Simple Summary
This study investigates the latest achievements, challenges, and future research directions of deep learning techniques for lung cancer and pulmonary nodule detection. Hopefully, these research findings will help scientists, investigators, and clinicians develop new and effective medical imaging tools to improve lung nodule diagnosis accuracy, sensitivity, and specificity.
Abstract
Medical imaging tools are essential in early-stage lung cancer diagnostics and the monitoring of lung cancer during treatment. Various medical imaging modalities, such as chest X-ray, magnetic resonance imaging, positron emission tomography, computed tomography, and molecular imaging techniques, have been extensively studied for lung cancer detection. These techniques have some limitations, including not classifying cancer images automatically, which is unsuitable for patients with other pathologies. It is urgently necessary to develop a sensitive and accurate approach to the early diagnosis of lung cancer. Deep learning is one of the fastest-growing topics in medical imaging, with rapidly emerging applications spanning medical image-based and textural data modalities. With the help of deep learning-based medical imaging tools, clinicians can detect and classify lung nodules more accurately and quickly. This paper presents the recent development of deep learning-based imaging techniques for early lung cancer detection.
1. Introduction
Lung cancer is the most frequent cancer and the cause of cancer death, with the highest morbidity and mortality in the United States [1]. In 2018, GLOBOCAN estimated approximately 2.09 million new cases and 1.76 million lung cancer-related deaths [2]. Lung cancer cases and deaths have increased significantly globally [2]. Approximately 85–88% of lung cancer cases are non-small cell lung carcinoma (NSCLS), and about 12–15% of lung cancer cases are small cell lung cancer (SCLC) [3]. Early lung cancer diagnosis and intervention are crucial to increase the overall 5-year survival rate due to the invasiveness and heterogeneity of lung cancer [4].
Over the past two decades, various medical imaging techniques, such as chest X-ray, positron emission tomography (PET), magnetic resonance imaging (MRI), computed tomography (CT), low-dose CT (LDCT), and chest radiograph (CRG), have been extensively investigated for lung nodule detection. Although CT is the golden standard imaging tool for lung nodule detection, it can only detect apparent lung cancer with high false-positive rates and produces harmful X-ray radiation [5]. LDCT has been proposed to reduce harmful radiation to detect lung cancer [6]. However, cancer-related deaths were concentrated in subjects undergoing LDCT. 2-deoxy-18F-fluorodeoxyglucose (18F-FDG) PET has been developed to improve the detection performance of lung cancer [7]. 18F-FDG PET produces semi-quantitative parameters of tumor glucose metabolism, which is helpful in the diagnosis of NSCLC [8]. However, 18F-FDG PET requires further evaluation of patients with NSCLC. Some new imaging techniques, such as magnetic induction tomography (MIT), have been developed for early-stage cancer cell detection [9]. However, this technique lacks clinical validation of human subjects.
Many computer-aided detection (CAD) systems have been extensively studied for lung cancer detection and classification [10,11]. Compared to trained radiologists, CAD systems provide better lung nodules and cancer detection performance in medical images. Generally, the CAD-based lung cancer detection system includes four steps: image processing, extraction of the region of interest (ROI), feature selection, and classification. Among these steps, feature selection and classification play the most critical roles in improving the accuracy and sensitivity of the CAD system, which relies on image processing to capture reliable features. However, benign and malignant nodule classification is a challenge. Many investigators have applied deep learning techniques to help radiologists make more accurate diagnoses [12,13,14,15]. Previous studies have confirmed that deep learning-based CAD systems can effectively improve the efficiency and accuracy of medical diagnosis, especially for diagnosing various common cancers, such as lung and breast cancers [16,17]. Deep learning-based CAD systems can automatically extract high-level features from original images using different network structures than traditional CAD systems. However, deep learning-based CAD systems have some limitations, such as low sensitivity, high FP, and time consumption. Therefore, a rapid, cost-effective, and highly sensitive deep learning-based CAD system for lung cancer prediction is urgently needed.
The deep learning-based lung imaging techniques research mainly includes pulmonary nodule detection, segmentation, and classification of benign and malignant pulmonary nodules. Researchers mainly focus on developing new network structures and loss functions to improve the performance of deep learning models. Several research groups have recently published review papers on deep learning techniques [18,19,20]. However, deep learning techniques have developed rapidly, and many new methods and applications have emerged every year. This research has appeared with content that previous studies cannot cover.
This paper presents recent achievements in lung cancer segmentation, detection, and classification using deep learning methods. This study highlights current state-of-the-art deep learning-based lung cancer detection methods. This paper also highlights recent achievements, relevant research challenges, and future research directions. The rest of the paper is structured as follows. Section 2 describes the currently available medical lung imaging techniques for lung cancer detection; Section 3 reviews some recently developed deep learning-based imaging techniques; Section 4 presents lung cancer prediction using deep learning techniques; Section 5 describes the current challenges and future research directions of deep learning-based lung imaging methods; and Section 6 concludes this study.
2. Lung Imaging Techniques
Medical imaging tools help radiologists diagnose lung disease. Among these medical imaging approaches, CT offers more advantages, including size, location, characterization, and lesion growth, which could identify lung cancer and nodule information. 4D CT provides more precise targeting of the administered radiation, which significantly impacts lung cancer management [21]. Lakshmanaprabu et al. [22] developed an automatic detection system based on linear discriminate analysis (LDA) and an optimal deep neural network (ODNN) to classify lung cancer in CT lung images. The LDA reduced the extracted image features to minimize the feature dimension. The ODNN was applied and optimized by a modified gravitational search algorithm to provide a more accurate classification result. Compared to CT, LDCT is more sensitive to early-stage lung nodules and cancer detection with reduced radiation. However, it does not help reduce lung cancer mortality. It is recommended that LDCT be carried out annually for high-risk smokers aged 55 to 74 [23].
PET produces much higher sensitivity and specificity for lung nodule detection than CT due to reactive or granulomatous nodal disease [24]. PET offers a good correlation with longer progression times and overall survival rates. 18F-FDG PET has been applied to diagnose solitary pulmonary nodules [25]. 18F-FDG PET is a crucial in-patient selection and advanced NSCLC for radical radiotherapy. PET-assisted radiotherapy offers more accuracy [26] and manages about 32% of patients with stage IIIA lung cancer [27]. 18F-FDG PET provides a significant response assessment in patients with NSCLC undergoing induction chemotherapy.
MRI is the most potent lung imaging tool without ionizing radiation, but it provides insufficient information with high costs and time-consuming limitations. It failed to detect about 10% of small lung nodules (4–8 mm in diameter) [28]. MRI with ultra-short echo time (UTE) can improve signal intensity and reduce lung susceptibility artifacts. MRI with UTE is sensitive for detecting small lung nodules (4–8 mm) [29]. MRI achieves a higher lung nodule detection rate than LDCT. MRI with different pulse sequences also improved lung nodule detection sensitivity. The authors investigated T1-weighted and T2-weighted MRI to detect small lung nodules [30,31]. Compared to 3T 1.5 MRI, 1.5T MRI is much easier to identify ground glass opacities [32]. Ground glass opacities were successfully detected in 75% of subjects with lung fibrosis who received 1.5T MRI with SSFP sequences [33]. MRI with T2-weighted fast spin echo provides similar or even better performance for ground glass infiltrate detection in immunocompromised subjects [34].
Several research groups have recently investigated the feasibility of using MIT for lung disease detection [35,36]. However, due to the lack of measurement systems, expensive computational electromagnetic models, low image resolution, and some other challenges, MIT technology still has a long way to go before it can be widely used as a commercial imaging tool in clinical conditions.
Medical imaging approaches play an essential strategy in early-stage lung cancer detection and improve the survival rate. However, these techniques have some limitations, including high false positives, and cannot detect lesions automatically. Several CAD systems have been developed for lung cancer detection [37,38]. As shown in Figure 1, a CAD-based lung nodule detection system [14] usually consists of three main phases: data collection and pre-processing, training, and testing. There are two types of CAD systems: the detection system identifies specific anomalies according to interest regions, and the diagnostic system analyses lesion information, such as type, severity, stage, and progression.
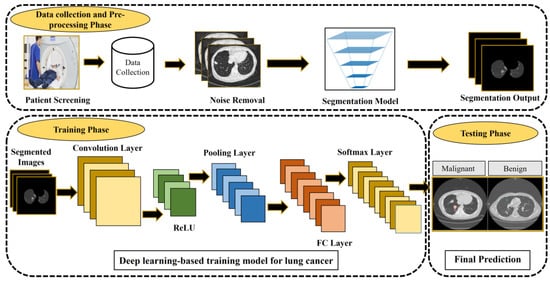
Figure 1.
CAD-based lung cancer detection system [14]. The figure is reused from reference [14]; no special permission is required to reuse all or part of articles published by MDPI, including figures and tables. For articles published under an open-access Creative Common CC BY license.
3. Deep Learning-Based Imaging Techniques
A deep learning-based CAD system has been reported as a promising tool for the automatic diagnosis of lung disease in medical imaging with significant accuracy [34,35,36]. The deep learning model is a neural network model with multiple levels of data representation. The deep learning approaches can be grouped into unsupervised, reinforcement, and supervised learning.
Unsupervised learning does not require user guidance, which analyzes the data and then sorts inherent similarities between the input data. Therefore, semi-supervised learning is a mixed model that can provide a win-win situation, even with different challenges. Semi-supervised learning techniques use both labeled and unlabeled data. With the help of labeled and unlabeled data, the accuracy of the decision boundary becomes much higher. Auto-Encoders (AE), Restricted Boltzmann Machines (RBM), and Generative Adversarial Networks (GAN) are good at clustering and nonlinear dimensionality reduction. A large amount of labeled data is usually required during training, which increases cost, time, and difficulty. Researchers have applied deep clustering to reduce labeling and make a more robust model [39,40].
Convolutional neural networks (CNN), deep convolutional neural networks (DCNN), and recurrent neural networks (RNN) are the most widely used unsupervised learning algorithms in medical images. CNN architecture is one of the most widely used supervised deep learning approaches for lesion segmentation and classification because less pre-processing is required. CNN architectures have recently been applied to medical images for image segmentation (such as Mask R-CNN [41]) and classification (such as AlexNet [42] and VGGNet [43]). DCNN architectures usually contain more layers with complex nonlinear relationships, which have been used for classification and regression with reasonable accuracy [44,45,46]. RNN architecture is a higher-order neural network that can accommodate the network output to re-input [47]. RNN applies the Elman network with feedback links from the hidden layer to the input layer, which has the potential to capture and exploit cross-slice variations to incorporate volumetric patterns of nodules. However, RNN has a vanishing gradient problem.
The reinforcement learning technique was first applied in Google Deep Mind in 2013 [48]. Since then, reinforcement learning approaches have been extensively investigated to improve lung cancer detection accuracy, sensitivity, and specificity. Semi-supervised learning approaches, such as deep reinforcement learning and generative adversarial networks, use labeled datasets.
Supervised learning usually involves a learning algorithm, and labels are assigned to the input data according to the labeling data during training. Various supervised deep learning approaches have been applied to CT images to identify abnormalities with anatomical localization. These approaches have some drawbacks, such as the large amount of labeled data required during training, the assumption of fixed network weights upon training completion, and the inability to be improved after training. Thus, a few-shot learning (FSL) model is developed to reduce data requirements during training.
4. Lung Cancer Prediction Using Deep Learning
This section presents recent achievements in lung cancer and nodule prediction using deep learning techniques. The processing includes image pre-processing, lung nodule segmentation, detection, and classification.
4.1. Imaging Pre-Processing Techniques and Evaluation
4.1.1. Pre-Processing Techniques
The pre-processed images are injected into a deep learning algorithm with specific architecture and training and tested on the image datasets. The image noise affects the precision of the final classifier. Several noise reduction approaches, such as median filter [48], Wiener filter [49], and non-local means filter [50], have been developed for pre-processing to improve accuracy and generalization performance. After denoising, a normalization method, such as min-max normalization, is required to rescale the images and reduce the complexity of image datasets.
4.1.2. Performance Metrics
Several performance metrics have been used to evaluate the performance of deep learning algorithms, including accuracy, precision, sensitivity, specificity, F1_score, error, mean squared error (MSE), receiver operation characteristic (ROC) curve, over-segmentation rate (OR), under-segmentation rate (UR), Dice similarity coefficient (DSC), Jaccard Score (JS), average symmetric surface distance (ASD), modified Hausdorff distance (MHD), and intersection over union (IoU).
Accuracy assesses the capability concerning the results with the existing information features. Sensitivity is helpful for evaluation when FN is high. Precision is an effective measurement index when FP is high. The F1_score is applied when the class distribution is uneven. ROC can tune detection sensitivity. The area under the receiver operating characteristic curve (AUC) has been used to evaluate the proposed deep learning model. Larger values of accuracy, precision, sensitivity, specificity, AUC, DSC, and JS, and smaller values of Error, UR, OR, and MHD indicate better performance of a deep learning-based algorithm.
These performance metrics can be computed using the following equations [51,52]:
where TP (true positive) denotes the number of correct positives; TN (true negative) indicates the number of correct negatives; FP (false positive) means the number of incorrect positives; FN (false negative) denotes the number of incorrect negatives; B is the target object region, A denotes ground truth dataset, and is the number of pixels in A; IoU refers to the percentage of the intersection to the union of the ground truth and predicted areas and is a metric for various object detection and semantic segmentation problems.
4.2. Datasets
Lung image datasets play an essential role in evaluating the performance of deep learning-based algorithms for lung nodule classification and detection. Table 1 shows publicly available lung images and clinical datasets for assessing nodule classification and detection performance.

Table 1.
Lung image dataset.
4.3. Lung Image Segmentation
Image segmentation aims to recognize the voxel information and external contour of the region of interest. In medical imaging, segmentation is mainly used to segment organs or lesions to quantitatively analyze relevant clinical parameters and provide further guidance for follow-up diagnosis and treatment. For example, target delineation is crucial for surgical image navigation and tumor radiotherapy guidance.
Lung segmentation plays a crucial role in medical images for lesion detection, including thorax extraction (removes artifacts) and lung extraction (identifies the left and right lungs). Several threshold techniques, such as the threshold method [69], iterative threshold [70], Otsu threshold [71], and adaptive threshold [72,73], have been investigated for lung segmentation. Few research groups have investigated segmentation methods based on region and 3D region growth [74,75]. Kass et al. [76] first introduced the active contour model, and Lan et al. [77] applied the active contour model for lung segmentation. These techniques are manual segmentation and have many disadvantages, such as being relatively slow, prone to human error, scarcity of ground truth, and class imbalance.
Several deep learning approaches have been investigated for lung segmentation. Wang et al. [78] developed a multi-view CNN (MV-CNN) for lung nodule segmentation, with an average DSC of 77.67% and an average ASD of 0.24 for the LIDC-IDRI dataset. Unlike conventional CNN, MV-CNN integrates multiple input images for lung nodule identification. However, it is difficult for MV-CNN to process 3D CT scans. Thus, a 3D CNN was developed to process volumetric patterns of cancerous nodules [79]. Sun et al. [80] designed a two-stage CAD system to segment lung nodules and FP reduction automatically. The first stage aims to identify and segment the nodules, and the second stage aims to reduce FP. The system was tested on the LIDC-IDRI dataset and evaluated by four experienced radiologists. The system obtained an average F1_score of 0.8501 for lung nodule segmentation.
In 2020, Cao et al. [81] developed a dual-branch residual network (DB-ResNet) that simultaneously captures the multi-view and multi-scale features of nodules. The proposed DB-ResNet was evaluated on the LIDC-IDRI dataset and achieved a DSC of 82.74%. Compared to trained radiologists, DB-ResNet provides a higher DSC.
In 2021, Banu et al. [82] proposed an attention-aware weight excitation U-Net (AWEU-Net) architecture in CT images for lung nodule segmentation. The architecture contains two stages: lung nodule detection based on fine-tuned Faster R-CNN and lung nodule segmentation based on the U-Net with position attention-aware weight excitation (PAWE) and channel attention-aware weight excitation (CAWE). The AWEU-Net obtained DSC of 89.79% and 90.35%, IoU of 82.34%, and 83.21% for the LUNA16 and LIDC-IDRI datasets, respectively.
Dutta [83] developed a dense recurrent residual CNN (Dense R2Unet) based on the U-Net and dense interconnections. The proposed method was tested on a lung segmentation dataset, and the results showed that the Dense R2UNet offers better segmentation performance than U-Net and ResUNet.
Table 2 shows the recently developed lung nodule segmentation techniques. Among these approaches, SVM systems obtained an accuracy range of 92.6–98.1%, CNN-based systems obtained a specificity range of 77.67–91%, ResNet models obtained a DSC range of 82.74–98.1%, and U-Net segmentation systems achieved an accuracy range of 82.2–99.27%, precision range of 46.61–98.2%, recall range of 21.43–96.33%, and F1_score range of 24.64–99.1%, respectively. The DenseNet201 system obtained an accuracy of 97%, a sensitivity of 96.2%, a specificity of 97.5%, an AUC of 0.968, and an F1_score of 96.1%. Several segmentation methods, including SVM, Dense R2UNet, 3D Attention U-Net, Dense R2UNet, Res BCDU-Net, U-Net FSL, U-Net CT, U-Net PET, U-Net PET/CT, CNN, and DenseNet201, achieved high accuracy results (over 94%).

Table 2.
Lung nodule segmentation approaches.
4.4. Lung Nodule Detection
Lung nodule detection is challenging because its shape, texture, and size vary greatly, and some non-nodules, such as blood vessels and fibrosis, have a similar appearance to lung nodules that often appear in the lungs. The processing includes two main steps: lung nodule detection and false-positive nodule reduction. Over the past few decades, researchers worldwide have extensively investigated machine learning and deep learning-based approaches for lung nodule detection. Chang et al. [106] applied the support vector machine (SVM) for nodules classification in ultrasound images. Nithila et al. [107] developed a lung nodule detection model based on heuristic search and particle clustering algorithms for network optimization. In 2005, Zhang et al. [108] developed a discrete-time cellular neural network (DTCNN) to detect small (2–10 mm) juxtapleural and non-pleural nodules in CT images. The method obtained a sensitivity of 81.25% at 8.29 FPs per scan for juxtapleural nodule detection and a sensitivity of 83.9% at 3.47 FPs per scan for non-pleural nodule detection.
Hwang et al. [109] investigated the relationship between CT and commercial CAD to detect lung nodules. They also studied LDCT images with three reconstruction kernels (B, C, and L) from 36 human subjects. The sensitivities of 82%, 88%, and 82% for the nodules of B, C, and L were obtained for all images. Experimental results showed that CAD sensitivity could be elevated by combining data from 2 different kernels without radiation exposure. Young et al. [110] studied the effects on the performance of a CAD-based nodule detection model by reducing the CT dose. The CAD system was evaluated on the NLST dataset and obtained sensitivities of 35%, 20%, and 42.5% at the initial dose, 50% dose, and 25% dose, respectively. Tajbakhsh et al. [111] studied massive training ANN (MTANN) and CNN for lung nodule detection and classification. MTANN and CNN obtained AUCs of 0.8806 and 0.7755, respectively. MTANN performs better than CNN for lung nodule detection and classification.
Liu et al. [112] developed a cascade CNN for lung nodule detection. The transfer learning model was applied to train the network to detect nodules, and a non-nodule filter was introduced to the detection network to reduce false positives (FP). The proposed architecture effectively reduces FP in the lung nodule detection system. Li et al. [65] developed a lung nodule detection method based on a faster R-CNN network and an FP reduction model in thoracic MR images. In this study, a faster R-CNN was developed to detect lung nodules, and an FP reduction model was developed to reduce FP. The method was tested on the FAHGMU dataset and obtained a sensitivity of 85.2%, with 3.47 FP per scan. Cao et al. [113] developed a two-stage CNN (TSCNN) model for lung nodule detection. In the first stage, a U-Net based on ResDense was applied to detect lung nodules. A 3D CNN-based ensemble learning architecture was proposed in the second stage to reduce false-positive nodules. The proposed model was compared with three existing models, including 3DDP-DenseNet, 3DDP-SeResNet, and 3DMBInceptionNet.
Several 3D CNN models have been developed for lung nodule detection [114,115,116]. Perez et al. [117] developed a 3D CNN to automatically detect lung cancer and tested the model on the LIDC-IDRI dataset. The experimental results showed that the proposed method provides a recall of 99.6% and an AUC of 0.913. Vipparla et al. [118] proposed a multi-patched 3D CNN with a hybrid fusion architecture for lung nodule detection with reduced FP. The method was tested on the LUNA16 dataset and achieved a competition performance metric (CPM) of 0.931. Dutande et al. [119] developed a 2D–3D cascaded CNN architecture and compared it with existing lung nodule detection and segmentation methods. The results showed that the 2D–3D cascaded CNN architecture obtained a DCM of 0.80 for nodule segmentation and a sensitivity of 90.01% for nodule detection. Luo et al. [120] developed a 3D sphere representation-based center-point matching detection network (SCPM-Net) consisting of sphere representation and center-point matching components. The SCPM-Net was tested on the LUNA16 dataset and achieved an average sensitivity of 89.2% at 7 FPs per image for lung nodule detection. Franck et al. [121] investigated the effects on the performance of deep learning image reconstruction (DLIR) techniques on lung nodule detection in chest CT images. In this study, up to 6 artificial nodules were located within the lung phantom. Images were generated using 50% ASIR-V and DLIR with low (DL-L), medium (DL-M), and high (DL-H) strengths. No statistically significant difference was obtained between these methods (p = 0.987, average AUC: 0.555, 0.561, 0.557, and 0.558 for ASIR-V, DL-L, DL-M, and DL-H).
Table 3 shows recently developed lung nodule detection approaches using deep learning techniques. Among these approaches, the co-learning feature fusion CNN obtained the best accuracy of 99.29%, which is higher than other lung nodule detection approaches. Several networks, including 3D Faster R-CNN with U-Net-like encoder, YOLOv2, YOLOv3, VGG-16, DTCNN-ELM, U-Net++, MIXCAPS, and ProCAN, obtained good accuracy (>90%) of lung nodule detection.

Table 3.
Lung nodule detection approaches.
4.5. Lung Nodule Classification
In recent years, investigators have studied various deep learning techniques to improve the performance of lung nodule classification [160,161,162,163,164,165,166,167,168,169,170,171,172,173]. The sensitivity and specificity of the SIFT-based classifier and SVM in the classification of pulmonary nodules reached 86% and 97% [160], 91.38%, and 89.56% [163], respectively. The accuracy, sensitivity, and specificity of multi-scale CNN and multi-crop CNN in lung nodule classification were 90.63%, 92.30%, and 89.47% [164], respectively, and 87%, 77%, and 93% [170], respectively. The accuracy of deep-level semantic networks and multi-scale CNN in lung nodule classification were 84.2% [167] and 86.84% [168], respectively. The CAD system developed by Cheng et al. [169] achieved the best accuracy of 95.6%, sensitivity of 92.4%, and specificity of 98.9% in the classification of pulmonary nodules.
The comparative study results showed that the sensitivity and specificity of CNN and DBN for pulmonary nodule classification are 73.40% and 73.30%, 82.20%, and 78.70%, respectively [165]. Another comparative study showed that the sensitivity and specificity of CNN and ResNet in the classification of nodules are 76.64% and 89.50%, 81.97%, and 89.38%, respectively [171]. The combined application of CNN and RNN achieved accuracy, sensitivity, and specificity of 94.78%, 94.66%, and 95.14%, respectively, in classifying pulmonary nodules [172].
In 2019, Zhang et al. [174] used an ensemble learner of multiple deep CNN in CT images and obtained a classification accuracy of 84% for the LIDC-IDRI dataset. The proposed classifier achieved better performance than other algorithms, such as SVM, multi-layer perceptron, and random forests.
Sahu et al. [175] proposed a lightweight multi-section CNN with a classification accuracy of 93.18% for the LIDC-IDRI dataset to improve accuracy. The proposed architecture could be applied to select the representative cross sections determining malignancy that facilitate the interpretation of the results.
Ali et al. [176] developed a system based on transferable texture CNN that consists of nine layers to extract features automatically and classify lung nodules. The proposed method achieved an accuracy of 96.69% ± 0.72%, with an error of 3.30% ± 0.72% and a recall of 97.19% ± 0.57%, respectively.
Marques et al. [177] developed a multi-task CNN to classify malignancy nodules with an AUC of 0.783. Thamilarasi et al. [178] proposed an automatic lung nodule classifier based on CNN with an accuracy of 86.67% for the JSRT dataset. Kawathekar et al. [179] developed a lung nodule classifier using a machine-learning technique with an accuracy of 94% and an F1_score of 92% for the LNDb dataset.
More recently, Radford et al. [180] proposed deep convolution GAN (DCGAN), Chuquicusma et al. [181] applied DCGAN to generate realistic lung nodules, and Zhao et al. [182] applied Forward and Backward GAN (F&BGAN) to classify lung nodules. The F&BGAN was evaluated on the LIDC-IDRI dataset and obtained the best accuracy of 95.24%, a sensitivity of 98.67%, a specificity of 92.47%, and an AUC of 0.98.
Table 4 shows the recently developed traditional and deep learning-based techniques for classifying lung nodules. Among these methods, CNN variants obtained an accuracy range of 83.4–99.6%, a specificity range of 73.3–95.17%, a sensitivity range of 73.3–96.85%, and an AUC range of 0.7755–0.9936, respectively. Several methods achieved high classification accuracy (>95%), including F&BGAN, Inception_ResNet_V2, ResNet152V2, ResNet152V2+GRU, CSO-CADLCC, ProCAN, Net121, ResNet50, DITNN, and optimal DBN with an opposition-based pity beetle algorithm. DCNN systems obtained a sensitivity of 89.3% [183] and an accuracy of 97.3% [184]. The classifier was developed based on the VGG19 and CNN models and achieved accuracy, sensitivity, specificity, recall, F1_score, AUC, and MCC above 98%.

Table 4.
Lung nodule classification approaches.
Forte et al. [209] recently conducted a systematic review and meta-analysis of the diagnostic accuracy of current deep learning approaches for lung cancer diagnosis. The pooled sensitivity and specificity of deep learning approaches for lung cancer detection were 93% and 68%, respectively. The results showed that AI plays an important role in medical imaging, but there are still many research challenges.
5. Challenges and Future Research Directions
This study extensively surveys papers published between 2014 and 2022. Table 2, Table 3 and Table 4 demonstrate that deep learning-based lung imaging systems have achieved high efficiency and state-of-the-art performance for lung nodule segmentation, detection, and classification using existing medical images. Compared to reinforcement and supervised learning techniques, unsupervised deep learning techniques (such as CNN, Faster R-CNN, Mask R-CNN, and U-Net) are more popular methods that have been used to develop convolutional networks for lung cancer detection and false-positive reduction.
Previous studies have shown that CT is the most widely used imaging tool in the CAD system for lung cancer diagnosis. Compared to 2D CNN, 3D CNN architectures provide more promising usefulness in obtaining representative features of malignant nodules. To this day, only a few works on 3D CNN for lung cancer diagnosis have been reported.
Deep learning techniques have achieved good performance in segmentation and classification. However, deep learning techniques still have many unsolved problems in lung cancer detection. First, clinicians have not fully acknowledged deep learning techniques for everyday clinical exercise due to the lack of standardized medical image acquisition protocols. The unification of the acquisition protocols could minimize it.
Second, deep learning techniques usually require massive annotated medical images by experienced radiologists to complete training tasks. However, it is costly and time consuming to collect an enormous annotated image dataset, even performed by experienced radiologists. Several methods were applied to overcome the scarcity of annotated data. For example, transfer learning is a possible way to solve the training problem of small samples. Another possible method is the computer synthesis of images, such as the generation of confrontation networks. Inadequate data will inevitably affect the accuracy and stability of predictions. Therefore, improving prediction accuracy using weak supervision, transfer learning, and multi-task learning with small labeled data is one of the future research directions.
Third, the clinical application of deep learning requires high interpretability, but current deep learning techniques cannot effectively explain the learned features. Many researchers have applied visualization and parameter analysis methods to explain deep learning models. However, there is still a certain distance from the interpretable imaging markers required by clinical requirements. Therefore, investigating the interpretable deep learning method will be a hot spot in the medical image field.
Fourth, developing the robustness of the prediction model is a challenging task. Most deep learning techniques work well only for a single dataset. The image of the same disease may vary significantly due to different acquisition parameters, equipment, time, and other factors. This led to poor robustness and generalization of existing deep learning models. Thus, improving the model structure and training methods by combining brain cognitive ideas and improving the generalization ability of deep learning is one of the key future directions.
Finally, some of the current literature has little translation into applicability in clinical practice due to the lack of experience of non-medical investigators in choosing more relevant clinical outcomes. Most deep learning techniques were developed by non-medical professionals with little or no oversight of radiologists, who, in practice, will use these resources when they become more widely available. As a result, some performance metrics, such as accuracy, AUC, and precision, which have little meaningful clinical application, continue to be used and are often the only summary outcomes reported by some studies. Instead, investigators should always strive to report more relevant clinical parameters, such as sensitivity and specificity, because they are independent of the prevalence of the disease and can be more easily translated into practice.
In the future, investigators should pay more attention to the following research directions: (1) develop new convolutional networks and loss functions to improve the performance; (2) weak supervised learning, using a large number of incomplete, inaccurate, and ambiguous annotation data in the existing medical records to achieve model training; (3) bring prior clinical knowledge into model training; (4) radiologists, computer scientists, and engineers need to work more closely to develop more realistic and sensitive models and add more meaning to the research field; (5) single disease identification to complete disease identification. In clinical examination, only a few cases need to solve one well-defined problem. For example, clinicians can detect pulmonary nodules in LDCT and check whether there are other abnormalities, such as emphysema. Solving multiple problems with one network will not reduce performance in specific tasks. In addition, deep learning can be explored in some areas where the medical mechanism is not precise, such as large-scale lung image analysis using deep learning, which is expected to make diagnosing lung diseases more objective.
6. Conclusions
This paper reviewed recent achievements in deep learning-based approaches for lung nodule segmentation, detection, and classification. CNN is one of the most widely used deep learning techniques for lung disease detection and classification, and CT image datasets are the most frequently used imaging datasets for training networks. The article review was based on recent publications (published in 2014 and later). Experimental and clinical trial results demonstrate that deep learning techniques can be superior to trained radiologists. Deep learning is expected to effectively improve lung nodule segmentation, detection, and classification. With this powerful tool, radiologists can interpret images more accurately. Deep learning algorithm has shown great potential in a series of tasks in the radiology department and has solved many medical problems. However, it still faces many difficulties, including large-scale clinical verification, patient privacy protection, and legal accountability. Despite these limitations, with the current trend and rapid development of the medical industry, deep learning is expected to generate a greater demand for accurate diagnosis and treatment in the medical field.
Funding
This research was funded by the International Science and Technology Cooperation Project of the Shenzhen Science and Technology Commission (GJHZ20200731095804014).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The author would like to thank the reviewers for their critical comments to improve the manuscript significantly.
Conflicts of Interest
The author declares no conflict of interest.
Correction Statement
This article has been republished with a minor correction to the reference 52. This change does not affect the scientific content of the article.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Bade, B.C.; Cruz, C. Lung cancer. Clin. Chest Med. 2020, 41, 1–24. [Google Scholar] [CrossRef]
- Stamatis, G.; Eberhard, W.; Pöttgen, C. Surgery after multimodality treatment for non-small-cell lung cancer. Lung Cancer 2004, 45, S107–S112. [Google Scholar] [CrossRef] [PubMed]
- Chiang, T.A.; Chen, P.H.; Wu, P.F.; Wang, T.N.; Chang, P.Y.; Ko, A.M.; Huang, M.S.; Ko, Y.C. Important prognostic factors for the long-term survival of lung cancer subjects in Taiwan. BMC Cancer 2008, 8, 324. [Google Scholar] [CrossRef] [PubMed]
- Journy, N.; Rehel, J.L.; Pointe, H.D.L.; Lee, C.; Brisse, H.; Chateil, J.F.; Caer-Lorho, S.; Laurier, D.; Bernier, M.O. Are the studies on cancer risk from ct scans biased by indication? Elements of answer from a large-scale cohort study in France. Br. J. Cancer 2015, 112, 1841–1842. [Google Scholar] [CrossRef]
- National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef]
- Ippolito, D.; Capraro, C.; Guerra, L.; De Ponti, E.; Messa, C.; Sironi, S. Feasibility of perfusion CT technique integrated into conventional (18) FDG/PET-CT studies in lung cancer patients: Clinical staging and functional information in a single study. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 156–165. [Google Scholar] [CrossRef]
- Park, S.Y.; Cho, A.; Yu, W.S.; Lee, C.Y.; Lee, J.G.; Kim, D.J.; Chung, K.Y. Prognostic value of total lesion glycolysis by F-18-FDG PET/CT in surgically resected stage IA non-small cell lung cancer. J. Nucl. Med. 2015, 56, 45–49. [Google Scholar] [CrossRef][Green Version]
- Griffiths, H. Magnetic induction tomography. Meas. Sci. Technol. 2011, 12, 1126–1131. [Google Scholar] [CrossRef]
- Brown, M.S.; Lo, P.; Goldin, J.G.; Barnoy, E.; Kim, G.H.J.; Mcnitt-Gray, M.F.; Aberle, D.R. Toward clinically usable CAD for lung cancer screening with computed tomography. Eur. Radiol. 2020, 30, 1822. [Google Scholar] [CrossRef]
- Roberts, H.C.; Patsios, D.; Kucharczyk, M.; Paul, N.; Roberts, T.P. The utility of computer-aided detection (CAD) for lung cancer screening using low-dose CT. Int. Congr. Ser. 2005, 1281, 1137–1142. [Google Scholar] [CrossRef]
- Abdul, L.; Rajasekar, S.; Lin, D.S.Y.; Venkatasubramania Raja, S.; Sotra, A.; Feng, Y.; Liu, A.; Zhang, B. Deep-lumen assay-human lung epithelial spheroid classification from brightfield images using deep learning. Lab A Chip 2021, 21, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Armato, S.G.I. Deep learning demonstrates potential for lung cancer detection in chest radiography. Radiology 2020, 297, 697–698. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Li, J.; Pei, Y.; Khurram, R.; Rehman, K.U.; Rasool, A.B. State-of-the-Art Challenges and Perspectives in Multi-Organ Cancer Diagnosis via Deep Learning-Based Methods. Cancers 2021, 13, 5546. [Google Scholar] [CrossRef]
- Riquelme, D.; Akhloufi, M.A. Deep Learning for Lung Cancer Nodules Detection and Classification in CT Scans. AI 2020, 1, 28–67. [Google Scholar] [CrossRef]
- Zhukov, T.A.; Johanson, R.A.; Cantor, A.B.; Clark, R.A.; Tockman, M.S. Discovery of distinct protein profiles specific for lung tumors and pre-malignant lung lesions by SELDI mass spectrometry. Lung Cancer 2003, 40, 267–279. [Google Scholar] [CrossRef]
- Zeiser, F.A.; Costa, C.; Ramos, G.; Bohn, H.C.; Santos, I.; Roehe, A.V. Deepbatch: A hybrid deep learning model for interpretable diagnosis of breast cancer in whole-slide images. Expert Syst. Appl. 2021, 185, 115586. [Google Scholar] [CrossRef]
- Mandal, M.; Vipparthi, S.K. An empirical review of deep learning frameworks for change detection: Model design, experimental frameworks, challenges and research needs. IEEE Trans. Intell. Transp. Syst. 2022, 23, 6101–6122. [Google Scholar] [CrossRef]
- Alireza, H.; Cheikh, M.; Annika, K.; Jari, V. Deep learning for forest inventory and planning: A critical review on the remote sensing approaches so far and prospects for further applications. Forestry 2022, 95, 451–465. [Google Scholar]
- Highamcatherine, F.; Highamdesmond, J. Deep learning. SIAM Rev. 2019, 32, 860–891. [Google Scholar]
- Latifi, K.; Dilling, T.J.; Feygelman, V.; Moros, E.G.; Stevens, C.W.; Montilla-Soler, J.L.; Zhang, G.G. Impact of dose on lung ventilation change calculated from 4D-CT using deformable image registration in lung cancer patients treated with SBRT. J. Radiat. Oncol. 2015, 4, 265–270. [Google Scholar] [CrossRef]
- Lakshmanaprabu, S.K.; Mohanty, S.N.; Shankar, K.; Arunkumar, N.; Ramirez, G. Optimal deep learning model for classification of lung cancer on CT images. Future Gener. Comput. Syst. 2019, 92, 374–382. [Google Scholar]
- Shim, S.S.; Lee, K.S.; Kim, B.T.; Chung, M.J.; Lee, E.J.; Han, J.; Choi, J.Y.; Kwon, O.J.; Shim, Y.M.; Kim, S. Non-small cell lung cancer: Prospective comparison of integrated FDG PET/CT and CT alone for preoperative staging. Radiology 2005, 236, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Ab, G.D.C.; Domínguez, J.F.; Bolton, R.D.; Pérez, C.F.; Martínez, B.C.; García-Esquinas, M.G.; Carreras Delgado, J.L. PET-CT in presurgical lymph node staging in non-small cell lung cancer: The importance of false-negative and false-positive findings. Radiologia 2017, 59, 147–158. [Google Scholar]
- Yaturu, S.; Patel, R.A. Metastases to the thyroid presenting as a metabolically inactive incidental thyroid nodule with stable size in 15 months. Case Rep. Endocrinol. 2014, 2014, 643986. [Google Scholar] [CrossRef][Green Version]
- Eschmann, S.M.; Friedel, G.; Paulsen, F.; Reimold, M.; Hehr, T.; Budach, W.; Langen, H.J.; Bares, R. 18F-FDG PET for assessment of therapy response and preoperative re-evaluation after neoadjuvant radio-chemotherapy in stage III non-small cell lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2007, 34, 463–471. [Google Scholar] [CrossRef]
- Lee, W.K.; Lau, E.W.; Chin, K.; Sedlaczek, O.; Steinke, K. Modern diagnostic and therapeutic interventional radiology in lung cancer. J. Thorac. Dis. 2013, 5, 511–523. [Google Scholar]
- Zurek, M.; Bessaad, A.; Cieslar, K.; Crémillieux, Y. Validation of simple and robust protocols for high-resolution lung proton MRI in mice. Magn. Reson. Med. 2010, 64, 401–407. [Google Scholar] [CrossRef]
- Burris, N.S.; Johnson, K.M.; Larson, P.E.Z.; Hope, M.D.; Nagle, S.K.; Behr, S.C.; Hope, T.A. Detection of small pulmonary nodules with ultrashort echo time sequences in oncology patients by using a PET/MR system. Radiology 2016, 278, 239–246. [Google Scholar] [CrossRef]
- Fink, C.; Puderbach, M.; Biederer, J.; Fabel, M.; Dietrich, O.; Kauczor, H.U.; Reiser, M.F.; Schönberg, S.O. Lung MRI at 1.5 and 3 tesla: Observer preference study and lesion contrast using five different pulse sequences. Investig. Radiol. 2007, 42, 377–383. [Google Scholar] [CrossRef]
- Cieszanowski, A.; Anyszgrodzicka, A.; Szeszkowski, W.; Kaczynski, B.; Maj, E.; Gornicka, B.; Grodzicki, M.; Grudzinski, I.P.; Stadnik, A.; Krawczyk, M.; et al. Characterization of focal liver lesions using quantitative techniques: Comparison of apparent diffusion coefficient values and T2 relaxation times. Eur. Radiol. 2012, 22, 2514–2524. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.; Tiddens, H.; Wild, J.M. Lung imaging in cystic fibrosis. Imaging Decis. MRI 2009, 13, 28–37. [Google Scholar] [CrossRef]
- Groth, M.; Henes, F.O.; Bannas, P.; Muellerleile, K.; Adam, G.; Regier, M. Intraindividual comparison of contrast-enhanced MRI and unenhanced SSFP sequences of stenotic and non-stenotic pulmonary artery diameters. Rofo 2011, 183, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Chong, A.L.; Chandra, R.V.; Chuah, K.C.; Roberts, E.L.; Stuckey, S.L. Proton density MRI increases detection of cervical spinal cord multiple sclerosis lesions compared with T2-weighted fast spin-echo. Am. J. Neuroradiol. 2016, 37, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Alzeibak, S.; Saunders, N.H. A feasibility study of in vivo electromagnetic imaging. Phys. Med. Biol. 1993, 38, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Merwa, R.; Hollaus, K.; Brunner, P.; Scharfetter, H. Solution of the inverse problem of magnetic induction tomography (MIT). Physiol. Meas. 2006, 26, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.L.; Gurupur, V.P.; Lin, H.; Martis, R.J. A novel fusion approach for early lung cancer detection using computer aided diagnosis techniques. J. Med. Imaging Health Inform. 2017, 7, 1841–1850. [Google Scholar] [CrossRef]
- Lu, H. Computer-aided diagnosis research of a lung tumor based on a deep convolutional neural network and global features. BioMed Res. Int. 2021, 2021, 5513746. [Google Scholar] [CrossRef] [PubMed]
- Standford.edu. Deep Learning Tutorial. Available online: http://deeplearning.stanford.edu/tutorial/ (accessed on 5 October 2022).
- Dauphin, Y.N.; Fan, A.; Auli, M.; Grangier, D. Language Modeling with Gated Convolutional Networks. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; Volume 70, pp. 933–941. [Google Scholar]
- Jeong, J.; Lei, Y.; Shu, H.K.; Liu, T.; Wang, L.; Curran, W.; Shu, H.-K.; Mao, H.; Yang, X. Brain tumor segmentation using 3D mask R-CNN for dynamic susceptibility contrast enhanced perfusion imaging. Med. Imaging Biomed. Appl. Mol. Struct. Funct. Imaging 2020, 65, 185009. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 6. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.K.; Lee, K.M. Accurate image super-resolution using very deep convolutional networks. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Liu, W.; Chen, W.; Wang, C.; Mao, Q.; Dai, X. Capsule Embedded ResNet for Image Classification. In Proceedings of the 2021 5th International Conference on Computer Science and Artificial Intelligence (CSAI 2021), Beijing, China, 4–6 December 2021. [Google Scholar]
- Guan, X.; Gao, W.; Peng, H.; Shu, N.; Gao, D.W. Image-based incipient fault classification of electrical substation equipment by transfer learning of deep convolutional neural network. IEEE Can. J. Electr. Comput. Eng. 2021, 45, 1–8. [Google Scholar] [CrossRef]
- Warin, K.; Limprasert, W.; Suebnukarn, S.; Jinaporntham, S.; Jantana, P. Performance of deep convolutional neural network for classification and detection of oral potentially malignant disorders in photographic images. Int. J. Oral Maxillofac. Surg. 2022, 51, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Magge, A.; Weissenbacher, D.; Sarker, A.; Scotch, M.; Gonzalez-Hernandez, G. Bi-directional recurrent neural network models for geographic location extraction in biomedical literature. Pac. Symp. Biocomput. 2019, 24, 100–111. [Google Scholar] [PubMed]
- Garg, J.S. Improving segmentation by denoising brain MRI images through interpolation median filter in ADTVFCM. Int. J. Comput. Trends Technol. 2013, 4, 187–188. [Google Scholar]
- Siddeq, M. De-noise color or gray level images by using hybred dwt with wiener filter. Hepato-Gastroenterology 2014, 61, 1308–1312. [Google Scholar]
- Rajendran, K.; Tao, S.; Zhou, W.; Leng, S.; Mccollough, C. Spectral prior image constrained compressed sensing reconstruction for photon-counting detector based CT using a non-local means filtered prior (NLM-SPICCS). Med. Phys. 2018, 6, 45. [Google Scholar]
- Powers, D.M.W. Evaluation: From precision, recall and f-measure to roc., informedness, markedness & correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Das, A.; Rajendra Acharya, U.; Panda, S.S.; Sabut, S. Deep learning-based liver cancer detection using watershed transform and Gaussian mixture model techniques. Cogn. Syst. Res. 2019, 54, 165–175. [Google Scholar] [CrossRef]
- Lung Image Database Consortium (LIDC). Available online: https://imaging.nci.nih.gov/ncia/login.jsf (accessed on 5 October 2022).
- Armato Samuel, G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. Data from LIDC-IDRI. 2015. Available online: https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI (accessed on 5 October 2022).
- Setio, A.A.A.; Traverso, A.; de Bel, T.; Berens, M.S.N.; van den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, Comparison, and Combination of Algorithms for Automatic Detection of Pulmonary Nodules in Computed Tomography Images: The LUNA16 Challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef]
- ELCAP Public Lung Image Database. 2014. Available online: http://www.via.cornell.edu/lungdb.html (accessed on 5 October 2022).
- Pedrosa, J.; Aresta, G.; Ferreira, C.; Rodrigues, M.; Leito, P.; Carvalho, A.S.; Rebelo, J.; Negrao, E.; Ramos, I.; Cunha, A.; et al. LNDb: A Lung Nodule Database on Computed Tomography. arXiv 2019, arXiv:1911.08434. [Google Scholar]
- Prasad, D.; Ujjwal, B.; Sanjay, T. LNCDS: A 2D-3D cascaded CNN approach for lung nodule classification, detection and segmentation. Biomed. Signal Process. Control. 2021, 67, 102527. [Google Scholar]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, A.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.; Matsiu, M.; Fujita, H.; Kodera, Y.; Doi, K. Development of a digital image database for chest radiographs with and without a lung nodule: Receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgen. 2000, 174, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.D.; Broodman, I.; Hoogsteden, H.; Luider, T.; Klaveren, R.V. Biomarker identification for early detection of lung cancer by proteomic techniques in the NELSON lung cancer screening trial. Cancer Res. 2008, 68, 3961. [Google Scholar]
- Van Ginneken, B.; Armato, S.G.; de Hoop, B.; van Amelsvoort-van de Vorst, S.; Duindam, T.; Niemeijer, M.; Murphy, K.; Schilham, A.; Retico, A.; Fantacci, M.E.; et al. Comparing and Combining Algorithms for Computer-Aided Detection of Pulmonary Nodules in Computed Tomography Scans: The ANODE09 Study. Med. Image Anal. 2010, 14, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Hospital, S.Z. A Trial to Evaluate the Impact of Lung-Protective Intervention in Patients Undergoing Esophageal Cancer Surgery; US National Library of Medicine: Bethesda, MD, USA, 2013. [Google Scholar]
- Armato Samuel, G., III; Hadjiiski, L.; Tourassi, G.D.; Drukker, K.; Giger, M.L.; Li, F.; Redmond, G.; Farahani, K.; Kirby, J.S.; Clarke, L.P. SPIE-AAPM-NCI Lung Nodule Classification Challenge Dataset. 2015. Available online: https://wiki.cancerimagingarchive.net/display/Public/SPIE-AAPM+Lung+CT+Challenge (accessed on 5 October 2022).
- Li, T.Y.; Li, S.P.; Zhang, Q.L. Protective effect of ischemic preconditioning on lung injury induced by intestinal ischemia/reperfusion in rats. Mil. Med. J. South China 2011, 25, 107–110. [Google Scholar]
- Li, Y.; Zhang, L.; Chen, H.; Yang, N. Lung nodule detection with deep learning in 3D thoracic MR images. IEEE Access 2019, 7, 37822–37832. [Google Scholar] [CrossRef]
- Aerts, H.; Velazquez, E.; Leijenaar, R.; Parmar, C.; Grossman, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Danish Lung Cancer Screening Trial (DLCST)—Full Text View—ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/show/NCT00496977 (accessed on 5 October 2022).
- Trial Summary—Learn—NLST—The Cancer Data Access System. Available online: https://biometry.nci.nih.gov/cdas/learn/nlst/trial-summary/ (accessed on 5 October 2022).
- Hu, S.; Hoffman, E.A.; Reinhardt, J.M. Accurate lung segmentation for accurate quantization of volumetric X-ray CT images. IEEE Trans. Med. Imaging 2001, 20, 490–498. [Google Scholar] [CrossRef]
- Dawoud, A. Lung segmentation in chest radiographs by fusing shape information in iterative thresholding. Comput. Vis. IET 2011, 5, 185–190. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Sys. Man Cyber. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Peng, T.; Wang, C.; Zhang, Y.; Wang, J. H-SegNet: Hybrid segmentation network for lung segmentation in chest radiographs using mask region-based convolutional neural network and adaptive closed polyline searching method. Phys. Med. Biol. 2022, 67, 075006. [Google Scholar] [CrossRef] [PubMed]
- Tseng, L.Y.; Huang, L.C. An adaptive thresholding method for automatic lung segmentation in CT images. In Proceedings of the IEEE Africon, Nairobi, Kenya, 23–25 September 2009; pp. 1–5. [Google Scholar]
- Dehmeshki, J.; Amin, H.; Valdivieso, M.; Ye, X. Segmentation of pulmonary nodules in thoracic CT scans: A region growing approach. IEEE Trans. Med. Imaging 2008, 27, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Fabijacska, A. The influence of pre-processing of CT images on airway tree segmentation using 3D region growing. In Proceedings of the 5th International Conference on Perspective Technologies and Methods in MEMS Design, Lviv, Ukraine, 22–24 April 2009. [Google Scholar]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active contour models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Lan, Y.; Xu, N.; Ma, X.; Jia, X. Segmentation of Pulmonary Nodules in Lung CT Images based on Active Contour Model. In Proceedings of the 14th IEEE International Conference on Intelligent Human-Machine Systems and Cybernetics (IHMSC), Hangzhou, China, 20–21 August 2022. [Google Scholar]
- Wang, S.; Zhou, M.; Olivier, G.; Tang, Z.C.; Dong, D.; Liu, Z.Y.; Tian, J. A Multi-view Deep Convolutional Neural Networks for Lung Nodule Segmentation. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 1752–1755. [Google Scholar]
- Hamidian, S.; Sahiner, B.; Petrick, N.; Pezeshk, A. 3D Convolutional Neural Network for Automatic Detection of Lung Nodules in Chest CT. Proc. SPIE Int. Soc. Opt. Eng. 2017, 10134, 1013409. [Google Scholar]
- Sun, X.F.; Lin, H.; Wang, S.Y.; Zheng, L.M. Industrial robots sorting system based on improved faster RCNN. Comput. Syst. Appl. 2019, 28, 258–263. [Google Scholar]
- Cao, H.C.; Liu, H.; Song, E.; Hung, C.C.; Ma, G.Z.; Xu, X.Y.; Jin, R.C.; Jianguo Lu, J.G. Dual-branch residual network for lung nodule segmentation. Appl. Soft Comput. 2020, 86, 105934. [Google Scholar] [CrossRef]
- Banu, S.F.; Sarker, M.; Abdel-Nasser, M.; Puig, D.; Raswan, H.A. AWEU-Net: An attention-aware weight excitation u-net for lung nodule segmentation. arXiv 2021, arXiv:2110.05144. [Google Scholar] [CrossRef]
- Dutta, K. Densely connected recurrent residual (DENSE R2UNET) convolutional neural network for segmentation of lung CT images. arXiv 2021, arXiv:2102.00663. [Google Scholar]
- Keshani, M.; Azimifar, Z.; Tajeripour, F.; Boostani, R. Lung nodule segmentation and recognition using SVM classifier and active contour modeling: A complete intelligent system. Comput. Biol. Med. 2013, 43, 287–300. [Google Scholar] [CrossRef]
- Qi, S.L.; Si, G.L.; Yue, Y.; Meng, X.F.; Cai, J.F.; Kang, Y. Lung nodule segmentation based on thoracic CT images. Beijing Biomed. Eng. 2014, 33, 29–34. [Google Scholar]
- Wang, X.P.; Wen, Z.; Ying, C. Tumor segmentation in lung CT images based on support vector machine and improved level set. Optoelectron. Lett. 2015, 11, 395–400. [Google Scholar] [CrossRef]
- Shen, S.; Bui, A.; Cong, J.J.; Hsu, W. An automated lung segmentation approach using bidirectional chain codes to improve nodule detection accuracy. Comput. Biol. Med. 2015, 57(C), 139–149. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.R.; Farag, A.; Le, L.; Turkbey, E.B.; Summers, R.M. Deep convolutional networks for pancreas segmentation in CT imaging. Proc. SPIE 2015, 9413, 94131G. [Google Scholar]
- Yip, S.; Chintan, P.; Daniel, B.; Jose, E.; Steve, P.; John, K.; Aerts, H.J.W.L. Application of the 3D slicer chest imaging platform segmentation algorithm for large lung nodule delineation. PLoS ONE 2017, 12, e0178944. [Google Scholar] [CrossRef]
- Firdouse, M.J.; Balasubramanian, M. A survey on lung segmentation methods. Adv. Comput. Sci. Technol. 2017, 10, 2875–2885. [Google Scholar]
- Khosravan, N.; Bagci, U. Semi-Supervised Multi-Task Learning for Lung Cancer Diagnosis. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 2018, 710–713. [Google Scholar]
- Tong, G.; Li, Y.; Chen, H.; Zhang, Q.; Jiang, H. Improved U-NET network for pulmonary nodules segmentation. Optik 2018, 174, 460–469. [Google Scholar] [CrossRef]
- Jiang, J.; Hu, Y.C.; Liu, C.J.; Darragh, H.; Hellmann, M.D.; Deasy, J.O.; Mageras, G.; Veeraraghavan, H. Multiple resolution residually connected feature streams for automatic lung tumor segmentation from CT images. IEEE Trans. Med. Imaging 2019, 38, 134–144. [Google Scholar] [CrossRef]
- Burlutskiy, N.; Gu, F.; Wilen, L.K.; Backman, M.; Micke, P. A deep learning framework for automatic diagnosis in lung cancer. In Proceedings of the 1st Conference on Medical Imaging with Deep Learning (MIDL 2018), Amsterdam, The Netherlands, 4–6 July 2018. [Google Scholar]
- Yan, H.; Lu, H.; Ye, M.; Yan, K.; Jin, Q. Improved Mask R-CNN for Lung Nodule Segmentation. In Proceedings of the 2019 10th International Conference on Information Technology in Medicine and Education (ITME), Qingdao, China, 23–25 August 2019; pp. 137–147. [Google Scholar]
- Xiao, Z.; Liu, B.; Geng, L.; Zhang, F.; Liu, Y. Segmentation of lung nodules using improved 3D-Unet neural network. Symmetry 2020, 12, 1787. [Google Scholar] [CrossRef]
- Kashyap, M.; Panjwani, N.; Hasan, M.; Huang, C.; Bush, K.; Dong, P.; Zaky, S.; Chin, A.; Vitzthum, L.; Loo, B.; et al. Deep learning based identification and segmentation of lung tumors on computed tomography images. Int. J. Radiat. Oncol. Biol. Phys. 2021, 111(3S), E92–E93. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, K.; Zha, M.; Qu, X.; Xiao, R. An effective deep neural network for lung lesions segmentation from COVID-19 CT images. IEEE Trans. Ind. Inform. 2021, 17, 6528–6538. [Google Scholar] [CrossRef]
- Zhang, M.; Li, H.; Pan, S.; Lyu, J.; Su, S. Convolutional neural networks based lung nodule classification: A surrogate-assisted evolutionary algorithm for hyperparameter optimization. IEEE Trans. Evol. Comput. 2021, 25, 869–882. [Google Scholar] [CrossRef]
- Jalali, Y.; Fateh, M.; Rezvani, M.; Abolghasemi, V.; Anisi, M.H. ResBCDU-Net: A deep learning framework for lung ct image segmentation. Sensors 2021, 21, 268. [Google Scholar] [CrossRef]
- Balaha, H.; Balaha, M.; Ali, H. Hybrid COVID-19 segmentation and recognition framework (HMB-HCF) using deep learning and genetic algorithms. Artif. Intell. Med. 2021, 119, 102156. [Google Scholar] [CrossRef]
- Lin, X.; Jiao, H.; Pang, Z.; Chen, H.; Wu, W.; Wang, X.; Xiong, L.; Chen, B.; Huang, Y.; Li, S.; et al. Lung cancer and granuloma identification using a deep learning model to extract 3-dimensional radiomics features in CT imaging. Clin. Lung Cancer 2021, 22, e756–e766. [Google Scholar] [CrossRef]
- Gan, W.; Wang, H.; Gu, H.; Duan, Y.; Xu, Z. Automatic segmentation of lung tumors on CT images based on a 2D & 3D hybrid convolutional neural network. Br. J. Radiol. 2021, 94, 20210038. [Google Scholar]
- Protonotarios, N.E.; Katsamenis, I.; Sykiotis, S.; Dikaios, N.; Kastis, G.A.; Chatziioannou, S.N.; Metaxas, M.; Doulamis, N.; Doulamis, A. A FEW-SHOT U-NET deep learning model for lung cancer lesion segmentation via PET/CT imaging. Biomed. Phys. Eng. Express 2022, 8, 025019. [Google Scholar] [CrossRef]
- Kim, H.M.; Ko, T.; Young, C.I.; Myong, J.P. Asbestosis diagnosis algorithm combining the lung segmentation method and deep learning model in computed tomography image. Int. J. Med. Inform. 2022, 158, 104667. [Google Scholar] [CrossRef]
- Chang, C.Y.; Chen, S.J.; Tsai, M.F. Application of support-vector-machine-based method for feature selection and classification of thyroid nodules in ultrasound images. Pattern Recognit. 2010, 43, 3494–3506. [Google Scholar] [CrossRef]
- Nithila, E.E.; Kumar, S.S. Segmentation of lung nodule in CT data using active contour model and Fuzzy C-mean clustering. Alex. Eng. J. 2016, 55, 2583–2588. [Google Scholar] [CrossRef]
- Zhang, X. Computer-Aided Detection of Pulmonary Nodules in Helical CT Images. Ph.D. Dissertation, The University of Iowa, Iowa City, IA, USA, 2005. [Google Scholar]
- Hwang, J.; Chung, M.J.; Bae, Y.; Shin, K.M.; Jeong, S.Y.; Lee, K.S. Computer-aided detection of lung nodules. J. Comput. Assist. Tomogr. 2010, 34, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Young, S.; Lo, P.; Kim, G.; Brown, M.; Hoffman, J.; Hsu, W.; Wahi-Anwar, W.; Flores, C.; Lee, G.; Noo, F.; et al. The effect of radiation dose reduction on computer-aided detection (CAD) performance in a low-dose lung cancer screening population. Med. Phys. 2017, 44, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Suzuki, K. Comparing two classes of end-to-end machine-learning models in lung nodule detection and classification: MTANNS VS. CNNS. Pattern Recognit. 2017, 63, 476–486. [Google Scholar] [CrossRef]
- Liu, X.; Hou, F.; Hong, Q.; Hao, A. Multi-view multi-scale CNNs for lung nodule type classification from CT images. Pattern Recognit. 2018, 77, 262–275. [Google Scholar] [CrossRef]
- Cao, H.C.; Liu, H.; Song, E.; Ma, G.Z.; Xu, X.Y.; Jin, R.C.; Liu, T.Y.; Hung, C.C. A Two-Stage Convolutional Neural Networks for Lung Nodule Detection. IEEE J. Biomed. Health Inform. 2020, 24, 2006–2015. [Google Scholar] [CrossRef]
- Alakwaa, W.; Nassef, M.; Badr, A. Lung cancer detection and classification with 3D convolutional neural network (3D-CNN). Lung Cancer 2017, 8, 409–417. [Google Scholar] [CrossRef]
- Anirudh, R.; Thiagarajan, J.J.; Bremer, T.; Kim, H. Lung nodule detection using 3D convolutional neural networks trained on weakly labeled data. In Proceedings of the Medical Imaging 2016: Computer-Aided Diagnosis, International Society for Optics and Photonics, San Diego, CA, USA, 27 February–3 March 2016; Volume 9785, pp. 1–6. [Google Scholar]
- Feng, Y.; Hao, P.; Zhang, P.; Liu, X.; Wu, F.; Wang, H. Supervoxel based weakly-supervised multi-level 3D CNNs for lung nodule detection and segmentation. J. Ambient. Intell. Humaniz. Comput. 2019. [Google Scholar] [CrossRef]
- Perez, G.; Arbelaez, P. Automated lung cancer diagnosis using three-dimensional convolutional neural networks. Med. Biol. Eng. Comput. 2020, 58, 1803–1815. [Google Scholar] [CrossRef]
- Vipparla, V.K.; Chilukuri, P.K.; Kande, G.B. Attention based multi-patched 3D-CNNs with hybrid fusion architecture for reducing false positives during lung nodule detection. J. Comput. Commun. 2021, 9, 1–26. [Google Scholar] [CrossRef]
- Dutande, P.; Baid, U.; Talbar, S. Deep residual separable convolutional neural network for lung tumor segmentation. Comput. Biol. Med. 2022, 141, 105161. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Song, T.; Wang, G.; Chen, J.; Chen, Y.; Li, K.; Metaxas, D.N.; Zhang, S. SCPM-Net: An anchor-free 3D lung nodule detection network using sphere representation and center points matching. Med. Image Anal. 2022, 75, 102287. [Google Scholar] [CrossRef] [PubMed]
- Franck, C.; Snoeckx, A.; Spinhoven, M.; Addouli, H.E.; Zanca, F. Pulmonary nodule detection in chest CT using a deep learning-based reconstruction algorithm. Radiat. Prot. Dosim. 2021, 195, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.; Chen, H.; Yu, L.; Qin, J.; Heng, P.A. Multi-level contextual 3D CNNs for false positive reduction in pulmonary nodule detection. IEEE Trans. Biomed. Eng. 2016, 64, 1558–1567. [Google Scholar] [CrossRef]
- Setio, A.; Ciompi, F.; Litjens, G.; Gerke, P.; Jacobs, C.; Riel, S.; Wille, M.M.; Naqibullah, M.; Sanchez, C.I.; van Ginneken, B. Pulmonary nodule detection in CT images: False positive reduction using multi-view convolutional networks. IEEE Trans. Med. Imaging 2016, 35, 1160–1169. [Google Scholar] [CrossRef] [PubMed]
- Mercy Theresa, M.; Subbiah Bharathi, V. CAD for lung nodule detection in chest radiography using complex wavelet transform and shearlet transform features. Indian J. Sci. Technol. 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Jin, T.; Hui, C.; Shan, Z.; Wang, X. Learning Deep Spatial Lung Features by 3D Convolutional Neural Network for Early Cancer Detection. In Proceedings of the 2017 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Sydney, Australia, 29 November–1 December 2017. [Google Scholar]
- Zhu, W.; Liu, C.; Fan, W.; Xie, X. DeepLung: Deep 3D dual path nets for automated pulmonary nodule detection and classification. arXiv 2017, arXiv:1709.05538. [Google Scholar]
- Eun, H.Y.; Kim, D.Y.; Jung, C.; Kim, C. Single-view 2D CNNs with Fully Automatic Non-nodule Categorization for False Positive Reduction in Pulmonary Nodule Detection. Comput. Methods Programs Biomed. 2018, 165, 215–224. [Google Scholar] [CrossRef]
- Ramachandran, S.; George, J.; Skaria, S.; Varun, V.V. Using yolo based deep learning network for real time detection and localization of lung nodules from low dose CT scans. In Proceedings of the SPIE 10575, Medical Imaging 2018: Computer-Aided Diagnosis, 105751I (2018). Houston, TX, USA, 27 February 2018. [Google Scholar] [CrossRef]
- Serj, M.F.; Lavi, B.; Hoff, G.; Valls, D.P. A deep convolutional neural network for lung cancer diagnostic. arXiv 2018, arXiv:1804.08170. [Google Scholar]
- Zhang, J.; Xia, Y.; Zeng, H.; Zhang, Y. Nodule: Combining constrained multi-scale log filters with densely dilated 3D deep convolutional neural network for pulmonary nodule detection. Neurocomputing 2018, 317, 159–167. [Google Scholar] [CrossRef]
- Schwyzer, M.; Ferraro, D.A.; Muehlematter, U.J.; Curioni-Fontecedro, A.; Messerli, M. Automated detection of lung cancer at ultralow dose PET/CT by deep neural networks—Initial results. Lung Cancer 2018, 126, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Gerard, S.E.; Patton, T.J.; Christensen, G.E.; Bayouth, J.E.; Reinhardt, J.M. Fissurenet: A deep learning approach for pulmonary fissure detection in CT images. IEEE Trans. Med. Imaging 2018, 38, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.S.; Kim, Y.S.; Plichta, K.; Allen, B.G.; Zhou, L.X.; Buatti, J.; Wu, X.D. Simultaneous cosegmentation of tumors in PET-CT images using deep fully convolutional networks. Med. Phys. 2019, 2, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.C.; Choi, J.S.; Suk, H.I. Multi-scale gradual integration CNN for false positive reduction in pulmonary nodule detection. Neural Netw. 2019, 115, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Masood, A.; Sheng, B.; Li, P.; Hou, X.; Wei, X.; Qin, J.; Feng, D. Computer-assisted decision support system in pulmonary cancer detection and stage classification on CT images. J. Biomed. Inform. 2018, 79, 117–128. [Google Scholar] [CrossRef]
- Nam, J.G.; Park, S.; Hwang, E.J.; Lee, J.H.; Jin, K.-N.; Lim, K.Y.; Vu, T.H.; Sohn, J.H.; Hwang, S.; Goo, J.M.; et al. Development and validation of deep learning-based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Clin. Infect. Dis. 2019, 69, 739–747. [Google Scholar] [CrossRef]
- Choi, W.; Oh, J.H.; Riyahi, S.; Liu, C.J.; Lu, W. Radiomics analysis of pulmonary nodules in low-dose CT for early detection of lung cancer. Med. Phys. 2018, 45, 1537–1549. [Google Scholar] [CrossRef]
- Tan, J.X.; Huo, Y.M.; Liang, Z.; Li, L. Expert knowledge-infused deep learning for automatic lung nodule detection. J. X-Ray Sci. Technol. 2019, 27, 17–35. [Google Scholar] [CrossRef]
- Ozdemir, O.; Russell, R.L.; Berlin, A.A. A 3D probabilistic deep learning system for detection and diagnosis of lung cancer using low-dose CT scans. IEEE Trans. Med. Imaging 2020, 39, 1419–1429. [Google Scholar] [CrossRef]
- Cha, M.J.; Chung, M.J.; Lee, J.H.; Lee, K.S. Performance of deep learning model in detecting operable lung cancer with chest radiographs. J. Thorac. Imaging 2019, 34, 86–91. [Google Scholar] [CrossRef]
- Pham, H.; Futakuchi, M.; Bychkov, A.; Furukawa, T.; Fukuoka, J. Detection of lung cancer lymph node metastases from whole-slide histopathologic images using a two-step deep learning approach. Am. J. Pathol. 2019, 189, 2428–2439. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Vilmun, B.M.; Carlsen, J.F.; Albrecht-Beste, E.; Lauridsen, C.A.; Nielsen, M.B.; Hansen, K.L. The performance of deep learning algorithms on automatic pulmonary nodule detection and classification tested on different datasets that are not derived from LIDC-IDRI: A systematic review. Diagnostics 2019, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jin, W.; Li, G.; Yin, C. Yolo v2 network with asymmetric convolution kernel for lung nodule detection of CT image. Chin. J. Biomed. Eng. 2019, 38, 401–408. [Google Scholar]
- Guo, T.; Xie, S.P. Automated segmentation and identification of pulmonary nodule images. Comput. Eng. Des. 2019, 40, 467–472. [Google Scholar]
- Huang, W.; Hu, L. Using a noisy U-net for detecting lung nodule candidates. IEEE Access 2019, 7, 67905–67915. [Google Scholar] [CrossRef]
- Gu, Y.; Lu, X.; Zhang, B.; Zhao, Y.; Zhou, T. Automatic lung nodule detection using multi-scale dot nodule-enhancement filter and weighted support vector machines in chest computed tomography. PLoS ONE 2019, 14, e0210551. [Google Scholar] [CrossRef]
- Kumar, A.; Fulham, M.J.; Feng, D.; Kim, J. Co-learning feature fusion maps from PET-CT images of lung cancer. IEEE Trans. Med. Imaging 2019, 39, 204–217. [Google Scholar] [CrossRef]
- Pesce, E.; Withey, S.; Ypsilantis, P.P.; Bakewell, R.; Goh, V.; Montana, G. Learning to detect chest radiographs containing pulmonary lesions using visual attention networks. Med. Image Anal. 2019, 53, 26–38. [Google Scholar] [CrossRef]
- Huang, X.; Lei, Q.; Xie, T.; Zhang, Y.; Hu, Z.; Zhou, Q. Deep transfer convolutional neural network and extreme learning machine for lung nodule diagnosis on CT images. Knowl. Based Syst. 2020, 204, 105230. [Google Scholar] [CrossRef]
- Zheng, S.; Cornelissen, L.J.; Cui, X.; Jing, X.; Ooijen, P. Deep convolutional neural networks for multiplanar lung nodule detection: Improvement in small nodule identification. Med. Phys. 2021, 48, 733–744. [Google Scholar] [CrossRef]
- Xu, X.; Wang, C.; Guo, J.; Gan, Y.; Yi, Z. MSCS-DEEPLN: Evaluating lung nodule malignancy using multi-scale cost-sensitive neural networks. Med. Image Anal. 2020, 65, 101772. [Google Scholar] [CrossRef] [PubMed]
- Yektai, H.; Manthouri, M. Diagnosis of lung cancer using multi-scale convolutional neural network. Biomed. Eng. Appl. Basis Commun. 2020, 32, 2050030. [Google Scholar] [CrossRef]
- Heuvelmans, M.A.; Ooijen, P.; Ather, S.; Silva, C.F.; Oudkerk, M. Lung cancer prediction by deep learning to identify benign lung nodules. Lung Cancer 2021, 154, 1–4. [Google Scholar] [CrossRef]
- Hsu, H.H.; Ko, K.H.; Chou, Y.C.; Wu, Y.C.; Chiu, S.H.; Chang, C.K.; Chang, W.C. Performance and reading time of lung nodule identification on multidetector CT with or without an artificial intelligence-powered computer-aided detection system. Clin. Radiol. 2021, 76, 626.e23. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Choi, E.J.; Chung, J.H.; Lee, K.W.; Oh, J.W. A DNA-derived phage nose using machine learning and artificial neural processing for diagnosing lung cancer. Biosens. Bioelectron. 2021, 194, 113567. [Google Scholar] [CrossRef] [PubMed]
- Afshar, P.; Naderkhani, F.; Oikonomou, A.; Rafiee, M.J.; Plataniotis, K.N. MIXCAPS: A capsule network-based mixture of experts for lung nodule malignancy prediction. Pattern Recognit. 2021, 116(August 2021), 107942. [Google Scholar] [CrossRef]
- Lai, K.D.; Nguyen, T.T.; Le, T.H. Detection of lung nodules on ct images based on the convolutional neural network with attention mechanism. Ann. Emerg. Technol. Comput. 2021, 5, 78–89. [Google Scholar] [CrossRef]
- Bu, Z.; Zhang, X.; Lu, J.; Lao, H.; Liang, C.; Xu, X.; Wei, Y.; Zeng, H. Lung nodule detection based on YOLOv3 deep learning with limited datasets. Mol. Cell. Biomech. 2022, 19, 17–28. [Google Scholar] [CrossRef]
- Al-Shabi, M.; Shak, K.; Tan, M. ProCAN: Progressive growing channel attentive non-local network for lung nodule classification. Pattern Recognit. 2022, 122, 108309. [Google Scholar] [CrossRef]
- Farag, A.; Ali, A.; Graham, J.; Farag, A.; Elshazly, S.; Falk, R. Evaluation of geometric feature descriptors for detection and classification of lung nodules in low dose CT scans of the chest. In Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, USA, 30 March–2 April 2011; pp. 169–172. [Google Scholar]
- Orozco, H.M.; Villegas, O.O.V.; Domínguez, H.J.O.; Domínguez, H.D.J.O.; Sanchez, V.G.C. Lung nodule classification in CT thorax images using support vector machines. In Proceedings of the 2013 12th Mexican International Conference on Artificial Intelligence (MICAI), Mexico City, Mexico, 24–30 November 2013; pp. 277–283. [Google Scholar]
- Krewer, H.; Geiger, B.; Hall, L.O.; Goldgof, D.B.; Gu, Y.; Tockman, M.; Gillies, R.J. Effect of texture features in computer aided diagnosis of pulmonary nodules in low-dose computed tomography. In Proceedings of the 2013 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Manchester, UK, 13–16 October 2013; pp. 3887–3891. [Google Scholar]
- Parveen, S.S.; Kavitha, C. Classification of lung cancer nodules using SVM Kernels. Int. J. Comput. Appl. 2014, 95, 975–8887. [Google Scholar]
- Dandıl, E.; Çakiroğlu, M.; Ekşi, Z.; Özkan, M.; Kurt, Ö.K.; Canan, A. Artificial neural network-based classification system for lung nodules on computed tomography scans. In Proceedings of the 2014 6th International Conference of Soft Computing and Pattern Recognition (SoCPar), Tunis, Tunisia, 11–14 August 2014; pp. 382–386. [Google Scholar]
- Hua, K.L.; Hsu, C.H.; Hidayati, S.C.; Hidayati, S.C.; Cheng, W.H.; Chen, Y.J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets Ther. 2015, 8, 2015–2022. [Google Scholar]
- Kumar, D.; Wong, A.; Clausi, D.A. Lung nodule classification using deep features in CT images. In Proceedings of the 2015 12th Conference on Computer and Robot Vision (CRV), Halifax, NS, Canada, 3–5 June 2015; pp. 133–138. [Google Scholar]
- Shen, S.; Han, S.X.; Aberle, D.R.; Bui, A.A.; Hsu, W. An interpretable deep hierarchical semantic convolutional neural network for lung nodule malignancy classification. Expert Syst. Appl. 2019, 128, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Zhou, M.; Yang, F.; Yang, C.; Tian, J. Muti-scale convolutional neural networks for lung nodule Classification. Inf. Process. Med. Imaging 2015, 24, 588–599. [Google Scholar] [PubMed]
- Cheng, J.Z.; Ni, D.; Chou, Y.H.; Qin, J.; Tiu, C.M.; Chang, Y.C.; Huang, C.S.; Shen, D.; Chen, C.M. Computer-aided diagnosis with deep learning architecture: Applications to breast lesions in US images and pulmonary nodules in CT scans. Sci. Rep. 2016, 6, 24454. [Google Scholar] [CrossRef]
- Kwajiri, T.L.; Tezukam, T. Classification of Lung Nodules Using Deep Learning. Trans. Jpn. Soc. Med. Biol. Eng. 2017, 55, 516–517. [Google Scholar]
- Shen, W.; Zhou, M.; Yang, F.; Yu, D.; Dong, D.; Yang, C.; Tian, J.; Zang, Y. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit. 2017, 61, 663–673. [Google Scholar] [CrossRef]
- Abbas, Q. Lung-deep: A computerized tool for detection of lung nodule patterns using deep learning algorithms detection of lung nodules patterns. Int. J. Adv. Comput. Sci. Appl. 2017, 8, 112–116. [Google Scholar] [CrossRef]
- Da Silva, G.L.F.; da Silva Neto, O.P.; Silva, A.C.; Gattass, M. Lung nodules diagnosis based on evolutionary convolutional neural network. Multimed. Tools Appl. 2017, 76, 19039–19055. [Google Scholar] [CrossRef]
- Zhang, B.; Qi, S.; Monkam, P.; Li, C.; Qian, W. Ensemble learners of multiple deep CNNs for pulmonary nodules classification using CT images. IEEE Access 2019, 7, 110358–110371. [Google Scholar] [CrossRef]
- Sahu, P.; Yu, D.; Dasari, M.; Hou, F.; Qin, H. A lightweight multi-section CNN for lung nodule classification and malignancy estimation. IEEE J. Biomed. Health Inform. 2019, 23, 960–968. [Google Scholar] [CrossRef]
- Ali, I.; Muzammil, M.; Ulhaq, D.I.; Khaliq, A.A.; Malik, S. Efficient lung nodule classification using transferable texture convolutional neural network. IEEE Access 2020, 8, 175859–175870. [Google Scholar] [CrossRef]
- Marques, S.; Schiavo, F.; Ferreira, C.A.; Pedrosa, J.; Cunha, A.; Campilho, A. A multi-task CNN approach for lung nodule malignancy classification and characterization. Expert Syst. Appl. 2021, 184, 115469.1–115469.9. [Google Scholar] [CrossRef]
- Thamilarasi, V.; Roselin, R. Automatic classification and accuracy by deep learning using CNN methods in lung chest x-ray images. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1055, 012099. [Google Scholar] [CrossRef]
- Kawathekar, I.D.; Areeckal, A.S. Performance analysis of texture characterization techniques for lung nodule classification. J. Phys. Conf. Ser. 2022, 2161, 012045. [Google Scholar] [CrossRef]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2016, arXiv:1511.06434. [Google Scholar]
- Chuquicusma, M.J.M.; Hussein, S.; Burt, J.; Bagci, U. How to fool radiologists with generative adversarial networks? A visual turing test for lung cancer diagnosis. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 240–244. [Google Scholar]
- Zhao, D.; Zhu, D.; Lu, J.; Luo, Y.; Zhang, G. Synthetic Medical Images Using F&BGAN for Improved Lung Nodules Classification by Multi-Scale VGG16. Symmetry 2018, 10, 519. [Google Scholar]
- Teramoto, A.; Yamada, A.; Kiriyama, Y.; Tsukamoto, T.; Fujita, H. Automated classification of benign and malignant cells from lung cytological images using deep convolutional neural network. Inform. Med. Unlocked 2019, 16, 100205. [Google Scholar] [CrossRef]
- Rani, K.V.; Jawhar, S.J. Superpixel with nanoscale imaging and boosted deep convolutional neural network concept for lung tumor classification. Int. J. Imaging Syst. Technol. 2020, 30, 899–915. [Google Scholar] [CrossRef]
- Kuruvilla, J.; Gunavathi, K. Lung cancer classification using neural networks for CT images. Comput. Methods Programs Biomed 2014, 113, 202–209. [Google Scholar] [CrossRef]
- Ciompi, F.; Chung, K.; Riel, S.V.; Setio, A.; Gerke, P.K.; Jacobs, C.; Scholten, E.T.; Schaefer-Prokop, C.; Wille, M.M.W.; Marchianò, A.; et al. Corrigendum: Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci. Rep. 2017, 7, 46878. [Google Scholar] [CrossRef]
- Nurtiyasari, D.; Rosadi, D.; Abdurakhman. The application of Wavelet Recurrent Neural Network for lung cancer classification. In Proceedings of the 2017 3rd International Conference on Science and Technology—Computer (ICST), Yogyakarta, Indonesia, 11–12 July 2017; pp. 127–130. [Google Scholar]
- De Carvalho Filho, A.O.; Silva, A.C.; de Paiva, A.C.; Nunes, R.A.; Gattass, M. Classification of patterns of benignity and malignancy based on CT using topology-based phylogenetic diversity index and convolutional neural network. Pattern Recognit. 2018, 81, 200–212. [Google Scholar] [CrossRef]
- Lindsay, W.; Wang, J.; Sachs, N.; Barbosa, E.; Gee, J. Transfer learning approach to predict biopsy-confirmed malignancy of lung nodules from imaging data: A pilot study. In Image Analysis for Moving Organ, Breast, and Thoracic Images; Springer: Berlin/Heidelberg, Germany, 2018; pp. 295–301. [Google Scholar]
- Keming, M.; Renjie, T.; Xinqi, W.; Weiyi, Z.; Haoxiang, W. Feature representation using deep autoencoder for lung nodule image classification. Complexity 2018, 3078374. [Google Scholar] [CrossRef]
- Matsuyama, E.; Tsai, D.Y. Automated classification of lung diseases in computed tomography images using a wavelet based convolutional neural network. J. Biomed. Sci. Eng. 2018, 11, 263–274. [Google Scholar] [CrossRef][Green Version]
- Sathyan, H.; Panicker, J.V. Lung Nodule Classification Using Deep ConvNets on CT Images. In Proceedings of the 2018 9th International Conference on Computing, Communication and Networking Technologies, Bengaluru, India, 10–12 July 2018; p. 18192544. [Google Scholar] [CrossRef]
- Xie, Y.; Xia, Y.; Zhang, J.; Song, Y.; Feng, D.; Fulham, M.; Cai, W. Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans. Med. Imaging 2019, 38, 991–1004. [Google Scholar] [CrossRef]
- Nasrullah, N.; Sang, J.; Alam, M.S.; Mateen, M.; Cai, B.; Hu, H. Automated lung nodule detection and classification using deep learning combined with multiple strategies. Sensors 2019, 19, 3722. [Google Scholar] [CrossRef]
- Shakeel, P.M.; Burhanuddin, M.A.; Desa, M.I. Lung cancer detection from CT image using improved profuse clustering and deep learning instantaneously trained neural networks. Measurement 2019, 145, 702–712. [Google Scholar] [CrossRef]
- Suresh, S.; Mohan, S. Roi-based feature learning for efficient true positive prediction using convolutional neural network for lung cancer diagnosis. Neural Comput. Appl. 2020, 32, 15989–16009. [Google Scholar] [CrossRef]
- Su, R.; Xie, W.; Tan, T. 2.75D convolutional neural network for pulmonary nodule classification in chest CT. arXiv 2020, arXiv:2002.04251. [Google Scholar]
- Zia, M.B.; Zhao, J.J.; Ning, X. Detection and classification of lung nodule in diagnostic CT: A TSDN method based on improved 3D-FASTER R-CNN and multi-scale multi-crop convolutional neural network. Int. J. Hybrid Inf. Technol. 2020, 13, 45–56. [Google Scholar] [CrossRef]
- Lin, C.J.; Li, Y.C. Lung nodule classification using taguchi-based convolutional neural networks for computer tomography images. Electronics 2020, 9, 1066. [Google Scholar] [CrossRef]
- Mmmap, A.; Sjj, B.; Gjm, C. Optimal deep belief network with opposition based pity beetle algorithm for lung cancer classification: A DBNOPBA approach. Comput. Methods Programs Biomed 2021, 199, 105902. [Google Scholar]
- Baranwal, N.; Doravari, P.; Kachhoria, R. Classification of histopathology images of lung cancer using convolutional neural network (CNN). arXiv 2021, arXiv:2112.13553. [Google Scholar]
- Shiwei, L.I.; Liu, D. Automated classification of solitary pulmonary nodules using convolutional neural network based on transfer learning strategy. J. Mech. Med. Biol. 2021, 21, 2140002. [Google Scholar]
- Arumuga Maria Devi, T.; Mebin Jose, V.I. Three Stream Network Model for Lung Cancer Classification in the CT Images. Open Comput. Sci. 2021, 11, 251–261. [Google Scholar] [CrossRef]
- Naik, A.; Edla, D.R.; Kuppili, V. Lung Nodule Classification on Computed Tomography Images Using Fractalnet. Wireless Pers Commun 2021, 119, 1209–1229. [Google Scholar] [CrossRef]
- Ibrahim, D.M.; Elshennawy, N.M.; Sarhan, A.M. Deep-chest: Multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Comput. Biol. Med. 2021, 132, 104348. [Google Scholar] [CrossRef]
- Fu, X.; Bi, L.; Kumar, A.; Fulham, M.; Kim, J. An attention-enhanced cross-task network to analyse lung nodule attributes in CT images. Pattern Recognit. 2022, 126, 108576. [Google Scholar] [CrossRef]
- Vaiyapuri, T.; Liyakathunisa; Alaskar, H.; Parvathi, R.; Pattabiraman, V.; Hussain, A. CAT Swarm Optimization-Based Computer-Aided Diagnosis Model for Lung Cancer Classification in Computed Tomography Images. Appl. Sci. 2022, 12, 5491. [Google Scholar] [CrossRef]
- Halder, A.; Chatterjee, S.; Dey, D. Adaptive morphology aided 2-pathway convolutional neural network for lung nodule classification. Biomed. Signal Process. Control 2022, 72, 103347. [Google Scholar] [CrossRef]
- Forte, G.C.; Altmayer, S.; Silva, R.F.; Stefani, M.T.; Libermann, L.L.; Cavion, C.C.; Youssef, A.; Forghani, R.; King, J.; Mohamed, T.-L.; et al. Deep Learning Algorithms for Diagnosis of Lung Cancer: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 3856. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).