Polygenic Risk Scores Associated with Tumor Immune Infiltration in Common Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Genotype Data and Imputation
2.3. Construction of PRSs
2.4. Categorizing Immune Traits from Tumor Tissues
2.5. Statistical Analysis
3. Results
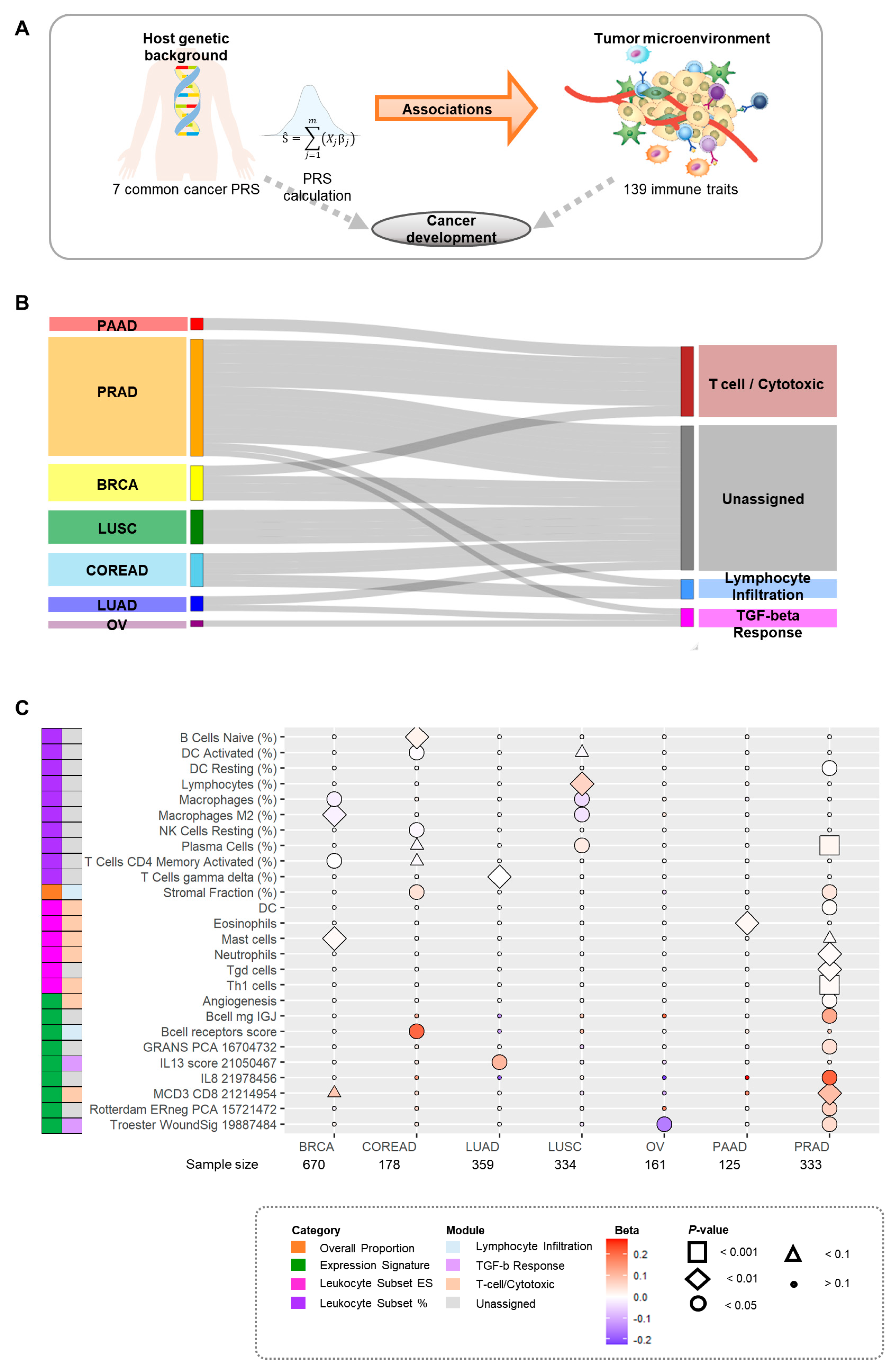
3.1. Study Design
3.2. Overall Associations between PRSs and Immune Traits
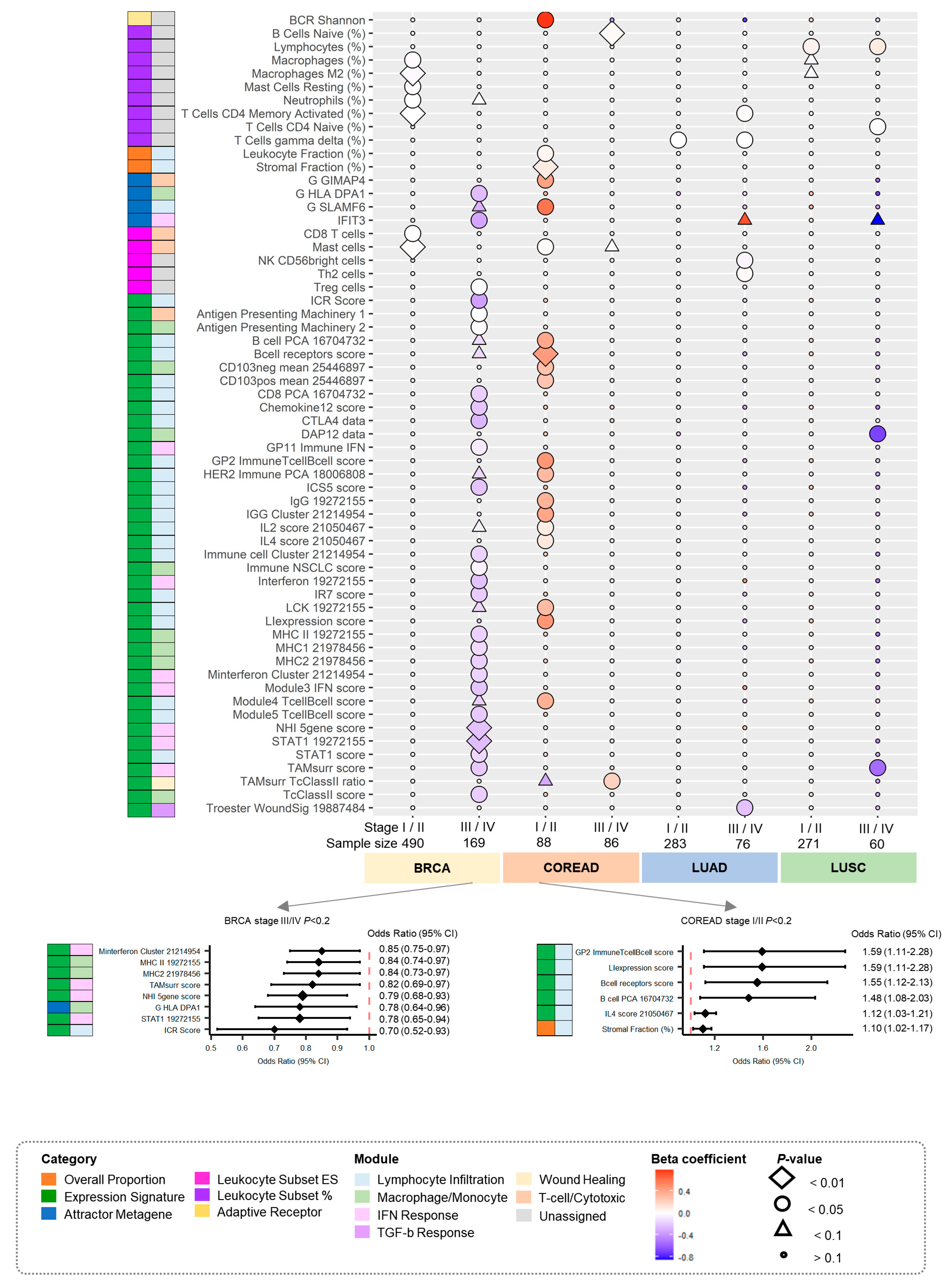
3.3. Associations by Stage between PRSs for Cancers and Immune Traits
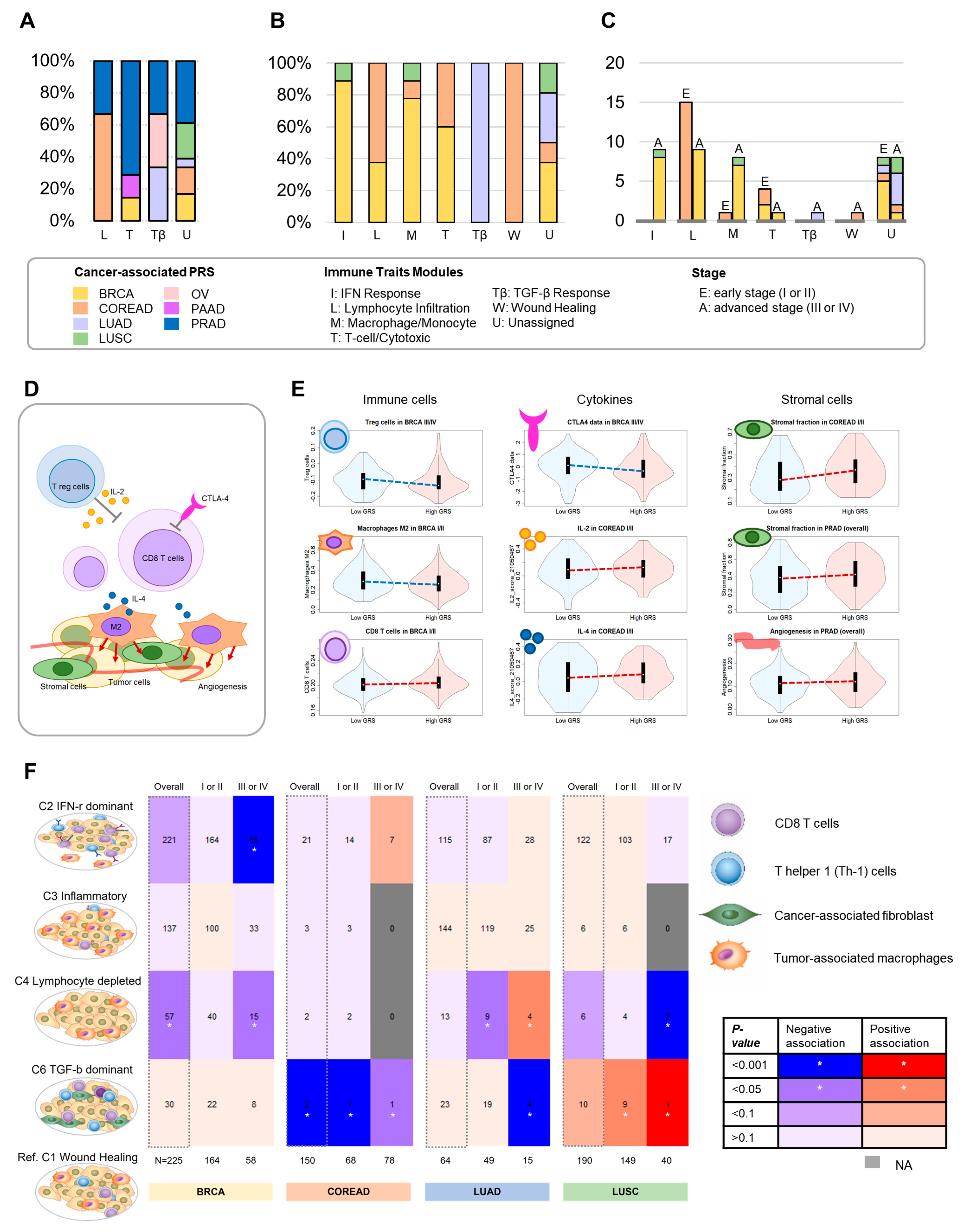
3.4. Overall Proportion of Associations for Immune Modules
3.5. Putative Predictive Biomarkers for Clinical Practice Associated with Antitumor Immune Responses
3.6. Immune Subtype Association Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phelan, C.M.; Kuchenbaecker, K.B.; Tyrer, J.P.; Kar, S.P.; Lawrenson, K.; Winham, S.J.; Dennis, J.; Pirie, A.; Riggan, M.J.; Chornokur, G.; et al. Identification of 12 New Susceptibility Loci for Different Histotypes of Epithelial Ovarian Cancer. Nat. Genet. 2017, 49, 680–691. [Google Scholar] [CrossRef] [PubMed]
- McKay, J.D.; Hung, R.J.; Han, Y.; Zong, X.; Carreras-Torres, R.; Christiani, D.C.; Caporaso, N.E.; Johansson, M.; Xiao, X.; Li, Y.; et al. Large-Scale Association Analysis Identifies New Lung Cancer Susceptibility Loci and Heterogeneity in Genetic Susceptibility across Histological Subtypes. Nat. Genet. 2017, 49, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; McGlynn, K.A.; Rajpert-De Meyts, E.; Bishop, D.T.; Chung, C.C.; Dalgaard, M.D.; Greene, M.H.; Gupta, R.; Grotmol, T.; Haugen, T.B.; et al. Meta-Analysis of Five Genome-Wide Association Studies Identifies Multiple New Loci Associated with Testicular Germ Cell Tumor. Nat. Genet. 2017, 49, 1141–1147. [Google Scholar] [CrossRef]
- Klein, A.P.; Wolpin, B.M.; Risch, H.A.; Stolzenberg-Solomon, R.Z.; Mocci, E.; Zhang, M.; Canzian, F.; Childs, E.J.; Hoskins, J.W.; Jermusyk, A.; et al. Genome-Wide Meta-Analysis Identifies Five New Susceptibility Loci for Pancreatic Cancer. Nat. Commun. 2018, 9, 556. [Google Scholar] [CrossRef]
- Schmit, S.L.; Edlund, C.K.; Schumacher, F.R.; Gong, J.; Harrison, T.A.; Huyghe, J.R.; Qu, C.; Melas, M.; Van Den Berg, D.J.; Wang, H.; et al. Novel Common Genetic Susceptibility Loci for Colorectal Cancer. J. Natl. Cancer Inst. 2019, 111, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ahearn, T.U.; Lecarpentier, J.; Barnes, D.; Beesley, J.; Qi, G.; Jiang, X.; O’Mara, T.A.; Zhao, N.; Bolla, M.K.; et al. Genome-Wide Association Study Identifies 32 Novel Breast Cancer Susceptibility Loci from Overall and Subtype-Specific Analyses. Nat. Genet. 2020, 52, 572–581. [Google Scholar] [CrossRef]
- Conti, D.V.; Darst, B.F.; Moss, L.C.; Saunders, E.J.; Sheng, X.; Chou, A.; Schumacher, F.R.; Olama, A.A.A.; Benlloch, S.; Dadaev, T.; et al. Trans-Ancestry Genome-Wide Association Meta-Analysis of Prostate Cancer Identifies New Susceptibility Loci and Informs Genetic Risk Prediction. Nat. Genet. 2021, 53, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Burnet, F.M. The Concept of Immunological Surveillance. Prog. Exp. Tumor Res. 1970, 13, 1–27. [Google Scholar] [CrossRef]
- Palomero, L.; Galván-Femenía, I.; de Cid, R.; Espín, R.; Barnes, D.R.; Cimba; Blommaert, E.; Gil-Gil, M.; Falo, C.; Stradella, A.; et al. Immune Cell Associations with Cancer Risk. iScience 2020, 23, 101296. [Google Scholar] [CrossRef]
- Ribatti, D. The Concept of Immune Surveillance against Tumors. The First Theories. Oncotarget 2017, 8, 7175–7180. [Google Scholar] [CrossRef]
- Thomas, L. On Immunosurveillance in Human Cancer. Yale J. Biol. Med. 1982, 55, 329–333. [Google Scholar] [PubMed]
- Wu, T.; Dai, Y. Tumor Microenvironment and Therapeutic Response. Cancer Lett. 2017, 387, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Ruan, G.; Ni, H.; Qin, H.; Chen, S.; Gu, X.; Shang, J.; Zhou, Y.; Tao, X.; Zheng, L. Tumor Immune Microenvironment and Its Related MiRNAs in Tumor Progression. Front. Immunol. 2021, 12, 624725. [Google Scholar] [CrossRef]
- Fanale, D.; Dimino, A.; Pedone, E.; Brando, C.; Corsini, L.R.; Filorizzo, C.; Fiorino, A.; Lisanti, M.C.; Magrin, L.; Randazzo, U.; et al. Prognostic and Predictive Role of Tumor-Infiltrating Lymphocytes (TILs) in Ovarian Cancer. Cancers 2022, 14, 4344. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Yang, F.; Yin, J.-Y.; Liu, Y.-Z.; Zhang, W.; Zhou, H.-H. Influence of Tumor Immune Infiltration on Immune Checkpoint Inhibitor Therapeutic Efficacy: A Computational Retrospective Study. Front. Immunol. 2021, 12, 685370. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Zhou, Y.; Ye, Z.; Xiong, J.; Lan, H.; Wang, F. Tumor-Infiltrating Lymphocytes in Colorectal Cancer: The Fundamental Indication and Application on Immunotherapy. Front. Immunol. 2021, 12, 808964. [Google Scholar] [CrossRef]
- Liu, S.-S.; Yang, Y.-Z.; Jiang, C.; Quan, Q.; Xie, Q.-K.; Wang, X.-P.; He, W.-Z.; Rong, Y.-M.; Chen, P.; Yang, Q.; et al. Comparison of Immunological Characteristics between Paired Mismatch Repair-Proficient and -Deficient Colorectal Cancer Patients. J. Transl. Med. 2018, 16, 195. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific Recruitment of Regulatory T Cells in Ovarian Carcinoma Fosters Immune Privilege and Predicts Reduced Survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef]
- Yang, C.; Lee, H.; Pal, S.; Jove, V.; Deng, J.; Zhang, W.; Hoon, D.S.B.; Wakabayashi, M.; Forman, S.; Yu, H. B Cells Promote Tumor Progression via STAT3 Regulated-Angiogenesis. PLoS ONE 2013, 8, e64159. [Google Scholar] [CrossRef]
- Shen, M.; Wang, J.; Ren, X. New Insights into Tumor-Infiltrating B Lymphocytes in Breast Cancer: Clinical Impacts and Regulatory Mechanisms. Front. Immunol. 2018, 9, 470. [Google Scholar] [CrossRef]
- Khan, Z.; Di Nucci, F.; Kwan, A.; Hammer, C.; Mariathasan, S.; Rouilly, V.; Carroll, J.; Fontes, M.; Ley Acosta, S.; Guardino, E.; et al. Polygenic Risk for Skin Autoimmunity Impacts Immune Checkpoint Blockade in Bladder Cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 12288–12294. [Google Scholar] [CrossRef] [PubMed]
- Sayaman, R.W.; Saad, M.; Thorsson, V.; Hu, D.; Hendrickx, W.; Roelands, J.; Porta-Pardo, E.; Mokrab, Y.; Farshidfar, F.; Kirchhoff, T.; et al. Germline Genetic Contribution to the Immune Landscape of Cancer. Immunity 2021, 54, 367–386.e8. [Google Scholar] [CrossRef] [PubMed]
- Shahamatdar, S.; He, M.X.; Reyna, M.A.; Gusev, A.; AlDubayan, S.H.; Van Allen, E.M.; Ramachandran, S. Germline Features Associated with Immune Infiltration in Solid Tumors. Cell Rep. 2020, 30, 2900–2908.e4. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Routh, E.D.; Pullikuth, A.; Jin, G.; Su, J.; Chou, J.W.; Hoadley, K.A.; Print, C.; Knowlton, N.; Black, M.A.; et al. Tumor Mutational Burden Is a Determinant of Immune-Mediated Survival in Breast Cancer. Oncoimmunology 2018, 7, e1490854. [Google Scholar] [CrossRef]
- Fuchsberger, C.; Abecasis, G.R.; Hinds, D.A. Minimac2: Faster Genotype Imputation. Bioinformatics 2015, 31, 782–784. [Google Scholar] [CrossRef]
- Howie, B.; Fuchsberger, C.; Stephens, M.; Marchini, J.; Abecasis, G.R. Fast and Accurate Genotype Imputation in Genome-Wide Association Studies through Pre-Phasing. Nat. Genet. 2012, 44, 955–959. [Google Scholar] [CrossRef]
- Cortes-Ciriano, I.; Lee, S.; Park, W.-Y.; Kim, T.-M.; Park, P.J. A Molecular Portrait of Microsatellite Instability across Multiple Cancers. Nat. Commun. 2017, 8, 15180. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.-H.; O’Reilly, P.F. Tutorial: A Guide to Performing Polygenic Risk Score Analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Morad, G.; Helmink, B.A.; Sharma, P.; Wargo, J.A. Hallmarks of Response, Resistance, and Toxicity to Immune Checkpoint Blockade. Cell 2022, 185, 576. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ali, M.; Corre, B.; Manry, J.; Barreiro, L.B.; Quach, H.; Boniotto, M.; Pellegrini, S.; Quintana-Murci, L. Functional Characterization of Naturally Occurring Genetic Variants in the Human TLR1-2-6 Gene Family. Hum. Mutat. 2011, 32, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Duffy, D.; Rouilly, V.; Libri, V.; Hasan, M.; Beitz, B.; David, M.; Urrutia, A.; Bisiaux, A.; Labrie, S.T.; Dubois, A.; et al. Functional Analysis via Standardized Whole-Blood Stimulation Systems Defines the Boundaries of a Healthy Immune Response to Complex Stimuli. Immunity 2014, 40, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Oosting, M.; Smeekens, S.P.; Jaeger, M.; Aguirre-Gamboa, R.; Le, K.T.T.; Deelen, P.; Ricaño-Ponce, I.; Schoffelen, T.; Jansen, A.F.M.; et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell 2016, 167, 1099–1110.e14. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; Chen-Harris, H.; Mayba, O.; Lianoglou, S.; Wuster, A.; Bhangale, T.; Khan, Z.; Mariathasan, S.; Daemen, A.; Reeder, J.; et al. Germline Genetic Polymorphisms Influence Tumor Gene Expression and Immune Cell Infiltration. Proc. Natl. Acad. Sci. USA 2018, 115, E11701–E11710. [Google Scholar] [CrossRef] [PubMed]
- Orrù, V.; Steri, M.; Sole, G.; Sidore, C.; Virdis, F.; Dei, M.; Lai, S.; Zoledziewska, M.; Busonero, F.; Mulas, A.; et al. Genetic Variants Regulating Immune Cell Levels in Health and Disease. Cell 2013, 155, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Patin, E.; Hasan, M.; Bergstedt, J.; Rouilly, V.; Libri, V.; Urrutia, A.; Alanio, C.; Scepanovic, P.; Hammer, C.; Jönsson, F.; et al. Natural Variation in the Parameters of Innate Immune Cells Is Preferentially Driven by Genetic Factors. Nat. Immunol. 2018, 19, 302–314. [Google Scholar] [CrossRef]
- Roederer, M.; Quaye, L.; Mangino, M.; Beddall, M.H.; Mahnke, Y.; Chattopadhyay, P.; Tosi, I.; Napolitano, L.; Terranova Barberio, M.; Menni, C.; et al. The Genetic Architecture of the Human Immune System: A Bioresource for Autoimmunity and Disease Pathogenesis. Cell 2015, 161, 387–403. [Google Scholar] [CrossRef]
- Urrutia, A.; Duffy, D.; Rouilly, V.; Posseme, C.; Djebali, R.; Illanes, G.; Libri, V.; Albaud, B.; Gentien, D.; Piasecka, B.; et al. Standardized Whole-Blood Transcriptional Profiling Enables the Deconvolution of Complex Induced Immune Responses. Cell Rep. 2016, 16, 2777–2791. [Google Scholar] [CrossRef]
- Chen, D.; Juko-Pecirep, I.; Hammer, J.; Ivansson, E.; Enroth, S.; Gustavsson, I.; Feuk, L.; Magnusson, P.K.E.; McKay, J.D.; Wilander, E.; et al. Genome-Wide Association Study of Susceptibility Loci for Cervical Cancer. J. Natl. Cancer Inst. 2013, 105, 624–633. [Google Scholar] [CrossRef]
- Michailidou, K.; Lindström, S.; Dennis, J.; Beesley, J.; Hui, S.; Kar, S.; Lemaçon, A.; Soucy, P.; Glubb, D.; Rostamianfar, A.; et al. Association Analysis Identifies 65 New Breast Cancer Risk Loci. Nature 2017, 551, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Sfanos, K.S.; Yegnasubramanian, S.; Nelson, W.G.; De Marzo, A.M. The Inflammatory Microenvironment and Microbiome in Prostate Cancer Development. Nat. Rev. Urol. 2018, 15, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Rao, X.; Lin, W. Immune Landscape and a Promising Immune Prognostic Model Associated with TP53 in Early-Stage Lung Adenocarcinoma. Cancer Med. 2021, 10, 806–823. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Zheng, Y.; Wang, Y.; Wang, Y.; Liang, N. Development and Validation of a Robust Immune-Related Prognostic Signature in Early-Stage Lung Adenocarcinoma. J. Transl. Med. 2020, 18, 380. [Google Scholar] [CrossRef]
- Wang, Q.; Gu, J.; Wang, L.; Chang, D.W.; Ye, Y.; Huang, M.; Roth, J.A.; Wu, X. Genetic Associations of T Cell Cancer Immune Response-Related Genes with T Cell Phenotypes and Clinical Outcomes of Early-Stage Lung Cancer. J. Immunother. Cancer 2020, 8, e000336. [Google Scholar] [CrossRef]
- Bao, X.; Shi, R.; Zhao, T.; Wang, Y. Immune Landscape and a Novel Immunotherapy-Related Gene Signature Associated with Clinical Outcome in Early-Stage Lung Adenocarcinoma. J. Mol. Med. 2020, 98, 805–818. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Woo, S.-R.; Zha, Y.; Spaapen, R.; Zheng, Y.; Corrales, L.; Spranger, S. Cancer Immunotherapy Strategies Based on Overcoming Barriers within the Tumor Microenvironment. Curr. Opin. Immunol. 2013, 25, 268–276. [Google Scholar] [CrossRef]
- Hegde, P.S.; Karanikas, V.; Evers, S. The Where, the When, and the How of Immune Monitoring for Cancer Immunotherapies in the Era of Checkpoint Inhibition. Clin. Cancer Res. 2016, 22, 1865–1874. [Google Scholar] [CrossRef]
- Hilmi, M.; Neuzillet, C.; Calderaro, J.; Lafdil, F.; Pawlotsky, J.-M.; Rousseau, B. Angiogenesis and Immune Checkpoint Inhibitors as Therapies for Hepatocellular Carcinoma: Current Knowledge and Future Research Directions. J. Immunother. Cancer 2019, 7, 333. [Google Scholar] [CrossRef]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ-Related MRNA Profile Predicts Clinical Response to PD-1 Blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-Tumor Genomic Biomarkers for PD-1 Checkpoint Blockade-Based Immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Bruni, D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity 2020, 52, 55–81. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lin, W.; Wen, W.; Huyghe, J.; Bien, S.; Cai, Q.; Harrison, T.; Chen, Z.; Qu, C.; Bao, J.; et al. Identifying Novel Susceptibility Genes for Colorectal Cancer Risk From a Transcriptome-Wide Association Study of 125,478 Subjects. Gastroenterology 2021, 160, 1164–1178.e6. [Google Scholar] [CrossRef] [PubMed]
- Franco, N.R.; Massi, M.C.; Ieva, F.; Manzoni, A.; Paganoni, A.M.; Zunino, P.; Veldeman, L.; Ost, P.; Fonteyne, V.; Talbot, C.J.; et al. Development of a Method for Generating SNP Interaction-Aware Polygenic Risk Scores for Radiotherapy Toxicity. Radiother. Oncol. 2021, 159, 241–248. [Google Scholar] [CrossRef]
- Distinct Genomic Landscapes in Early-Onset and Late-Onset Endometrial Cancer—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/35108035/ (accessed on 8 November 2022).
| Cancer Type a | Abbreviation | SNPs b | Samples c |
|---|---|---|---|
| Breast cancer | BRCA | 324 | 670 (25.2%) |
| Colorectal adenocarcinoma | COREAD | 129 | 178 (48.3%) |
| Lung adenocarcinoma | LUAD | 19 | 359 (21.2%) |
| Lung squamous cell carcinoma | LUSC | 17 | 334 (18.0%) |
| Ovarian serous cystadenocarcinoma | OV | 26 | 161 (—) |
| Pancreatic adenocarcinoma | PAAD | 21 | 125 (6.4%) |
| Prostate adenocarcinoma | PRAD | 257 | 333 (—) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, J.; Kim, J.S.; Sung, H.J.; Chen, Y.-W.; Chen, Z.; Wen, W.; Shu, X.-o.; Guo, X. Polygenic Risk Scores Associated with Tumor Immune Infiltration in Common Cancers. Cancers 2022, 14, 5571. https://doi.org/10.3390/cancers14225571
Choi J, Kim JS, Sung HJ, Chen Y-W, Chen Z, Wen W, Shu X-o, Guo X. Polygenic Risk Scores Associated with Tumor Immune Infiltration in Common Cancers. Cancers. 2022; 14(22):5571. https://doi.org/10.3390/cancers14225571
Chicago/Turabian StyleChoi, Jungyoon, Jung Sun Kim, Hwa Jung Sung, Yu-Wei Chen, Zhishan Chen, Wanqing Wen, Xiao-ou Shu, and Xingyi Guo. 2022. "Polygenic Risk Scores Associated with Tumor Immune Infiltration in Common Cancers" Cancers 14, no. 22: 5571. https://doi.org/10.3390/cancers14225571
APA StyleChoi, J., Kim, J. S., Sung, H. J., Chen, Y.-W., Chen, Z., Wen, W., Shu, X.-o., & Guo, X. (2022). Polygenic Risk Scores Associated with Tumor Immune Infiltration in Common Cancers. Cancers, 14(22), 5571. https://doi.org/10.3390/cancers14225571






