EMT-Related Genes Have No Prognostic Relevance in Metastatic Colorectal Cancer as Opposed to Stage II/III: Analysis of the Randomised, Phase III Trial FIRE-3 (AIO KRK 0306; FIRE-3)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Patients
2.3. Gene-Expression Analysis
2.4. EMT-Related Dataset
2.5. Outcomes
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Model | Hyperparameter | Searching Range |
| Random Survival Forest | Minimum Node Size | (1, 50) |
| Number of variables to possibly split at in each node | (0, 1) | |
| Fraction of observations to sample | (0.1, 1) | |
| Sample with replacement | (TRUE, FALSE) | |
| Splitting rule | (maxstat, logrank) | |
| Cox (regularised) | Elastic net mixing parameter alpha; penalty parameter is trained internally via 10-fold CV | (0, 1) |
| Gradient Boosting Tree | Maximum depth of a tree | (1, 20) |
| Subsample ratio of the training instances | (0.1, 1.0) | |
| Number of rounds | (10, 5000) | |
| Step size shrinkage | (0.01, 1.0) | |
| Subsample ratio of columns when constructing each tree | (0.01, 1.0) | |
| Minimum loss reduction required to make a further partition on a leaf node of the tree | (0.01, 3) | |
| The way new nodes are added to the tree | (Depth-wise, Loss-guide) |
References
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Manfredi, S.; Lepage, C.; Hatem, C.; Coatmeur, O.; Faivre, J.; Bouvier, A.M. Epidemiology and management of liver metastases from colorectal cancer. Ann. Surg. 2006, 244, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, S.; Bouvier, A.M.; Lepage, C.; Hatem, C.; Dancourt, V.; Faivre, J. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br. J. Surg. 2006, 93, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Elferink, M.A.G.; de Jong, K.P.; Klaase, J.M.; Siemerink, E.J.; de Wilt, J.H.W. Metachronous metastases from colorectal cancer: A population-based study in North-East Netherlands. Int. J. Colorectal Dis. 2015, 30, 205–212. [Google Scholar] [CrossRef]
- Brandi, G.; de Lorenzo, S.; Nannini, M.; Curti, S.; Ottone, M.; Dall’Olio, F.G.; Barbera, M.; Pantaleo, M.A.; Biasco, G. Adjuvant chemotherapy for resected colorectal cancer metastases: Literature review and meta-analysis. World J. Gastroenterol. 2016, 22, 519–533. [Google Scholar] [CrossRef]
- Colorectal Cancer Early Detection, Diagnosis, and Staging; American Cancer Society: Atlanta, GA, USA, 2022.
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; van Krieken, J.H.; Aderka, D.; Aguilar, E.A.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef]
- Benson, B.; Venook, A.P.; Cederquist, L.; Chan, E.M.; Chen, Y.; Cooper, H.S.; Fichera, A.; Grem, J.L.; Grothey, A.; Hochster, H.S.; et al. Journal of the national comprehensive cancer network, colon cancer, version 1.2021, NCCN clinical practice guidelines in oncology. NCCN Clin. Pract. Guidel. Oncol. 2021, 19, 329–359. [Google Scholar]
- Leitlinienprogramm Onkologie (Deutsche Krebsgesellschaft, Deutsche Krebshilfe and L. Onkologie). AWMF Regist. 021/007OL; S3-Leitlinie Kolorektales Karzinom; Langversion 2.1. 2019; pp. 1–328.
- Yoshino, T.; Arnold, D.; Taniguchi, H.; Pentheroudakis, G.; Yamazaki, K.; Xu, R.H.; Kim, T.W.; Ismail, F.; Tan, I.B.; Yeh, K.H.; et al. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: A JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann. Oncol. 2018, 29, 44–70. [Google Scholar] [CrossRef]
- Ren, H.; Bösch, F.; Pretzsch, E.; Jacob, S.; Westphalen, C.B.; Holch, J.W.; Werner, J.; Angele, M.K. Identification of an EMT-related gene signature predicting recurrence in stage II/III colorectal cancer—A retrospective study in 1780 patients. Ann. Surg. 2022, 276, 1–25. [Google Scholar] [CrossRef]
- Cao, M.L.H.; Xu, E.; Liu, H.; Wan, L. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol. Resarch Pract. 2015, 211, 557–569. [Google Scholar] [CrossRef]
- Pretzsch, E.; Bösch, F.; Neumann, J.; Ganschow, P.; Bazhin, A.; Guba, M.; Werner, J.; Angele, M.K. Mechanisms of metastasis in colorectal cancer and metastatic organotropism: Hematogenous versus peritoneal spread. J. Oncol. 2019, 2019, 7407190. [Google Scholar] [CrossRef] [PubMed]
- Stintzing, S.; Wirapati, P.; Lenz, H.-J.; Neureiter, D.; von Weikersthal, L.F.; Decker, T.; Kiani, A.; Kaiser, F.; Al-Batran, S.; Heintges, T.; et al. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann. Oncol. 2019, 30, 1796–1803. [Google Scholar] [CrossRef]
- Findlay, V.J.; Wang, C.; Watson, D.K.; Camp, E.R. Epithelial-to-mesenchymal transition and the cancer stem cell phenotype: Insights from cancer biology with therapeutic implications for colorectal cancer. Cancer Gene Ther. 2014, 21, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhao, E. Development and validation of a robust epithelial-mesenchymal transition (EMT)-related prognostic signature for hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101587. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Xiao, Y.; Li, J.; Hong, L.; Zhang, J.; Pei, M.; Lin, J.; Liu, S.; Wu, X.; Xiang, L.; et al. Identification of an EMT-related gene signature for predicting overall survival in gastric cancer. Front. Genet. 2021, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, V.; von Weikersthal, L.F.; Decker, T.; Kiani, A.; Vehling-Kaiser, U.; Al-Batran, S.-E.; Heintges, T.; Lerchenmüller, C.; Kahl, C.; Seipelt, G.; et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014, 15, 1065–1075. [Google Scholar] [CrossRef]
- Stintzing, S.; Modest, D.; Rossius, L.; Lerch, M.; von Weikersthal, L.F.; Decker, T.; Kiani, A.; Vehling-Kaiser, U.; Al-Batran, S.; Heintges, T.; et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): A post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol. 2016, 17, 1426–1434. [Google Scholar] [CrossRef]
- Stintzing, S.; Miller-Phillips, L.; Moddest, D.; von Weikersthal, L.F.; Decker, T.; Kiani, A.; Vehling-Kaiser, U.; Al-Batran, S.; Heintges, T. Impact of BRAF and RAS mutations on first-line efficacy of FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab: Analysis of the FIRE-3 (AIO KRK-0306) study. Eur. J. Cancer 2017, 79, 50–60. [Google Scholar] [CrossRef]
- Stahler, A.; Stintzing, S.; von Einem, J.C.; Westphalen, C.B.; Heinrich, K.; Krämer, N.; Michl, M.; Modest, D.P.; von Weikersthal, L.F.; Decker, T.; et al. Single-nucleotide variants, tumour mutational burden and microsatellite instability in patients with metastatic colorectal cancer: Next-generation sequencing results of the FIRE-3 trial. Eur. J. Cancer 2020, 137, 250–259. [Google Scholar] [CrossRef]
- Modest, D.P.; Heinemann, V.; Folprecht, G.; Denecke, T.; Pratschke, J.; Lang, H.; Bemelmans, M.; Becker, T.; Rentsch, M.; Seehofer, D.; et al. Factors that influence conversion to resectability and survival after resection of metastases in RAS WT metastatic colorectal cancer (mCRC): Analysis of FIRE-3- AIOKRK0306. Ann. Surg. Oncol. 2020, 27, 2389–2401. [Google Scholar] [CrossRef]
- Stahler, A.; Heinemann, V.; Holch, J.W.; Einem, J.C.; Westphalen, C.B.; Heinrich, K.; Schlieker, L.; Jelas, I.; Alig, A.H.S.; Fischer, L.E.; et al. Mutational profiles of metastatic colorectal cancer treated with FOLFIRI plus cetuximab or bevacizumab before and after secondary resection (AIO KRK 0306; FIRE-3). Int. J. Cancer 2021, 149, 1935–1943. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, E.L.; Meier, P. Nonparametric estimation from incomplete samples. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed]
- MWright, N.; Ziegler, A. Ranger: A fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the KDD ’16: 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Graf, E.; Schmoor, C.; Sauerbrei, W.; Schumacher, M. Assessment and comparison of prognostic classification schemes for survival data. Stat. Med. 1999, 18, 2529–2545. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Sonabend, R.; Király, F.J.; Bender, A.; Bischl, B.; Lang, M. mlr3proba: An R package for machine learning in survival analysis. Bioinformatics 2021, 37, 2789–2791. [Google Scholar] [CrossRef]
- Lang, M.; Binder, M.L.M.; Richter, J.; Schratz, P.; Pfisterer, F.; Coores, S.; Au, Q.; Casalicchio, G.; Kotthoff, L.; Bischl, B.; et al. mlr3: A modern object-oriented machine learning framework in R. J. Open Source Softw. 2019, 4, 1903. [Google Scholar] [CrossRef]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef]
- Mittal, V. Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. Mech. Dis. 2018, 13, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Schlicker, A.; Ellappalayam, A.; Beumer, I.J.; Snel, M.H.J.; Mittempergher, L.; Diosdado, B.; Dreezen, C.; Tian, S.; Salazar, R.; Loupakis, F.; et al. Investigating the concordance in molecular subtypes of primary colorectal tumors and their matched synchronous liver metastasis. Int. J. Cancer 2020, 147, 2303–2315. [Google Scholar] [CrossRef] [PubMed]
| Sub Category | Frequency | Percent (%) | |
|---|---|---|---|
| Total valid records | 350 | 100 | |
| Gender | Female | 113 | 32.28 |
| Male | 237 | 67.71 | |
| Type of treatment | Cetuximab | 165 | 47.14 |
| Bevacizumab | 185 | 52.86 | |
| Tumour location | Left | 273 | 78.00 |
| Right | 77 | 22.00 | |
| Solitary liver metastasis | Yes | 116 | 33.14 |
| No | 234 | 66.86 | |
| BRAFV600E | Wt | 265 | 75.71 |
| Mut | 20 | 5.71 | |
| Not tested | 65 | 18.57 | |
| Overall survival (OS) | Censored | 37 | 10.57 |
| Dead | 313 | 89.43 | |
| Progression-free survival (PFS) | Censored | 21 | 6.00 |
| Progression or dead | 329 | 94.00 |
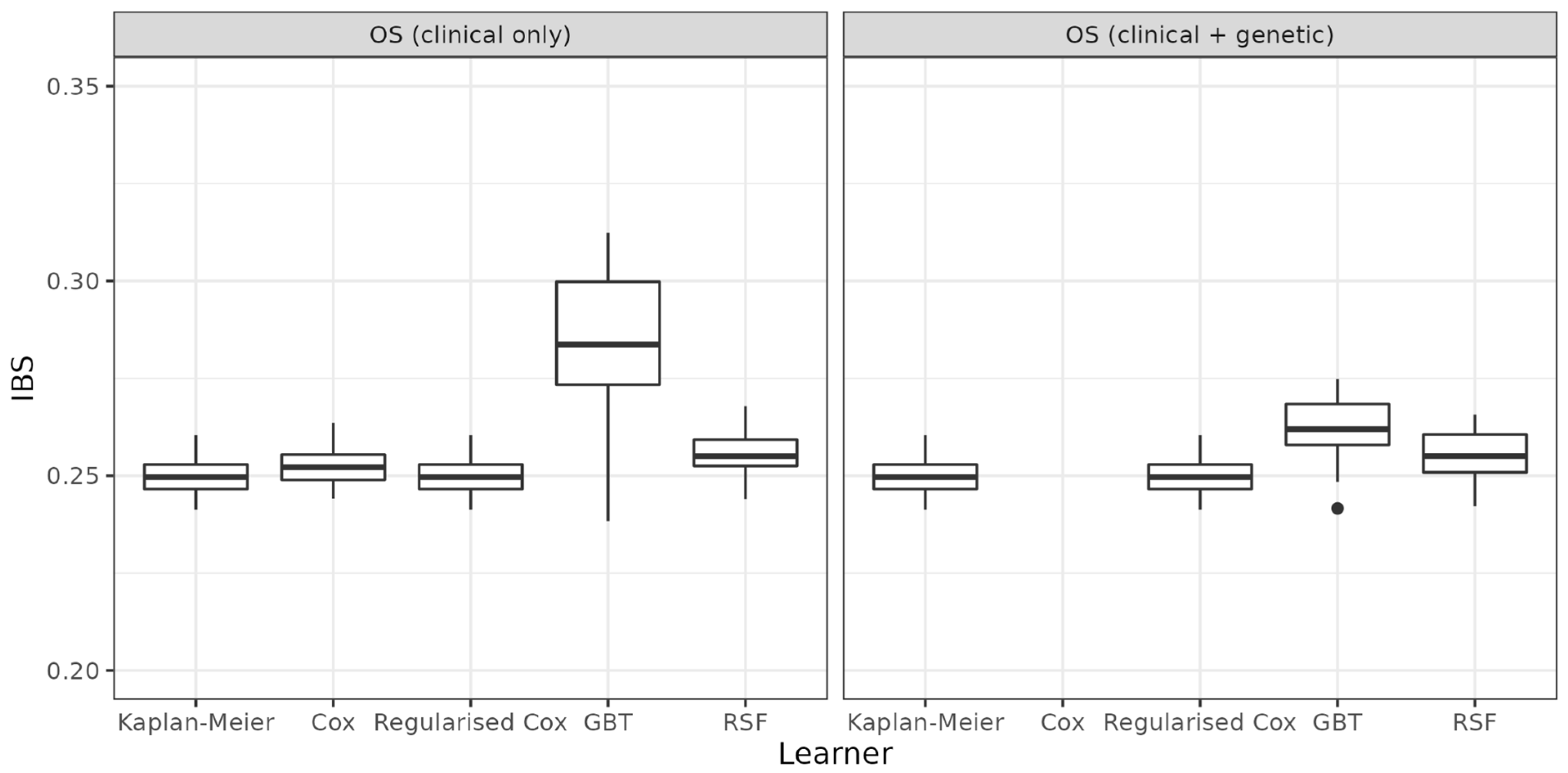
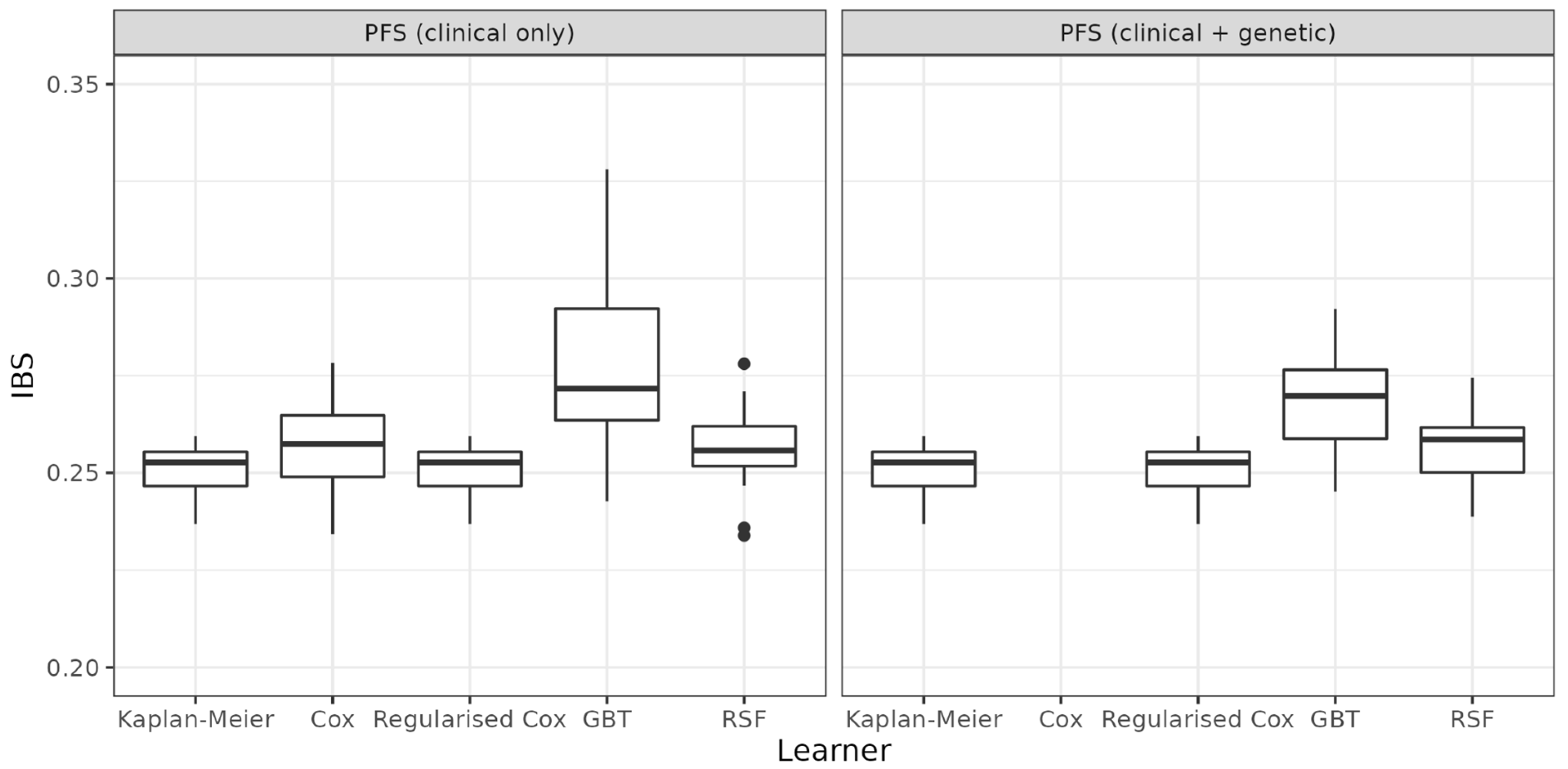
| Task | Learner | Mean (sd) | Median |
|---|---|---|---|
| OS (clinical only) | Kaplan-Meier | 0.25 (0.005) | 0.2496 |
| Cox | 0.253 (0.006) | 0.2522 | |
| Regularised Cox | 0.25 (0.005) | 0.2496 | |
| GBT | 0.284 (0.019) | 0.2837 | |
| RSF | 0.256 (0.007) | 0.2550 | |
| OS (clinical + genetic) | Kaplan-Meier | 0.250 (0.005) | 0.2496 |
| Cox | — | — | |
| Regularised Cox | 0.25 (0.005) | 0.2496 | |
| GBT | 0.262 (0.009) | 0.2619 | |
| RSF | 0.255 (0.007) | 0.2551 | |
| PFS (clinical only) | Kaplan-Meier | 0.251 (0.007) | 0.2527 |
| Cox | 0.257 (0.013) | 0.2574 | |
| Regularised Cox | 0.251 (0.007) | 0.2527 | |
| GBT | 0.276 (0.021) | 0.2717 | |
| RSF | 0.256 (0.011) | 0.2557 | |
| PFS (clinical + genetic) | Kaplan-Meier | 0.251 (0.007) | 0.2527 |
| Cox | — | — | |
| Regularised Cox | 0.251 (0.007) | 0.2527 | |
| GBT | 0.268 (0.013) | 0.2697 | |
| RSF | 0.258 (0.009) | 0.2585 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pretzsch, E.; Heinemann, V.; Stintzing, S.; Bender, A.; Chen, S.; Holch, J.W.; Hofmann, F.O.; Ren, H.; Bösch, F.; Küchenhoff, H.; et al. EMT-Related Genes Have No Prognostic Relevance in Metastatic Colorectal Cancer as Opposed to Stage II/III: Analysis of the Randomised, Phase III Trial FIRE-3 (AIO KRK 0306; FIRE-3). Cancers 2022, 14, 5596. https://doi.org/10.3390/cancers14225596
Pretzsch E, Heinemann V, Stintzing S, Bender A, Chen S, Holch JW, Hofmann FO, Ren H, Bösch F, Küchenhoff H, et al. EMT-Related Genes Have No Prognostic Relevance in Metastatic Colorectal Cancer as Opposed to Stage II/III: Analysis of the Randomised, Phase III Trial FIRE-3 (AIO KRK 0306; FIRE-3). Cancers. 2022; 14(22):5596. https://doi.org/10.3390/cancers14225596
Chicago/Turabian StylePretzsch, Elise, Volker Heinemann, Sebastian Stintzing, Andreas Bender, Shuo Chen, Julian Walter Holch, Felix Oliver Hofmann, Haoyu Ren, Florian Bösch, Helmut Küchenhoff, and et al. 2022. "EMT-Related Genes Have No Prognostic Relevance in Metastatic Colorectal Cancer as Opposed to Stage II/III: Analysis of the Randomised, Phase III Trial FIRE-3 (AIO KRK 0306; FIRE-3)" Cancers 14, no. 22: 5596. https://doi.org/10.3390/cancers14225596
APA StylePretzsch, E., Heinemann, V., Stintzing, S., Bender, A., Chen, S., Holch, J. W., Hofmann, F. O., Ren, H., Bösch, F., Küchenhoff, H., Werner, J., & Angele, M. K. (2022). EMT-Related Genes Have No Prognostic Relevance in Metastatic Colorectal Cancer as Opposed to Stage II/III: Analysis of the Randomised, Phase III Trial FIRE-3 (AIO KRK 0306; FIRE-3). Cancers, 14(22), 5596. https://doi.org/10.3390/cancers14225596







