, A.; Risch, T.; Martinez, V.R.; Schütte, M.; Warnatz, H.-J.; Scheidegger, N.; Mirkowska, P.; Temperli, M.; et al. The hematopoietic stem cell marker VNN2 is associated with chemoresistance in pediatric B-cell precursor ALL. Blood Adv. 2020, 4, 4052–4064. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Kurota, Y.; Kato, T.; Ito, H.; Araki, A.; Nara, H.; Saitoh, S.; Tanaka, N.; Tsuchiya, N.; Asao, H. GPI-80 Augments NF-κB Activation in Tumor Cells. Int. J. Mol. Sci. 2021, 22, 12027. [Google Scholar] [CrossRef]
- Soler, D.C.; Kerstetter-Fogle, A.; Young, A.B.; Rayman, P.; Finke, J.H.; Debanne, S.M.; Cooper, K.D.; Barnholtz-Sloan, J.; Sloan, A.E.; McCormick, T.S. Healthy myeloid-derived suppressor cells express the surface ectoenzyme Vanin-2 (VNN2). Mol. Immunol. 2022, 142, 1–10. [Google Scholar] [CrossRef]
- Kato, T.; Takeda, Y.; Ito, H.; Kurota, Y.; Yamagishi, A.; Sakurai, T.; Naito, S.; Araki, A.; Nara, H.; Asao, H.; et al. GPI-80 as a Useful Index for Myeloid Cell Heterogeneity and a Potential Prognostic Biomarker for Metastatic Renal Cell Carcinoma. Tohoku J. Exp. Med. 2019, 249, 203–212. [Google Scholar] [CrossRef]


| Variables | N = 212 | Responders (N = 65) | Non-Responders (N = 147) | p-Value * |
|---|---|---|---|---|
| Sex (females), n (%) | 68 (32.1) | 16 (24.6) | 52 (35.4) | 0.17 |
| Age (years), median (IQR) | 62 (70.3–49) | 64 (72–57) | 60 (69–47.5) | 0.04 * |
| Histological subtype, n (%) | ||||
| Acral Melanoma | 5 (2.4) | 1 (1.5) | 4 (2.7) | 1.00 |
| Cutaneous Melanoma | 154 (72.6) | 37 (56.9) | 117 (79.6) | 0.001 * |
| Melanoma of Unknown primary | 7 (3.3) | 1 (1.5) | 6 (4.1) | 0.68 |
| Unknown | 46 (21.7) | 26 (40) | 20 (13.6) | 0.00 |
| Immunotherapy Type, n (%) | ||||
| Anti-CTLA-4 | 174 (82.1) | 44 (67.7) | 130 (88.4) | 0.001 * |
| Anti-PD-1 | 38 (17.9) | 21 (32.3) | 17 (11.6) | |
| Overall survival status, n (%) | ||||
| Living-1 | 135 (63.7) | 13 (20) | 122 (83) | 0.00 * |
| Diseased-0 | 77 (36.3) | 52 (80) | 25 (17) | |
| Log2(TMB+1), median (IQR) | 3.4 (4.5–2.2) | 4.1 (5–3.1) | 3.16 (4.3–1.9) | 0.00 * |
| FGA, median (IQR) | 0.3 (0.5–0.2) | 0.3 (0.5–0.2) | 0.4 (0.5–0.2) | 0.21 |
| Training set, n (%) | 169 (80) | - | - | |
| Testing set, n (%) | 43 (20) | - | - |
| Model | Mean Bootstrap Estimate | 95% CI | 10-Fold CV | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|---|---|
| RFC7 | 0.73 | 0.60–0.86 | 0.67 | NR: 0.81 R: 0.57 | NR: 0.91 R: 0.36 | NR: 0.85 R: 0.44 | 0.71 |
| TMB alone | 0.67 | 0.50–0.84 | 0.61 | NR: 0.76 R: 0.43 | NR: 0.73 R: 0.46 | NR: 0.75 R: 0.44 | 0.68 |
| RFC-Seq | 0.75 | 0.54–0.93 | 0.67 | NR: 0.79 R: 1.0 | NR: 1.0 R: 0.38 | NR: 0.88 R: 0.55 | 0.87 |
| GSTA3 alone | 0.83 | 0.62–1.00 | 0.65 | NR: 0.75 R: 0.43 | NR: 0.79 R: 0.38 | NR: 0.77 R: 0.40 | 0.80 |
| RFC-Surv | 0.76 | 0.54–0.91 | 0.68 | NR: 0.78 R: 0.75 | NR: 0.95 R: 0.38 | NR: 0.86 R: 0.50 | 0.76 |
| VNN2 alone | 0.84 | 0.63–1.00 | 0.70 | NR: 0.76 R: 0.76 | NR: 0.89 R: 0.44 | NR: 0.82 R: 0.53 | 0.82 |
| RFC16 | 0.77 | 0.58–0.92 | 0.67 | NR: 0.77 R: 0.60 | NR: 0.89 R: 0.38 | NR: 0.83 R: 0.46 | 0.88 |
| Variable | Category | N (%) | Univariate | Multivariate | ||
|---|---|---|---|---|---|---|
| HR (95% CI) | p-Value * | HR (95% CI) | p-Value | |||
| Age | (>66) | 49 (55.7) | - | - | - | - |
| (<66) | 39 (44.3) | 1.09 (0.63–1.88) | 0.76 | - | - | |
| Sex | Female | 34 (38.6) | - | - | - | - |
| Male | 54 (61.4) | 0.61 (0.35–1.06) | 0.08 | - | - | |
| CD86 | High | 67 (75.3) | - | - | - | - |
| Low | 22 (24.7) | 1.96 (1.09–3.51) | 0.02 * | 0.83 (0.36–1.92) | 0.67 | |
| FCRL6 | High | 64 (71.9) | - | - | - | - |
| Low | 25 (28.1) | 1.95 (1.09–3.49) | 0.03 * | 0.61 (0.24–1.56) | 0.30 | |
| PARP15 | High | 67 (76.1) | - | - | - | - |
| Low | 21 (23.6) | 2.19 (1.17–4.09) | 0.01 * | 1.40 (0.60–3.26) | 0.43 | |
| ZNF831 | High | 60 (68.2) | - | - | - | - |
| Low | 28 (31.8) | 2.62 (1.48–4.64) | <0.01 * | 0.95 (0.29–3.11) | 0.94 | |
| LCK | High | 63 (71.6) | - | - | - | - |
| Low | 25 (28.4) | 3.03 (1.72–5.33) | <0.001 * | 4.33 (0.91–20.52) | 0.07 | |
| VNN2 | High | 63 (71.6) | - | - | - | - |
| Low | 25 (28.4) | 2.49 (1.41–4.42) | <0.01 * | 1.69 (0.76–3.77) | 0.20 | |
| IL4I1 | High | 55 (62.5) | - | - | - | - |
| Low | 33 (37.5) | 1.76 (1.02–3.05) | 0.04 * | 1.03 (0.50–2.21) | 0.94 | |
| CCL5 | High | 65 (73.9) | - | - | - | - |
| Low | 23 (26.1) | 2.46 (1.38–4.38) | <0.01 * | 0.78 (0.23–2.62) | 0.69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, Y.B.; Al-Bzour, A.N.; Ababneh, O.E.; Abushukair, H.M.; Saeed, A. Genomic and Transcriptomic Predictors of Response to Immune Checkpoint Inhibitors in Melanoma Patients: A Machine Learning Approach. Cancers 2022, 14, 5605. https://doi.org/10.3390/cancers14225605
Ahmed YB, Al-Bzour AN, Ababneh OE, Abushukair HM, Saeed A. Genomic and Transcriptomic Predictors of Response to Immune Checkpoint Inhibitors in Melanoma Patients: A Machine Learning Approach. Cancers. 2022; 14(22):5605. https://doi.org/10.3390/cancers14225605
Chicago/Turabian StyleAhmed, Yaman B., Ayah N. Al-Bzour, Obada E. Ababneh, Hassan M. Abushukair, and Anwaar Saeed. 2022. "Genomic and Transcriptomic Predictors of Response to Immune Checkpoint Inhibitors in Melanoma Patients: A Machine Learning Approach" Cancers 14, no. 22: 5605. https://doi.org/10.3390/cancers14225605
APA StyleAhmed, Y. B., Al-Bzour, A. N., Ababneh, O. E., Abushukair, H. M., & Saeed, A. (2022). Genomic and Transcriptomic Predictors of Response to Immune Checkpoint Inhibitors in Melanoma Patients: A Machine Learning Approach. Cancers, 14(22), 5605. https://doi.org/10.3390/cancers14225605






