Construction of Oxidative Stress-Related Genes Risk Model Predicts the Prognosis of Uterine Corpus Endometrial Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Differential Gene Expression Screening
2.3. Functional Enrichment Analysis of DEGs
2.4. The Prognostic Values of the Oxidative Stress-Related DEGs
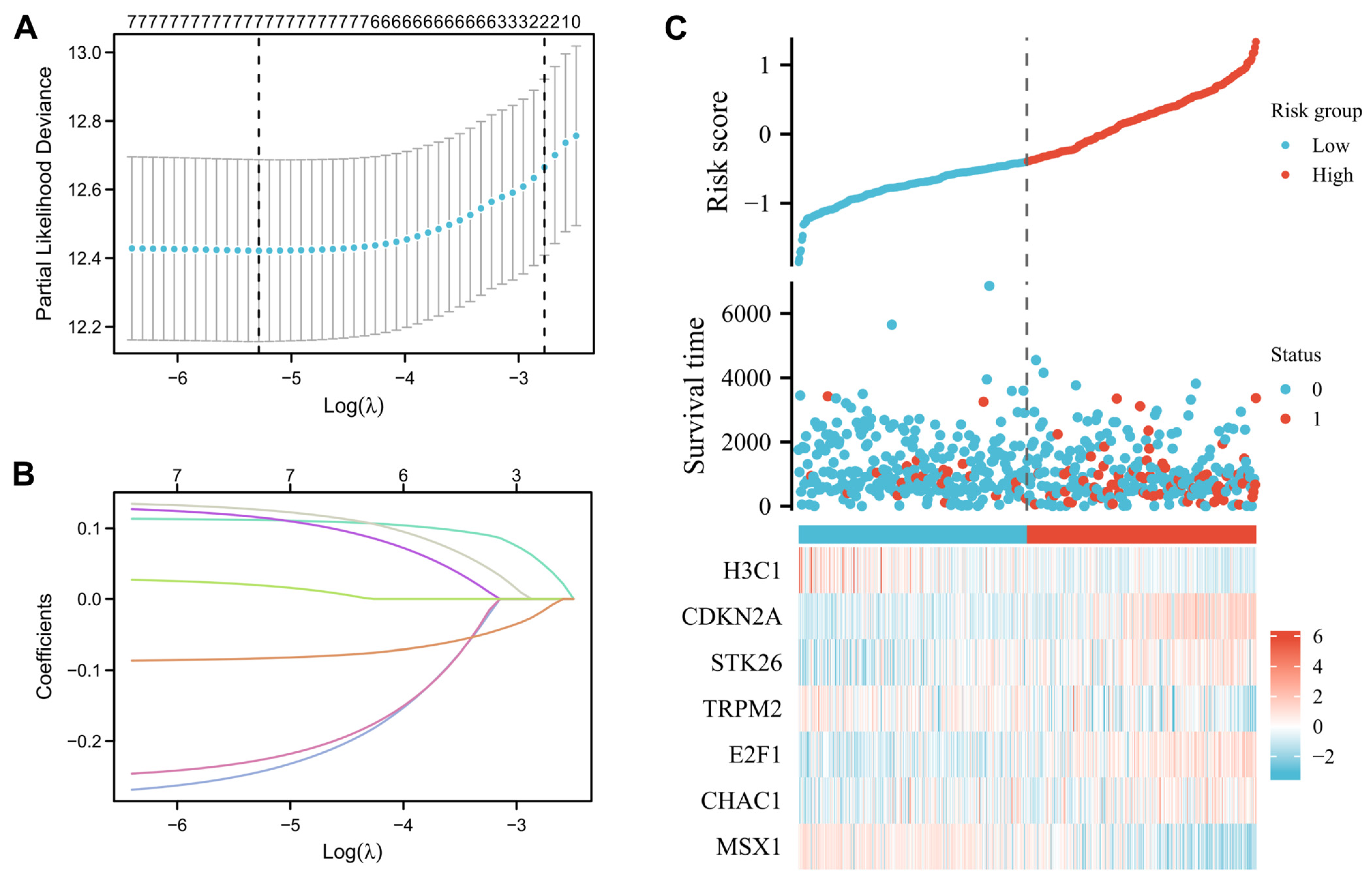
2.5. Development of a Prognostic Gene Signature Based on the Oxidative Stress-Related DEGs
2.6. Somatic Mutation Analysis between Two Subgroups
2.7. Immune Landscape Differences between Two Subgroups
2.8. Differential Analysis of Immune Cell Infiltration and Immune Checkpoint between Two Subgroups
2.9. Verification of Prognosis-Related 7-OSRGs Expression
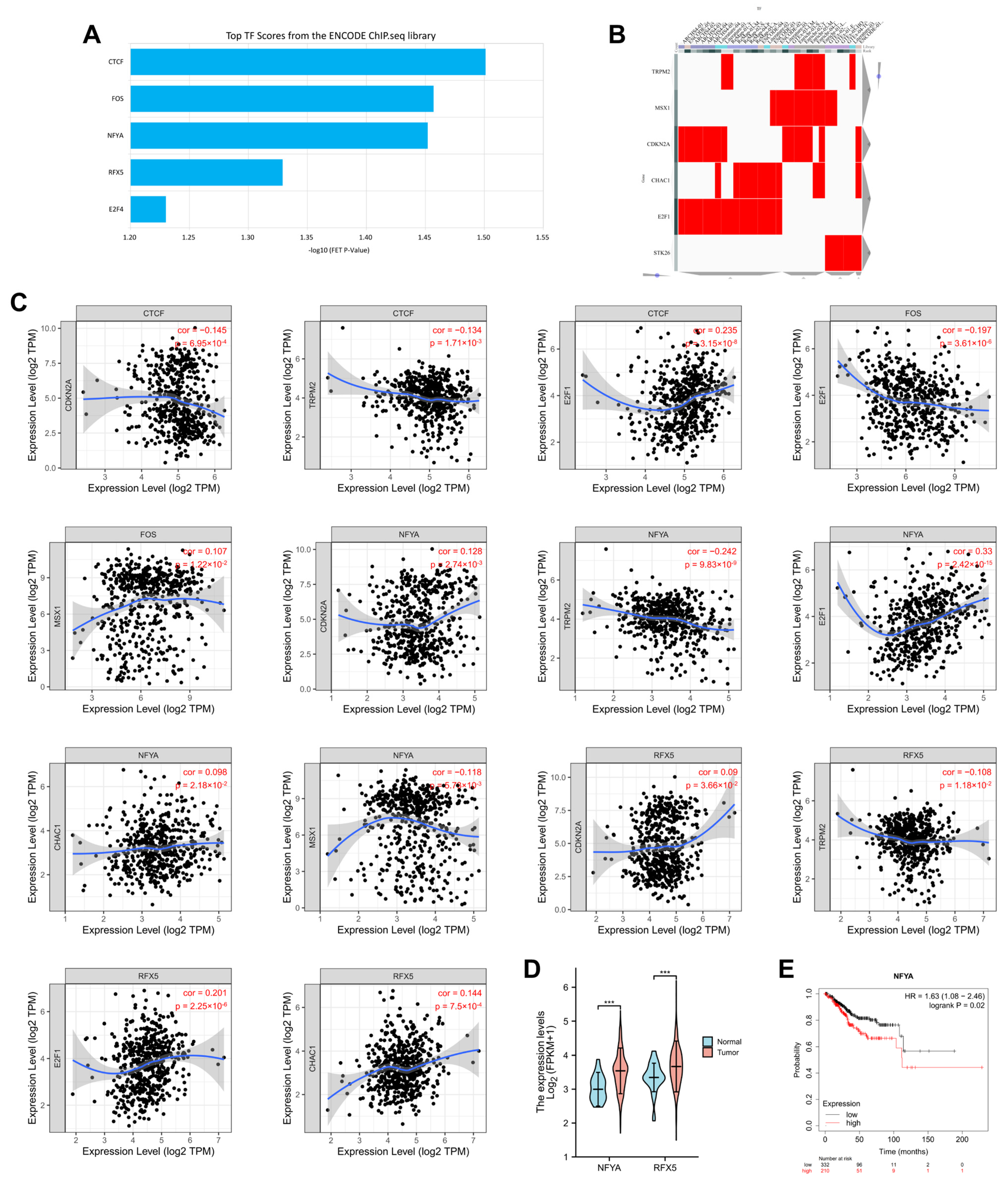
2.10. Prediction of Transcription Factors of 7 OSRGs in UCEC
3. Results
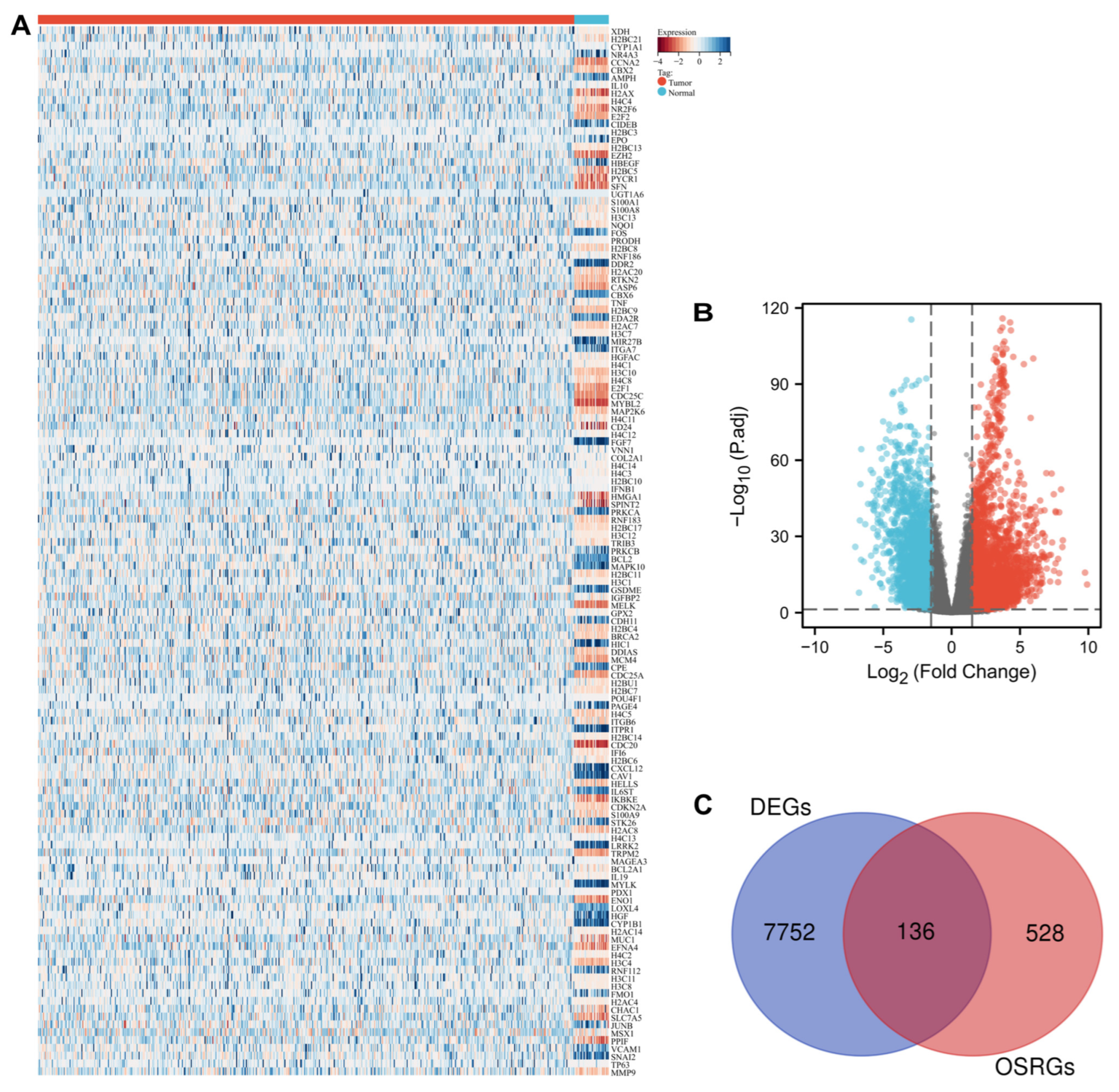
3.1. Identification of Oxidative Stress-Related DEGs in UCEC
3.2. Functional Analysis of Differentially Expressed OSRGs
3.3. Identification of OSR DEGs Related to the Prognosis in UCEC
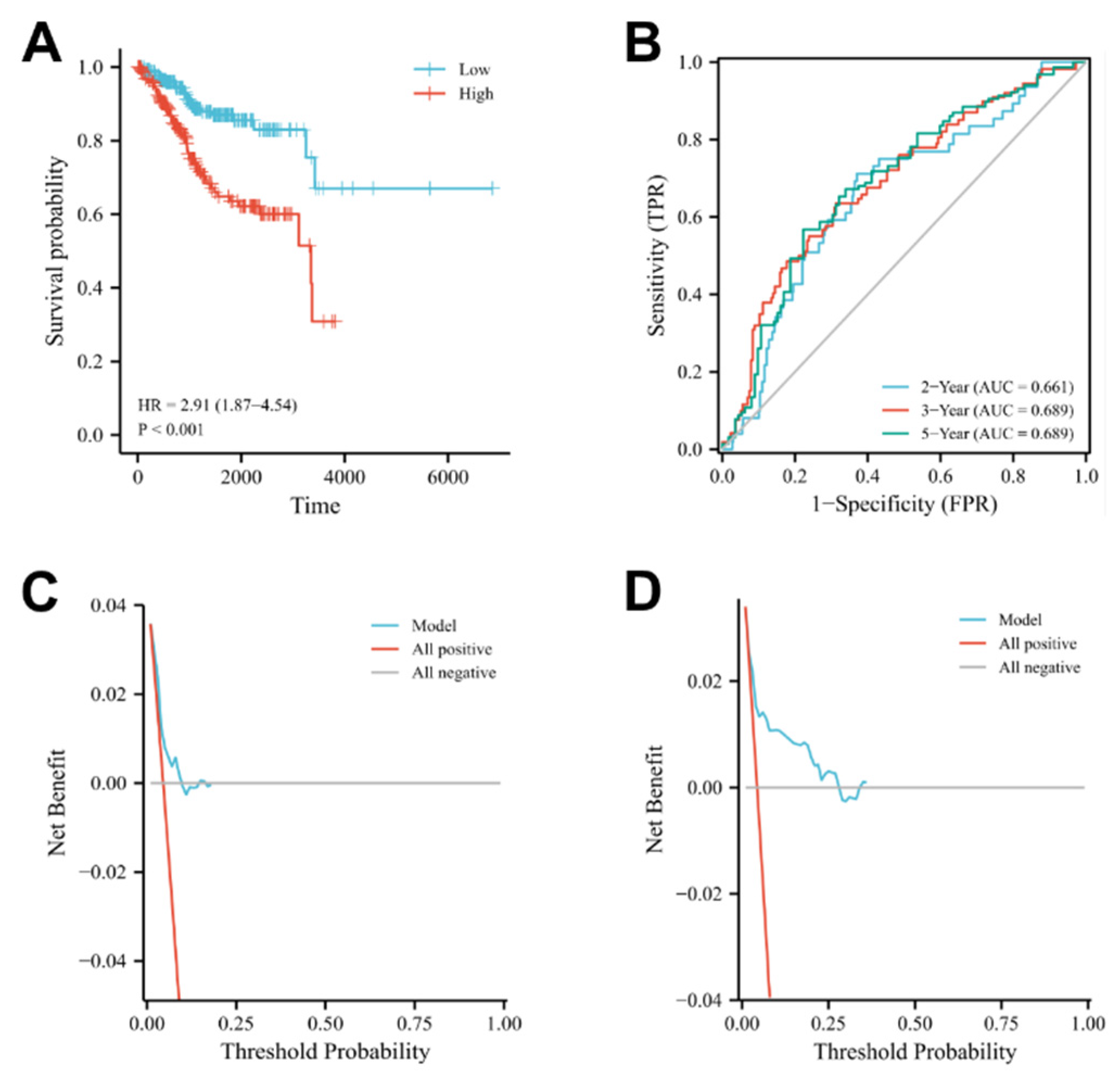
3.4. Construction and Prognostic Value of Differentially Expressed OSRGs
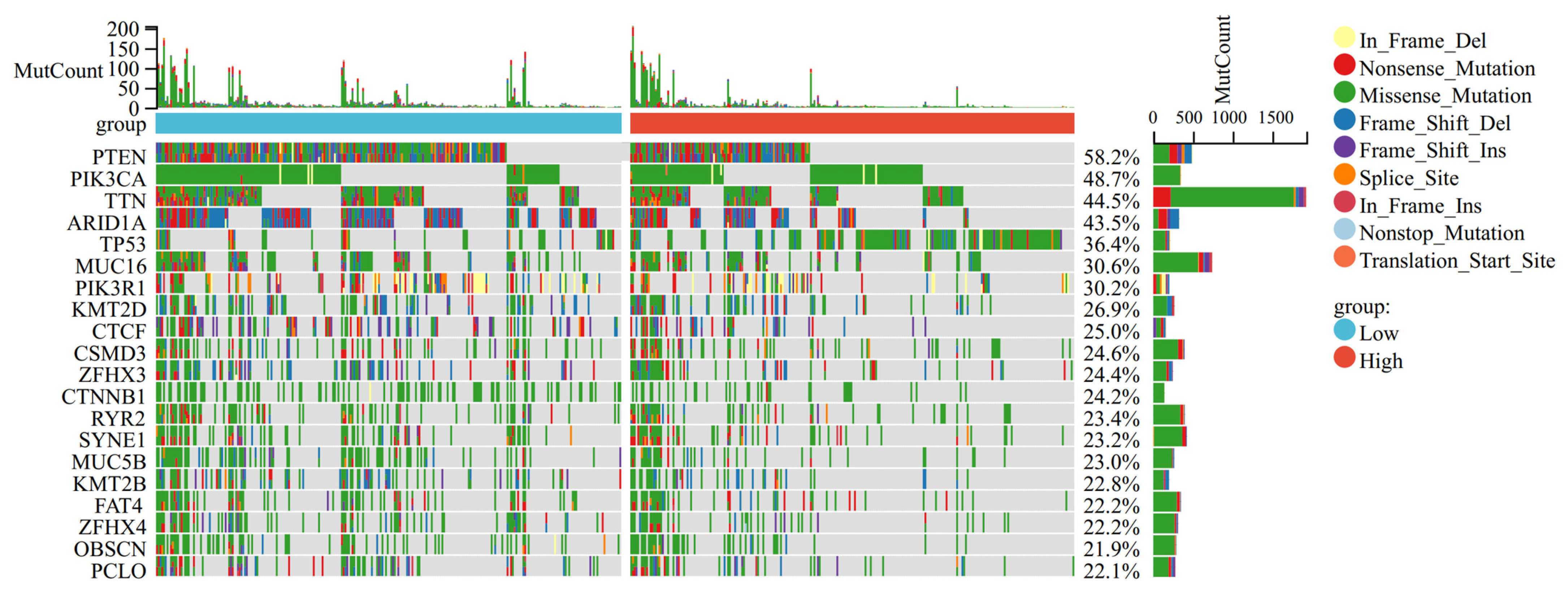
3.5. Mutation Landscape Associated with OSRGs Risk Scores
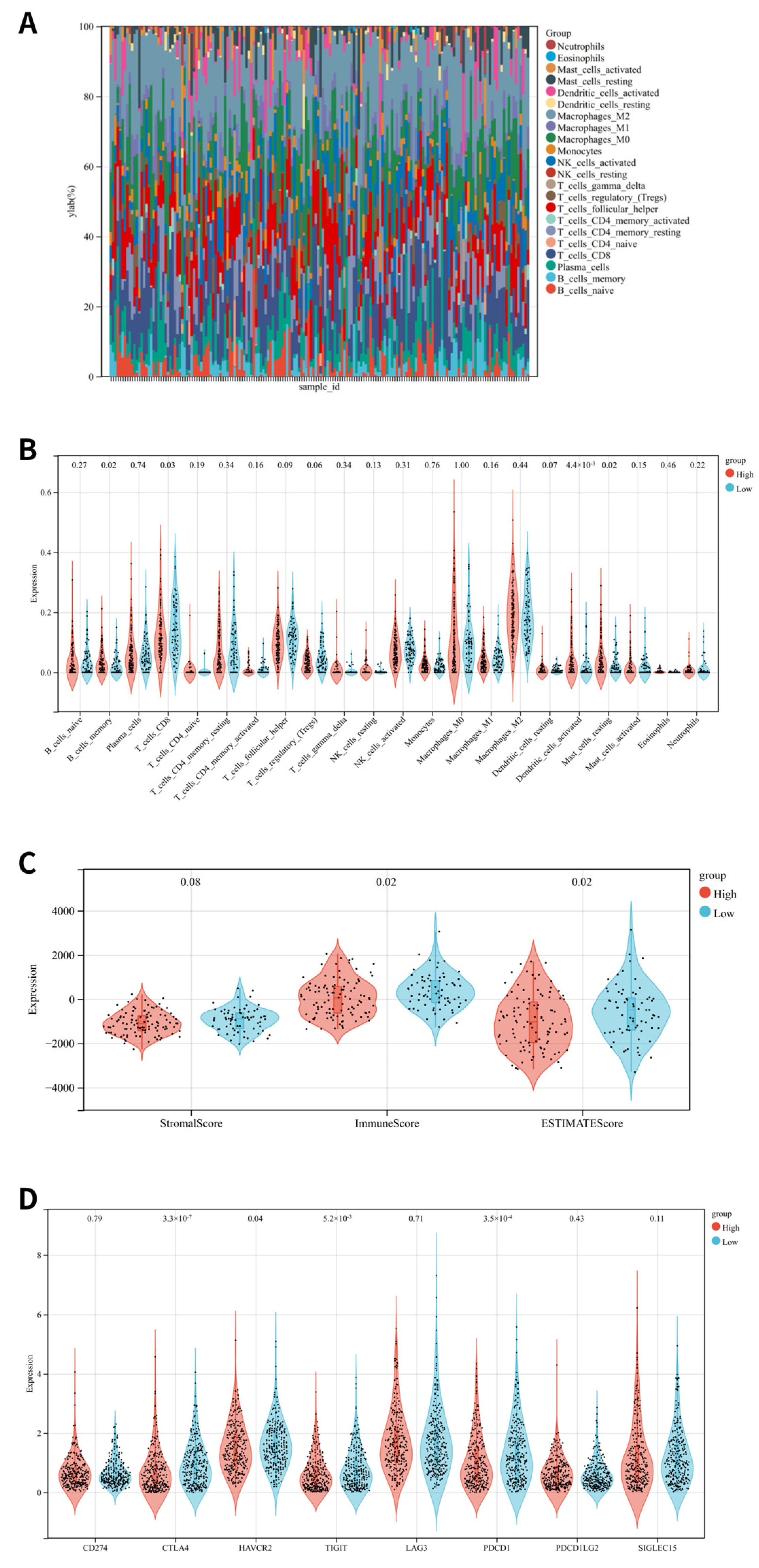
3.6. Immune Status Analysis
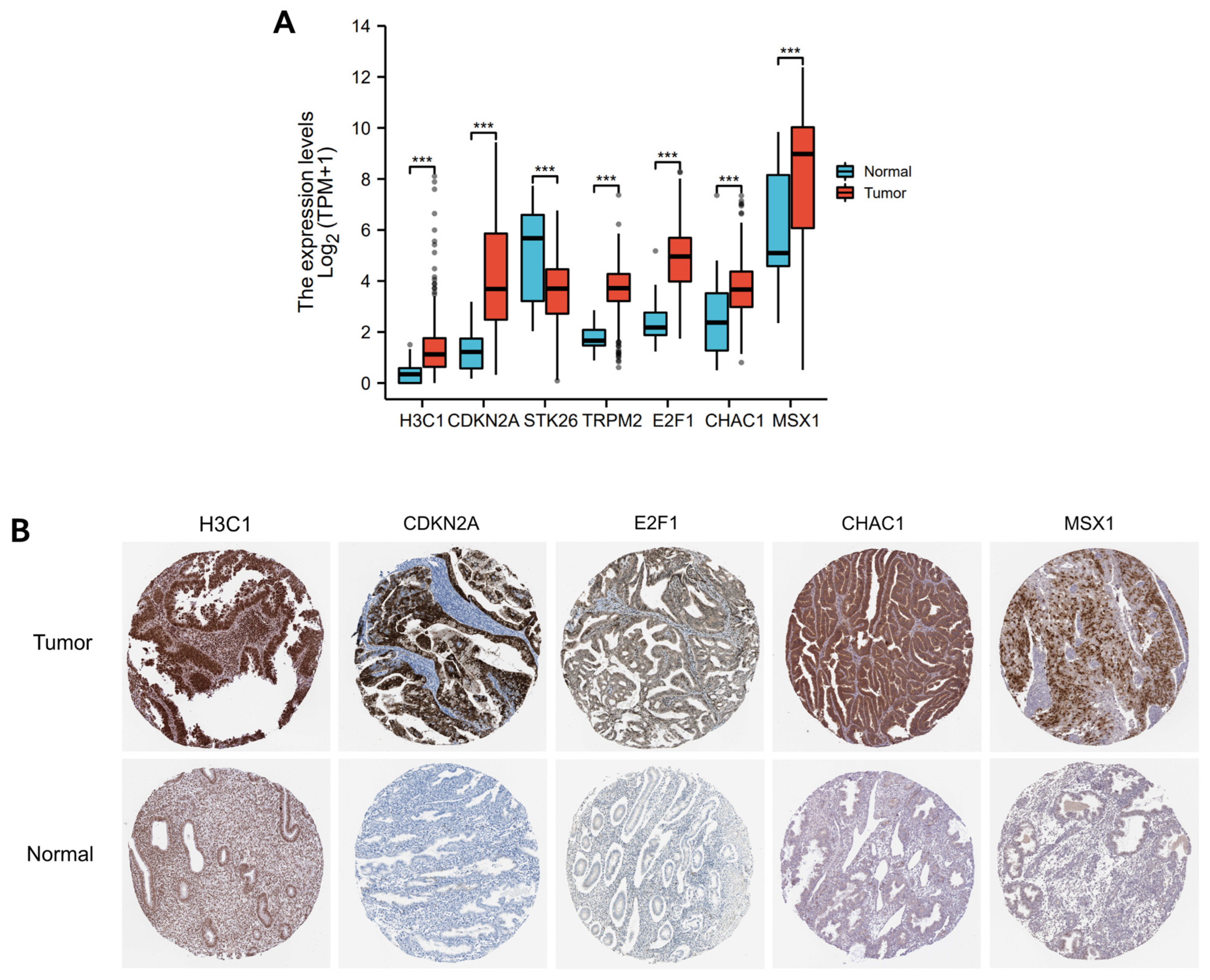
3.7. Immunohistochemistry Verification of 7 OSRGs Expression
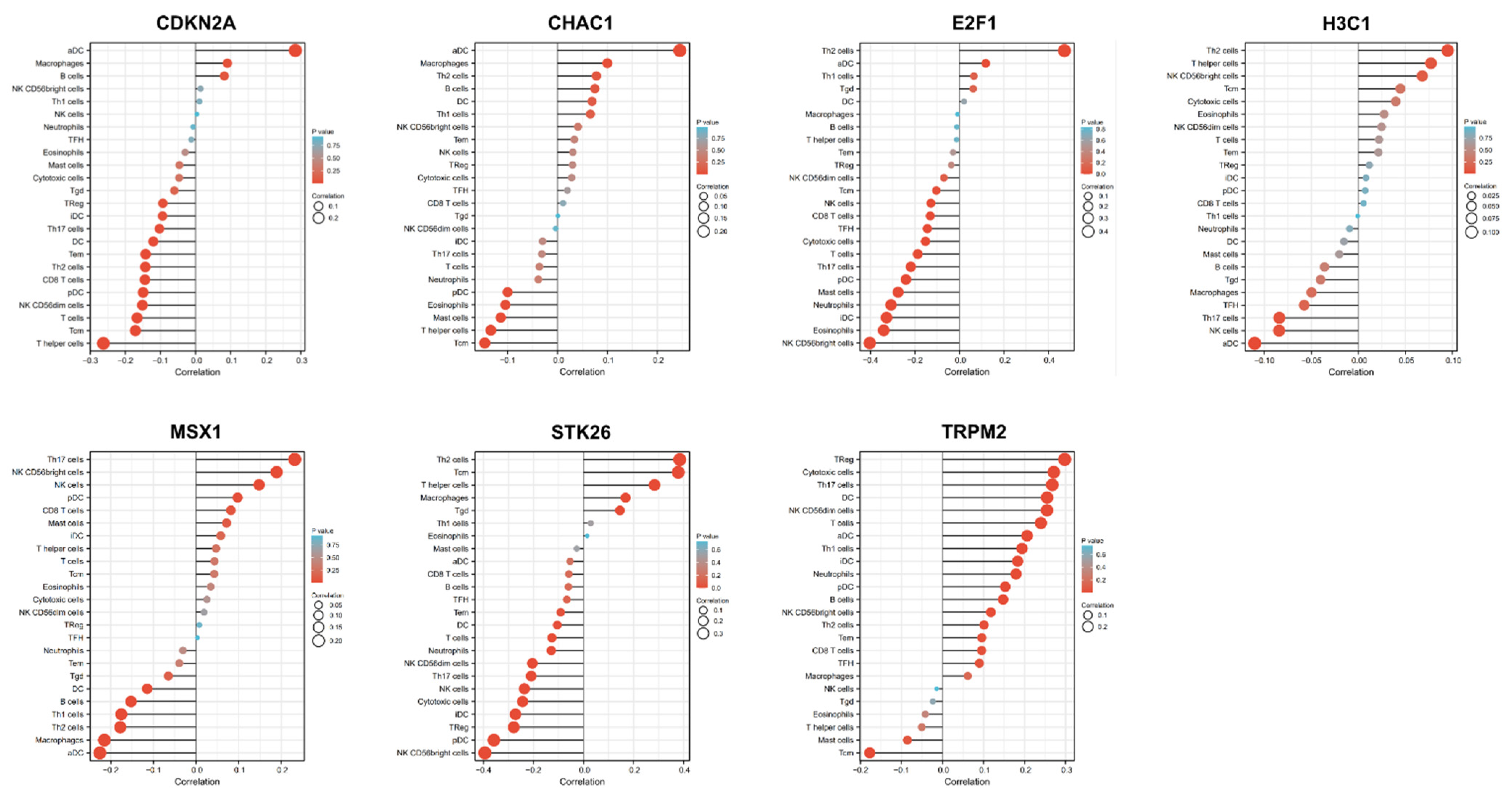
3.8. Identification of Transcription Factors of 7 OSRGs in UCEC
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Mara, T.A.; Glubb, D.M.; Amant, F.; Annibali, D.; Ashton, K.; Attia, J.; Auer, P.L.; Beckmann, M.W.; Black, A.; Bolla, M.K.; et al. Identification of nine new susceptibility loci for endometrial cancer. Nat. Commun. 2018, 9, 3166. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-Y.; Werner, H.M.; Li, J.; Westin, S.N.; Lu, Y.; Halle, M.K.; Trovik, J.; Salvesen, H.B.; Mills, G.B.; Liang, H. Integrative Protein-Based Prognostic Model for Early-Stage Endometrioid Endometrial Cancer. Clin. Cancer Res. 2016, 22, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Urick, M.E.; Bell, D.W. Clinical actionability of molecular targets in endometrial cancer. Nat. Cancer 2019, 19, 510–521. [Google Scholar] [CrossRef] [PubMed]
- De Boer, S.M.; Powell, M.E.; Mileshkin, L.; Katsaros, D.; Bessette, P.; Haie-Meder, C.; Ottevanger, P.B.; Ledermann, J.A.; Khaw, P.; Colombo, A.; et al. Adjuvant chemoradiotherapy versus radiotherapy alone for women with high-risk endometrial cancer (PORTEC-3): Final results of an international, open-label, multicentre, randomised, phase 3 trial. Lancet Oncol. 2018, 19, 295–309. [Google Scholar] [CrossRef]
- Soumerai, T.E.; Donoghue, M.T.; Bandlamudi, C.; Srinivasan, P.; Chang, M.T.; Zamarin, D.; Cadoo, K.A.; Grisham, R.N.; O’Cearbhaill, R.E.; Tew, W.P.; et al. Clinical Utility of Prospective Molecular Characterization in Advanced Endometrial Cancer. Clin. Cancer Res. 2018, 24, 5939–5947. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Berghe, T.V.; Vandenabeele, P.; Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef]
- Canli, Ö.; Alankuş, Y.B.; Grootjans, S.; Vegi, M.N.; Hültner, L.; Hoppe, P.S.; Schroeder, T.; Vandenabeele, P.; Bornkamm, G.W.; Greten, F.R. Glutathione peroxidase 4 prevents necroptosis in mouse erythroid precursors. Blood 2016, 127, 139–148. [Google Scholar] [CrossRef]
- Sun, Y.; Ai, J.Z.; Jin, X.; Liu, L.R.; Lin, T.H.; Xu, H.; Wei, Q.; Yang, L. IL-8 protects prostate cancer cells from GSK-3beta-induced oxidative stress by activating the mTOR signaling pathway. Prostate 2019, 79, 1180–1190. [Google Scholar] [CrossRef]
- Klaunig, J.E.; Kamendulis, L.M.; Hocevar, B.A. Oxidative Stress and Oxidative Damage in Carcinogenesis. Toxicol. Pathol. 2010, 38, 96–109. [Google Scholar] [CrossRef]
- Huo, L.; Li, C.W.; Huang, T.H.; Lam, Y.C.; Xia, W.; Tu, C.; Chang, W.C.; Hsu, J.L.; Lee, D.F.; Nie, L.; et al. Activation of Keap1/Nrf2 signaling pathway by nuclear epidermal growth factor receptor in cancer cells. Am. J. Transl. Res. 2014, 6, 649–663. [Google Scholar]
- Korbecki, J.; Baranowska-Bosiacka, I.; Gutowska, I.; Chlubek, D. The effect of reactive oxygen species on the synthesis of prostanoids from arachidonic acid. J. Physiol. Pharmacol. 2013, 64, 409–421. [Google Scholar] [PubMed]
- Matsuzawa, A.; Ichijo, H. Redox control of cell fate by MAP kinase: Physiological roles of ASK1-MAP kinase pathway in stress signaling. Biochim. Biophys. Acta BBA Gen. Subj. 2008, 1780, 1325–1336. [Google Scholar] [CrossRef] [PubMed]
- Barrera, G. Oxidative Stress and Lipid Peroxidation Products in Cancer Progression and Therapy. ISRN Oncol. 2012, 2012, 137289. [Google Scholar] [CrossRef] [PubMed]
- Jelic, M.; Mandic, A.; Maricic, S.; Srdjenovic, B. Oxidative stress and its role in cancer. J. Cancer Res. Ther. 2021, 17, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Liu, X.-L.; Liu, H.-S.; Luo, X.-Y.; Yuan, Y.; Ji, Y.-M.; Liu, T.; Guo, J.-L.; Zhang, J. The Risk Model Based on the Three Oxidative Stress-Related Genes Evaluates the Prognosis of LAC Patients. Oxidative Med. Cell. Longev. 2022, 2022, 4022896. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.; Feser, R.; Nault, B.; Hunter, S.; Maiti, S.; Ugwuagbo, K.C.; Majumder, M. miR526b and miR655 Induce Oxidative Stress in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4039. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, W.; Zhang, J.; You, Z.; Hu, T.; Shao, G.; Zhang, Z.; Xu, Z.; Yu, X. TWIST2 inhibits EMT and induces oxidative stress in lung cancer cells by regulating the FGF21-mediated AMPK/mTOR pathway. Exp. Cell Res. 2021, 405, 112661. [Google Scholar] [CrossRef]
- Punnonen, R.; Kudo, R.; Punnonen, K.; Hietanen, E.; Kuoppala, T.; Kainulainen, H.; Sato, K.; Ahotupa, M. Activities of antioxidant enzymes and lipid peroxidation in endometrial cancer. Eur. J. Cancer 1993, 29, 266–269. [Google Scholar] [CrossRef]
- Monge, M.; Colas, E.; Doll, A.; Gil-Moreno, A.; Castellvi, J.; Diaz, B.; Gonzalez, M.; Lopez-Lopez, R.; Xercavins, J.; Carreras, R.; et al. Proteomic approach to ETV5 during endometrial carcinoma invasion reveals a link to oxidative stress. Carcinogenesis 2009, 30, 1288–1297. [Google Scholar] [CrossRef]
- Carvalho, L.F.P.; Samadder, A.N.; Agarwal, A.; Fernandes, L.F.C.; Abrão, M.S. Oxidative stress biomarkers in patients with endometriosis: Systematic review. Arch. Gynecol. Obstet. 2012, 286, 1033–1040. [Google Scholar] [CrossRef]
- Turkyilmaz, E.; Yildirim, M.; Cendek, B.D.; Baran, P.; Alisik, M.; Dalgaci, F.; Yavuz, A.F. Evaluation of oxidative stress markers and intra-extracellular antioxidant activities in patients with endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 199, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Iwabuchi, T.; Yoshimoto, C.; Shigetomi, H.; Kobayashi, H. Oxidative Stress and Antioxidant Defense in Endometriosis and Its Malignant Transformation. Oxidative Med. Cell. Longev. 2015, 2015, 848595. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Yang, N.; Wang, S.; Liu, W.; Zhang, D.; Wang, J.; Huang, B.; Li, X. Identifying the Predictive Role of Oxidative Stress Genes in the Prognosis of Glioma Patients. Med. Sci. Monit. 2021, 27, 934161. [Google Scholar] [CrossRef]
- Guo, C.W.; Alexander, M.; Dib, Y.; Lau, P.K.; Weppler, A.M.; Au-Yeung, G.; Lee, B.; Khoo, C.; Mooney, D.; Joshi, S.B.; et al. A closer look at immune-mediated myocarditis in the era of combined checkpoint blockade and targeted therapies. Eur. J. Cancer 2019, 124, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Brooks, R.A.; Fleming, G.F.; Lastra, R.R.; Lee, N.K.; Moroney, J.W.; Son, C.H.; Tatebe, K.; Veneris, J.L. Current recommendations and recent progress in endometrial cancer. CA Cancer J. Clin. 2019, 69, 258–279. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [PubMed]
- Raffone, A.; Travaglino, A.; Raimondo, D.; Neola, D.; Renzulli, F.; Santoro, A.; Insabato, L.; Casadio, P.; Zannoni, G.F.; Zullo, F.; et al. Prognostic value of myometrial invasion and TCGA groups of endometrial carcinoma. Gynecol. Oncol. 2021, 162, 401–406. [Google Scholar] [CrossRef]
- Raffone, A.; Travaglino, A.; Cerbone, M.; Gencarelli, A.; Mollo, A.; Insabato, L.; Zullo, F. Diagnostic Accuracy of Immunohistochemistry for Mismatch Repair Proteins as Surrogate of Microsatellite Instability Molecular Testing in Endometrial Cancer. Pathol. Oncol. Res. 2020, 26, 1417–1427. [Google Scholar] [CrossRef]
- Santoro, A.; Angelico, G.; Travaglino, A.; Inzani, F.; Arciuolo, D.; Valente, M.; D’Alessandris, N.; Scaglione, G.; Fiorentino, V.; Raffone, A.; et al. New Pathological and Clinical Insights in Endometrial Cancer in View of the Updated ESGO/ESTRO/ESP Guidelines. Cancers 2021, 13, 2623. [Google Scholar] [CrossRef]
- Troisi, J.; Mollo, A.; Lombardi, M.; Scala, G.; Richards, S.M.; Symes, S.J.K.; Travaglino, A.; Neola, D.; de Laurentiis, U.; Insabato, L.; et al. The Metabolomic Approach for the Screening of Endometrial Cancer: Validation from a Large Cohort of Women Scheduled for Gynecological Surgery. Biomolecules 2022, 12, 1229. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, W.; Xu, H.; Hu, M.; Guo, X.; Jia, W.; Liu, G.; Li, J.; Cui, P.; Lager, S.; et al. Hyperandrogenism and insulin resistance-induced fetal loss: Evidence for placental mitochondrial abnormalities and elevated reactive oxygen species production in pregnant rats that mimic the clinical features of polycystic ovary syndrome. J. Physiol. 2019, 597, 3927–3950. [Google Scholar] [CrossRef] [PubMed]
- Shan, H.; Luo, R.; Guo, X.; Li, R.; Ye, Z.; Peng, T.; Liu, F.; Yang, Z. Abnormal Endometrial Receptivity and Oxidative Stress in Polycystic Ovary Syndrome. Front. Pharmacol. 2022, 13, 904942. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Gu, F.; Jie, H.; Ding, C.; Zhao, Q.; Wang, Q.; Zhou, C. Early miscarriage rate in lean polycystic ovary syndrome women after euploid embryo transfer—A matched-pair study. Reprod. Biomed. Online 2017, 35, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Wagener, F.A.D.T.G.; Volk, H.-D.; Willis, D.; Abraham, N.G.; Soares, M.P.; Adema, G.J.; Figdor, C.G. Different Faces of the Heme-Heme Oxygenase System in Inflammation. Pharmacol. Rev. 2003, 55, 551–571. [Google Scholar] [CrossRef]
- Lu, J.-J.; Abudukeyoumu, A.; Zhang, X.; Liu, L.-B.; Li, M.-Q.; Xie, F. Heme oxygenase 1: A novel oncogene in multiple gynecological cancers. Int. J. Biol. Sci. 2021, 17, 2252–2261. [Google Scholar] [CrossRef]
- Li, J.; Karim, M.A.; Che, H.; Geng, Q.; Miao, D. Deletion of p16 prevents estrogen deficiency-induced osteoporosis by inhibiting oxidative stress and osteocyte senescence. Am. J. Transl. Res. 2020, 12, 672–683. [Google Scholar]
- Che, H.; Li, J.; Li, Y.; Ma, C.; Liu, H.; Qin, J.; Dong, J.; Zhang, Z.; Xian, C.J.; Miao, D.; et al. p16 deficiency attenuates intervertebral disc degeneration by adjusting oxidative stress and nucleus pulposus cell cycle. eLife 2020, 9, 52570. [Google Scholar] [CrossRef]
- Fidalgo, M.; Guerrero, A.; Fraile, M.; Iglesias, C.; Pombo, C.M.; Zalvide, J. Adaptor Protein Cerebral Cavernous Malformation 3 (CCM3) Mediates Phosphorylation of the Cytoskeletal Proteins Ezrin/Radixin/Moesin by Mammalian Ste20-4 to Protect Cells from Oxidative Stress. J. Biol. Chem. 2012, 287, 11556–11565. [Google Scholar] [CrossRef]
- Miller, B.A.; Cheung, J.Y. TRPM2 protects against tissue damage following oxidative stress and ischaemia-reperfusion. J. Physiol. 2016, 594, 4181–4191. [Google Scholar] [CrossRef]
- Malko, P.; Jiang, L.-H. TRPM2 channel-mediated cell death: An important mechanism linking oxidative stress-inducing pathological factors to associated pathological conditions. Redox Biol. 2020, 37, 101755. [Google Scholar] [CrossRef]
- Tai, L.; Huang, C.J.; Choo, K.B.; Cheong, S.K.; Kamarul, T. Oxidative Stress Down-Regulates MiR-20b-5p, MiR-106a-5p and E2F1 Expression to Suppress the G1/S Transition of the Cell Cycle in Multipotent Stromal Cells. Int. J. Med. Sci. 2020, 17, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.D.; Lee, Y.-J.; Liu, K.; Li, G.; Lin, F.-T.; Lin, W.-C. E2F1 sumoylation as a protective cellular mechanism in oxidative stress response. Proc. Natl. Acad. Sci. USA 2020, 117, 14958–14969. [Google Scholar] [CrossRef] [PubMed]
- Williams, Z.J.; Velez-Irizarry, D.; Gardner, K.; Valberg, S.J. Integrated proteomic and transcriptomic profiling identifies aberrant gene and protein expression in the sarcomere, mitochondrial complex I, and the extracellular matrix in Warmblood horses with myofibrillar myopathy. BMC Genom. 2021, 22, 438. [Google Scholar] [CrossRef] [PubMed]
- Di Tucci, C.; Capone, C.; Galati, G.; Iacobelli, V.; Schiavi, M.C.; Di Donato, V.; Muzii, L.; Panici, P.B. Immunotherapy in endometrial cancer: New scenarios on the horizon. J. Gynecol. Oncol. 2019, 30, e46. [Google Scholar] [CrossRef]
- Pan, X.; Li, D.; Huo, J.; Kong, F.; Yang, H.; Ma, X. LINC01016 promotes the malignant phenotype of endometrial cancer cells by regulating the miR-302a-3p/miR-3130-3p/NFYA/SATB1 axis. Cell Death Dis. 2018, 9, 303. [Google Scholar] [CrossRef]
- Bezzecchi, E.; Ronzio, M.; Mantovani, R.; Dolfini, D. NF-Y Overexpression in Liver Hepatocellular Carcinoma (HCC). Int. J. Mol. Sci. 2020, 21, 9157. [Google Scholar] [CrossRef]
- Bezzecchi, E.; Ronzio, M.; Dolfini, D.; Mantovani, R. NF-YA Overexpression in Lung Cancer: LUSC. Genes 2019, 10, 937. [Google Scholar] [CrossRef]
- Bezzecchi, E.; Bernardini, A.; Ronzio, M.; Miccolo, C.; Chiocca, S.; Dolfini, D.; Mantovani, R. NF-Y Subunits Overexpression in HNSCC. Cancers 2021, 13, 3019. [Google Scholar] [CrossRef]
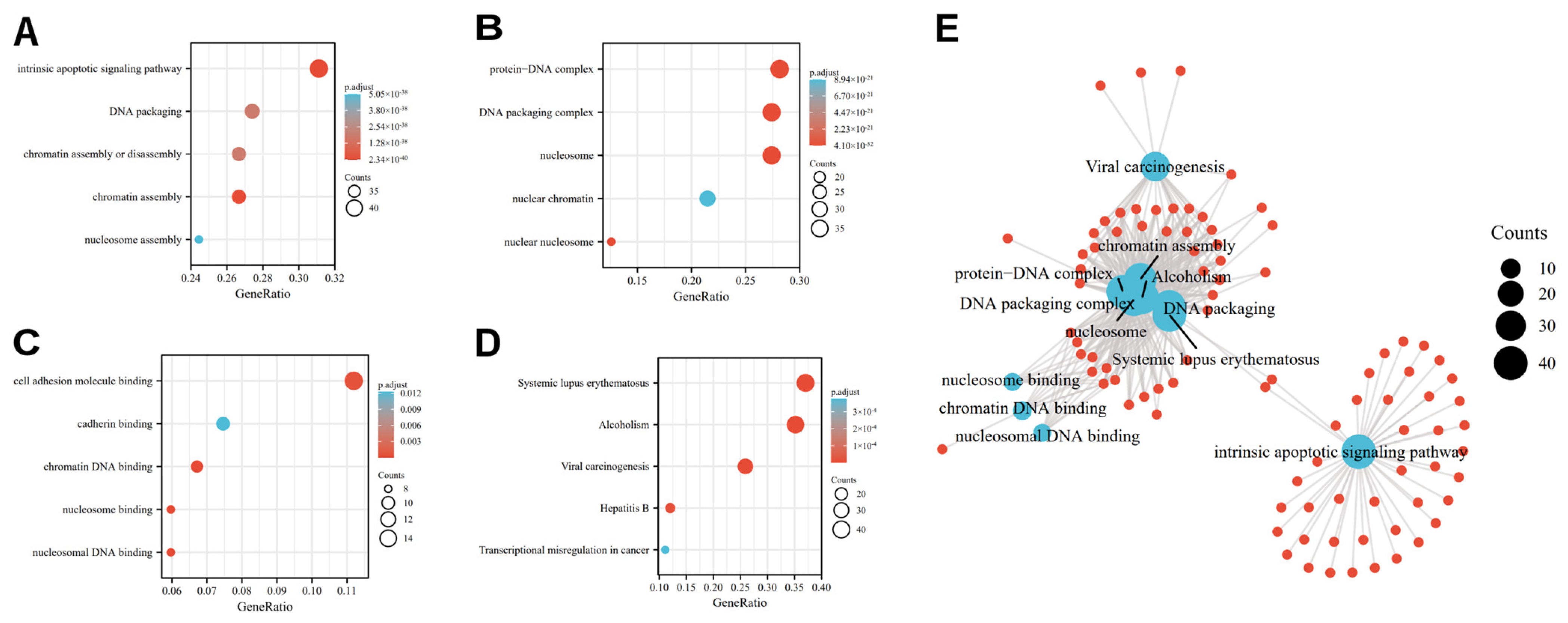

| Ontology | ID | Description | Gene Ratio | Bg Ratio | p Value | P. Adjust | Q Value |
|---|---|---|---|---|---|---|---|
| BP | GO:0031497 | chromatin assembly | 36/135 | 165/18,670 | 7.75 × 10−44 | 2.34 × 10−40 | 1.77 × 10−40 |
| BP | GO:0097193 | intrinsic apoptotic signaling pathway | 42/135 | 289/18,670 | 2.09 × 10−43 | 3.15 × 10−40 | 2.39 × 10−40 |
| BP | GO:0006323 | DNA packaging | 37/135 | 210/18,670 | 2.34 × 10−41 | 1.81 × 10−38 | 1.37 × 10−38 |
| CC | GO:0000786 | Nucleosome | 37/135 | 107/19,717 | 1.93 × 10−54 | 4.10 × 10−52 | 3.81 × 10−52 |
| CC | GO:0044815 | DNA packaging complex | 37/135 | 115/19,717 | 4.67 × 10−53 | 4.98 × 10−51 | 4.62 × 10−51 |
| CC | GO:0032993 | protein-DNA complex | 38/135 | 202/19,717 | 1.53 × 10−44 | 1.08 × 10−42 | 1.01 × 10−42 |
| MF | GO:0031492 | nucleosomal DNA binding | 8/134 | 55/17,697 | 7.90 × 10−9 | 2.63 × 10−6 | 2.30 × 10−6 |
| MF | GO:0031491 | nucleosome binding | 8/134 | 85/17,697 | 2.58 × 10−7 | 3.32 × 10−5 | 2.89 × 10−5 |
| MF | GO:0031490 | chromatin DNA binding | 9/134 | 119/17,697 | 2.99 × 10−7 | 3.32 × 10−5 | 2.89 × 10−5 |
| KEGG | hsa05322 | Systemic lupus erythematosus | 40/108 | 136/8076 | 6.16 × 10−45 | 1.32 × 10−42 | 9.01 × 10−43 |
| KEGG | hsa05034 | Alcoholism | 38/108 | 187/8076 | 8.11 × 10−36 | 8.72 × 10−34 | 5.93 × 10−34 |
| KEGG | hsa05203 | Viral carcinogenesis | 28/108 | 204/8076 | 3.05 × 10−21 | 2.18 × 10−19 | 1.49 × 10−19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Yu, M.; Zhang, T. Construction of Oxidative Stress-Related Genes Risk Model Predicts the Prognosis of Uterine Corpus Endometrial Cancer Patients. Cancers 2022, 14, 5572. https://doi.org/10.3390/cancers14225572
Liu Q, Yu M, Zhang T. Construction of Oxidative Stress-Related Genes Risk Model Predicts the Prognosis of Uterine Corpus Endometrial Cancer Patients. Cancers. 2022; 14(22):5572. https://doi.org/10.3390/cancers14225572
Chicago/Turabian StyleLiu, Qin, Minghua Yu, and Tao Zhang. 2022. "Construction of Oxidative Stress-Related Genes Risk Model Predicts the Prognosis of Uterine Corpus Endometrial Cancer Patients" Cancers 14, no. 22: 5572. https://doi.org/10.3390/cancers14225572
APA StyleLiu, Q., Yu, M., & Zhang, T. (2022). Construction of Oxidative Stress-Related Genes Risk Model Predicts the Prognosis of Uterine Corpus Endometrial Cancer Patients. Cancers, 14(22), 5572. https://doi.org/10.3390/cancers14225572






