The Role of a Natural Amphibian Skin-Based Peptide, Ranatensin, in Pancreatic Cancers Expressing Dopamine D2 Receptors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Drugs and Chemicals
2.2. Receptor Binding Assay Ex Vivo
2.2.1. Animals
2.2.2. Preparation of Brain Samples for Binding Assays
2.2.3. Receptor Binding Assays
Functional [35S]GTPγS Binding Experiments
Competitive Binding Experiments
2.3. Hemolytic Activity In Vitro
2.4. Cell Culture Preparation and In Vitro Experiments
2.4.1. Enzyme-Linked Immunosorbent Assay
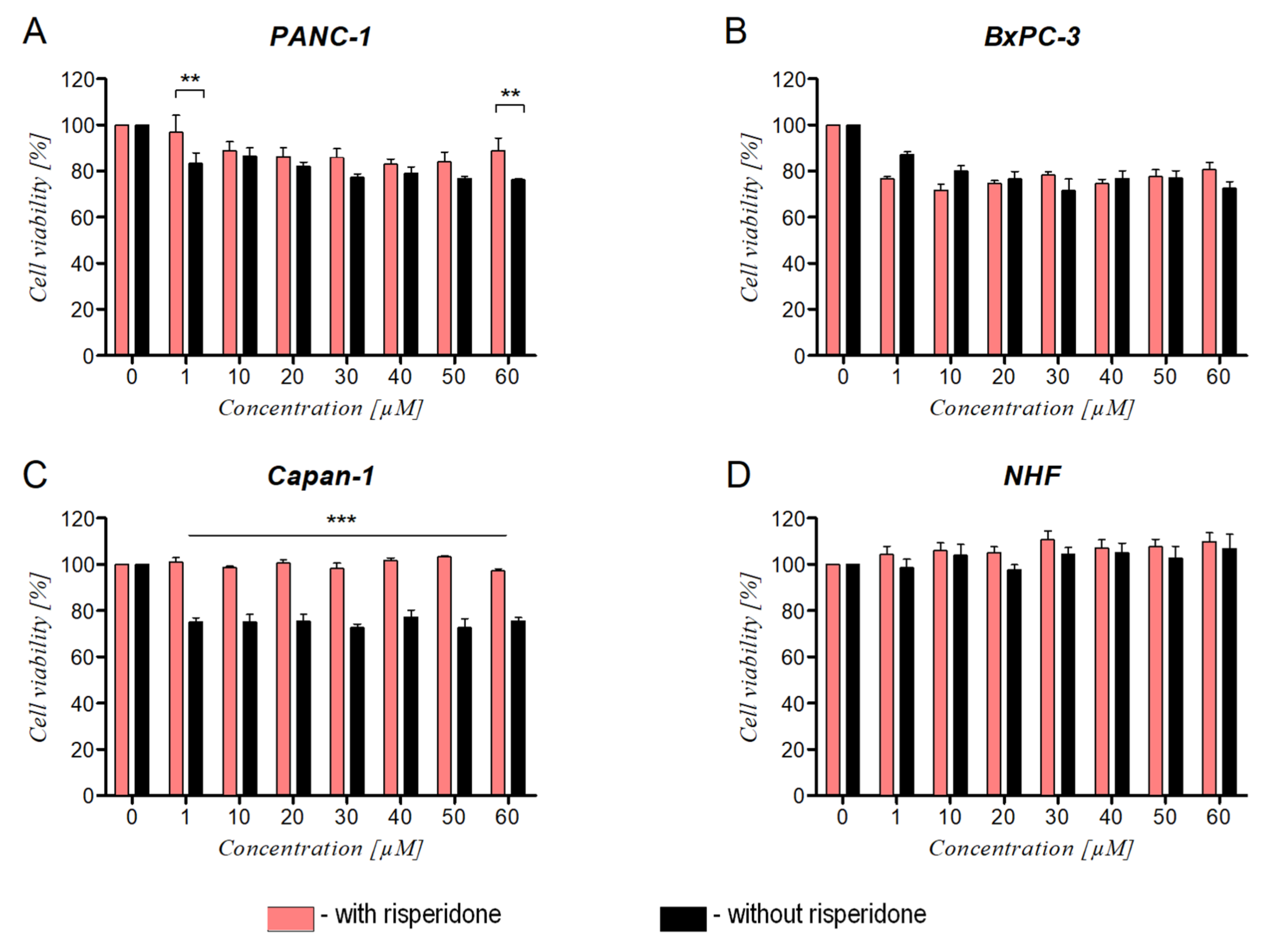
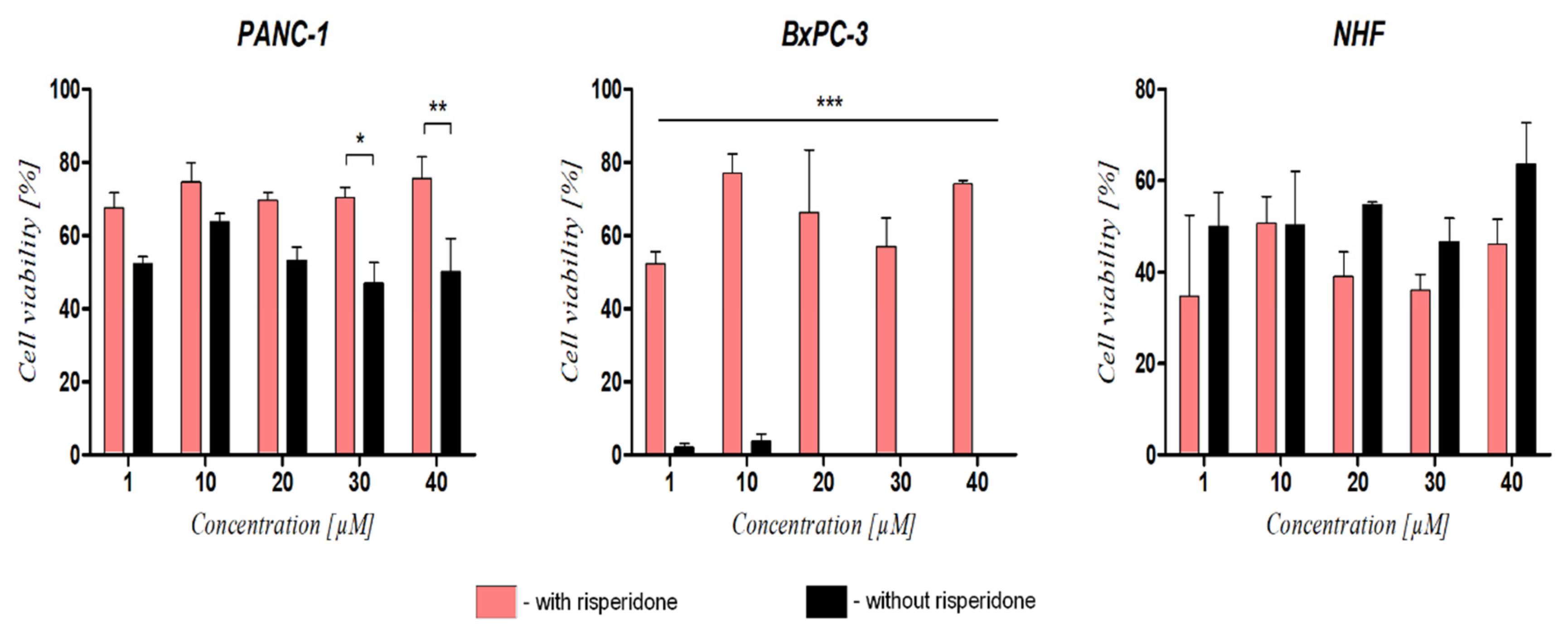
2.4.2. Cell Viability
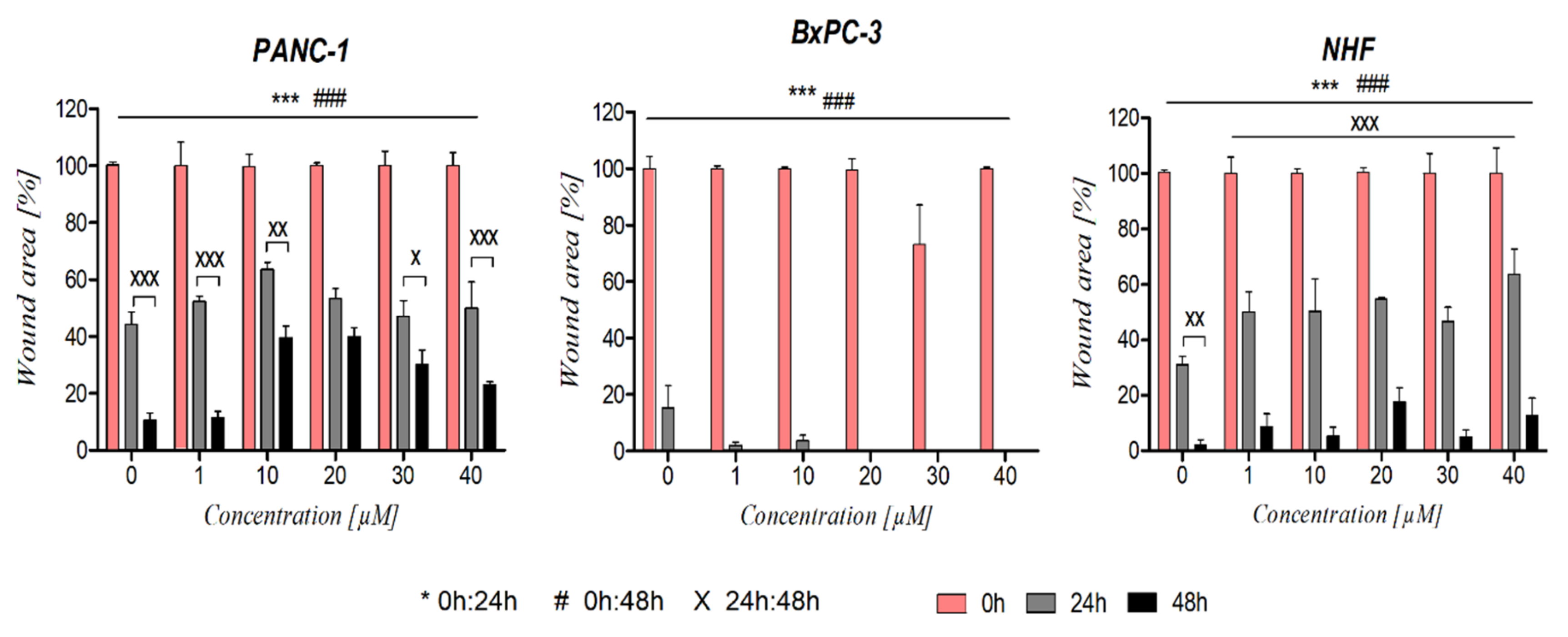
2.4.3. Cell Migration (Scratch Assay)
2.4.4. Western Blot
2.4.5. Statistical Analysis
3. Results
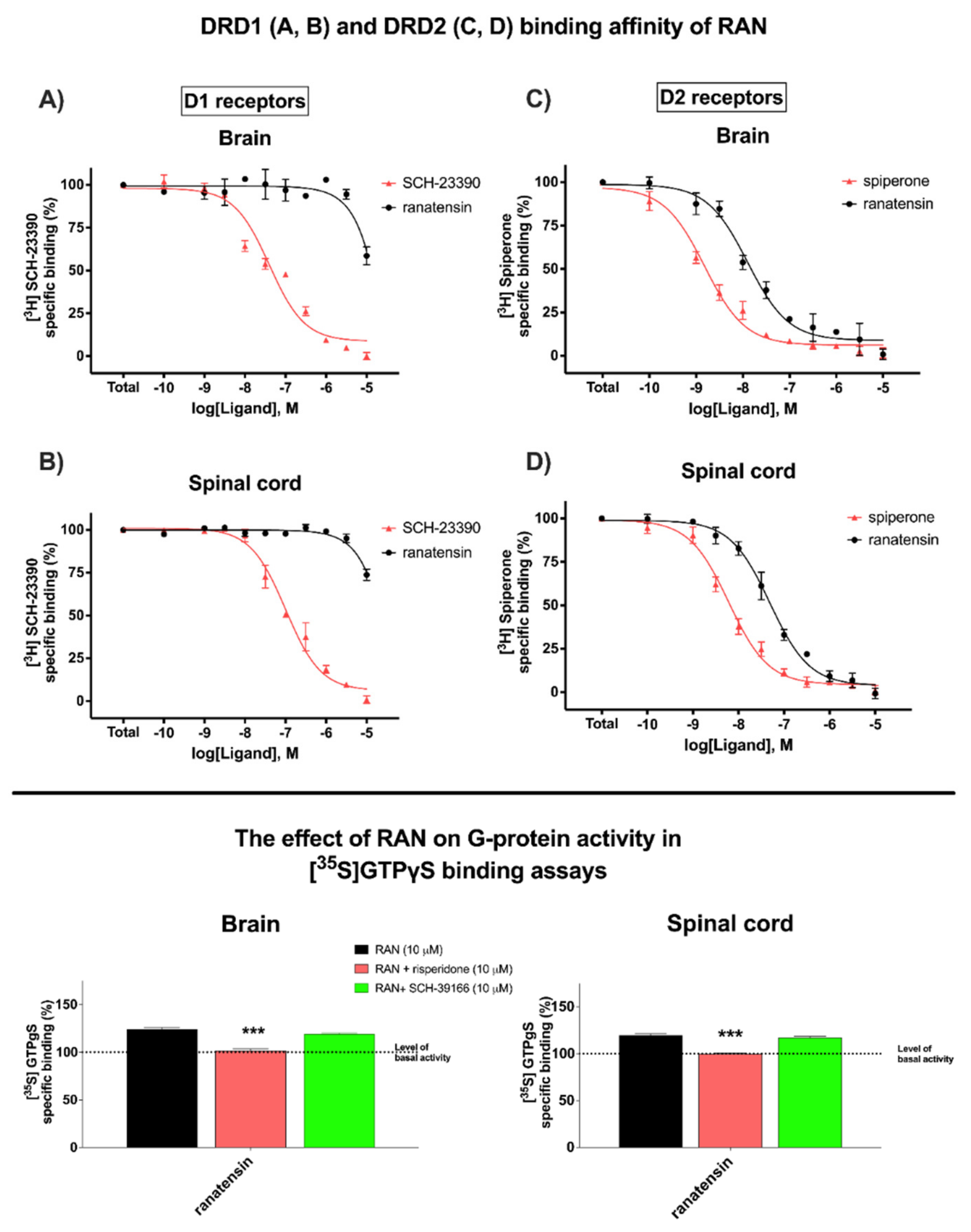
3.1. Ranatensin Affinity and Potency to Dopamine DRD1 and DRD2 Receptors
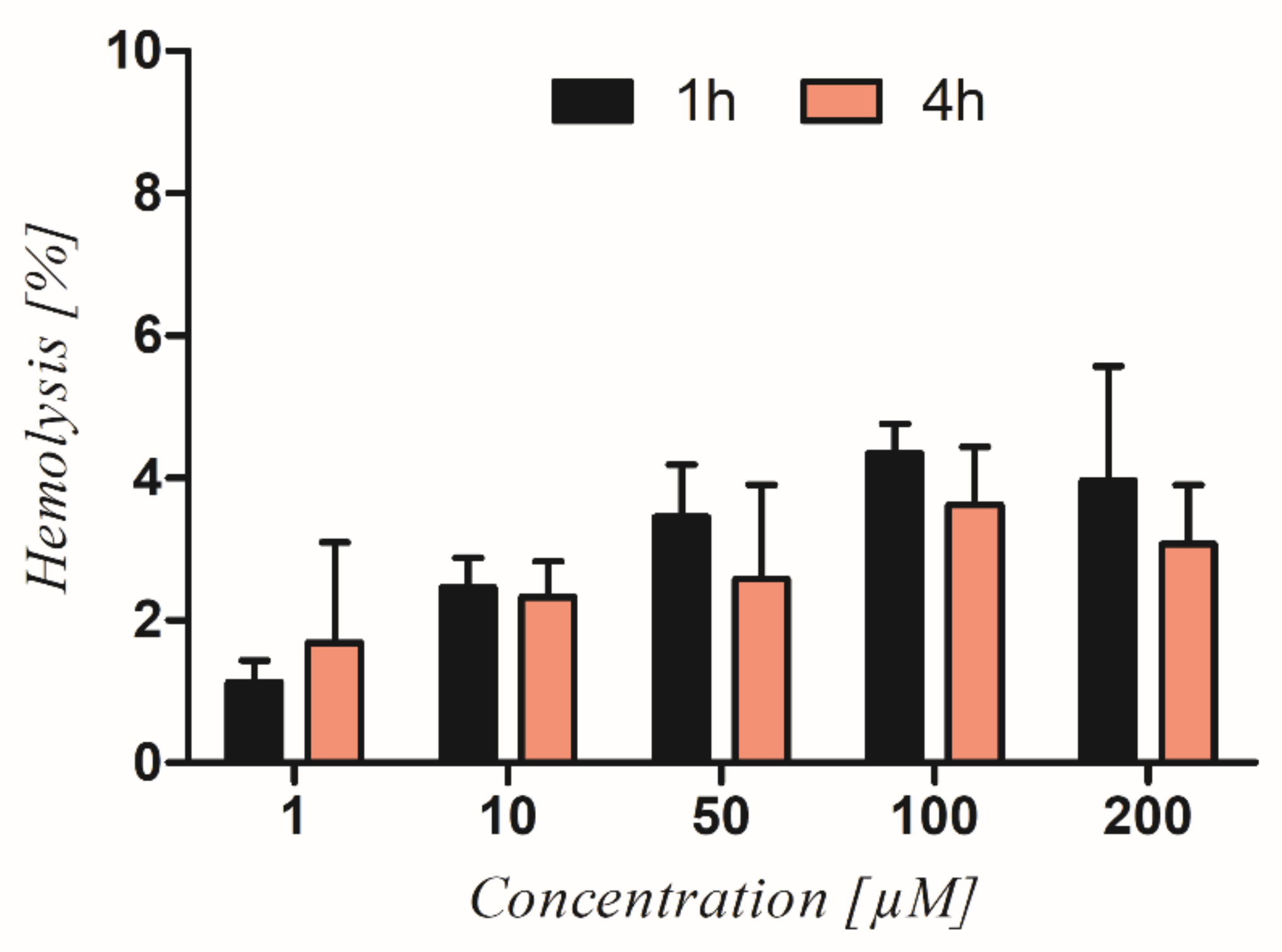
3.2. Hemolytic Activity of Ranatensin in Terms of Its Potent Toxicity
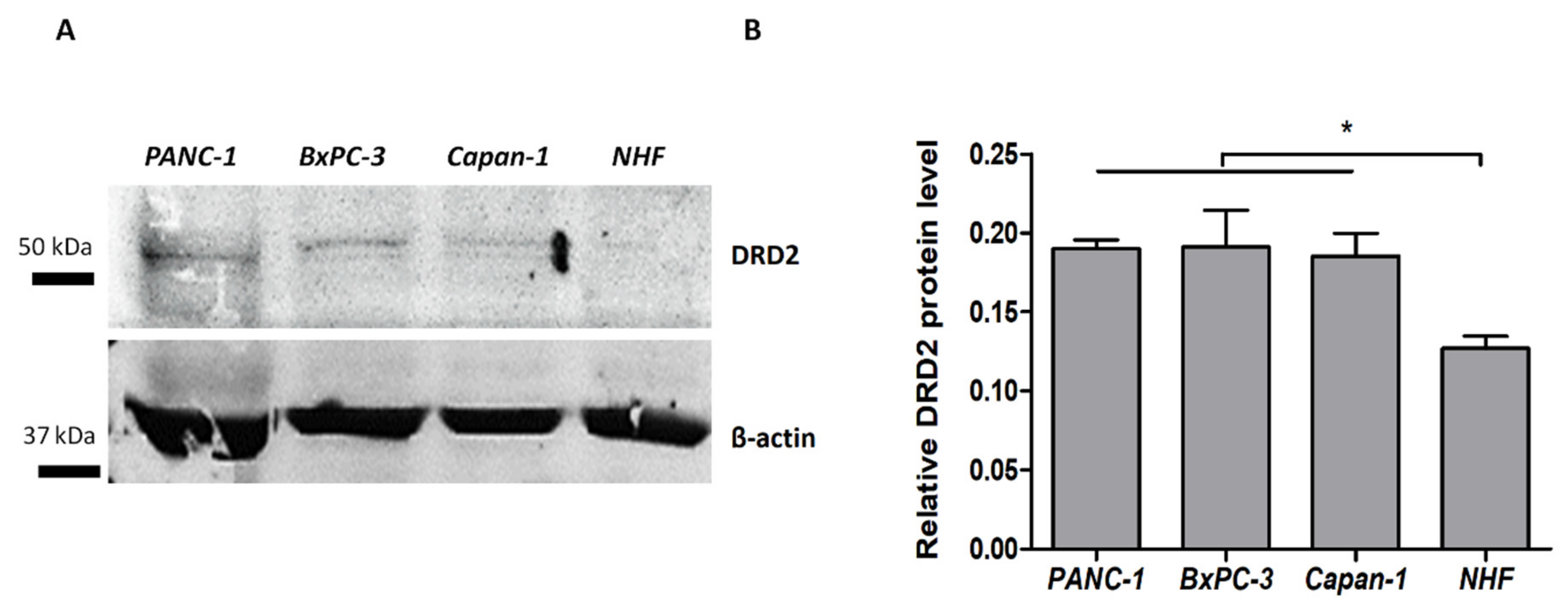
3.3. Dopamine D2 Receptors Expression in Pancreatic Cancer Cells
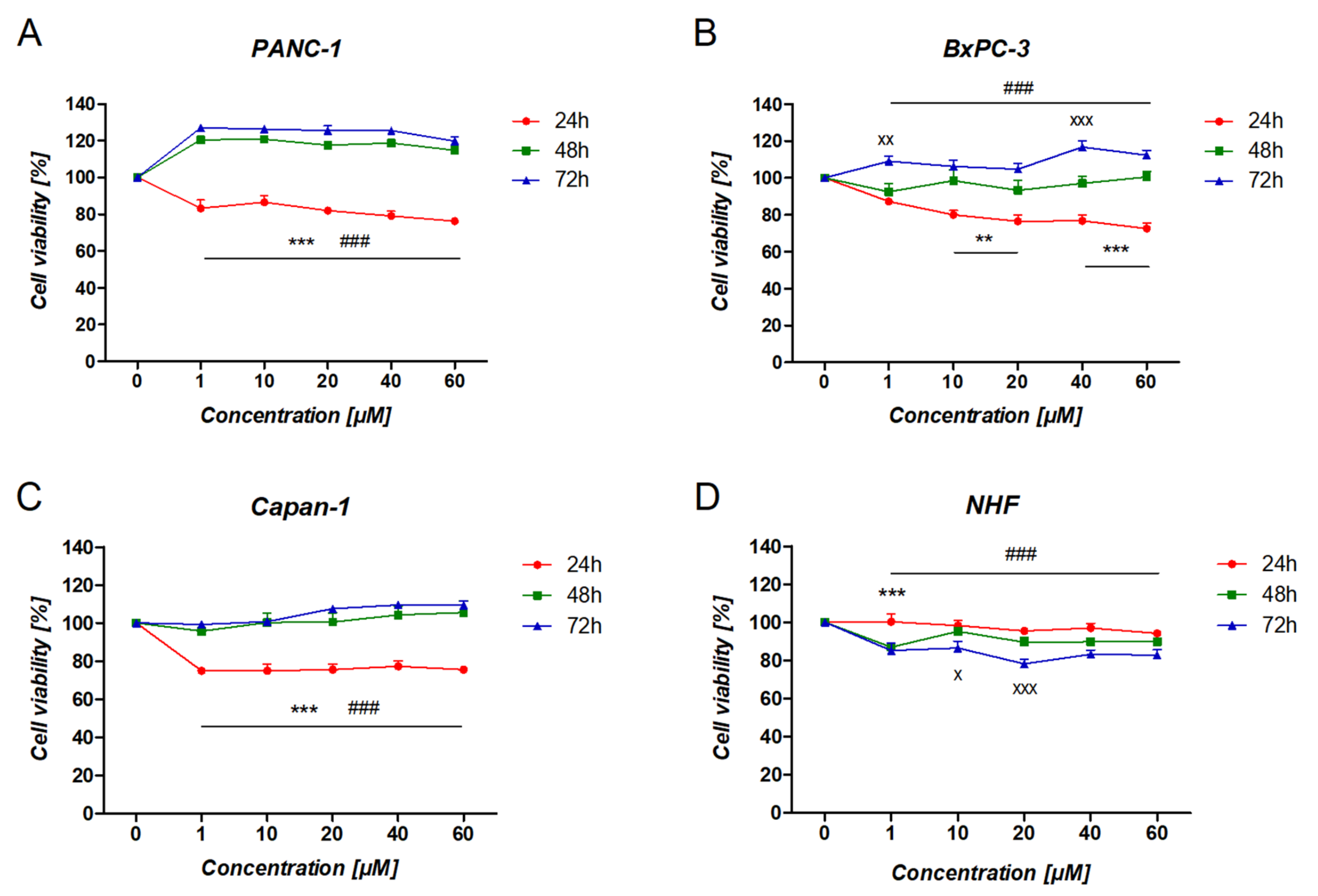
3.4. The Effect of Ranatensin on Pancreatic Cancer Cells
3.5. The Effect of DRD2 Blockage on Ranatensin Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Springfeld, C.; Jäger, D.; Büchler, M.W.; Strobel, O.; Hackert, T.; Palmer, D.H.; Neoptolemos, J.P. Chemotherapy for pancreatic cancer. Presse Med. 2019, 48, e159–e174. [Google Scholar] [CrossRef] [PubMed]
- Saung, M.T.; Zheng, L. Current standards of chemotherapy for pancreatic cancer. Clin. Ther. 2017, 39, 2125–2134. [Google Scholar] [CrossRef]
- Liang, Y.; Li, H.; Gan, Y.; Tu, H. Shedding light on the role of neurotransmitters in the microenvironment of pancreatic cancer. Front. Cell Dev. Biol. 2021, 30, 688953. [Google Scholar] [CrossRef] [PubMed]
- Schuller, H.M. Neurotransmission and cancer: Implications for prevention and therapy. Anticancer Drugs 2008, 19, 655–671. [Google Scholar] [CrossRef]
- Weissenrieder, J.S.; Neighbors, J.D.; Mailman, R.B.; Hohl, R.J. Cancer and the dopamine D2 receptor: A pharmacological perspective. J. Pharmacol. Exp. Ther. 2019, 370, 111–126. [Google Scholar] [CrossRef]
- Rosas-Cruz, A.; Salinas-Jazmín, N.; Velázquez, M.A.V. Dopamine receptors in cancer: Are they valid therapeutic targets? Technol. Cancer Res. Treat. 2021, 20, 15330338211027913. [Google Scholar] [CrossRef]
- Gingrich, J.A.; Caron, M.G. Recent advances in the molecular biology of dopamine receptors. Annu. Rev. Neurosci. 1993, 16, 299–321. [Google Scholar] [CrossRef]
- Wang, Z.; Wen, P.; Hu, B.; Cao, S.; Shi, X.; Guo, W.; Zhang, S. Dopamine and dopamine receptor D1 as a novel favourable biomarker for hepatocellular carcinoma. Cancer Cell Int. 2021, 30, 586. [Google Scholar] [CrossRef]
- Jandaghi, P.; Najafabadi, H.S.; Bauer, A.S.; Papadakis, A.I.; Fassan, M.; Hall, A.; Monast, A.; von Knebel Doeberitz, M.; Neoptolemos, J.P.; Costello, E.; et al. Expression of DRD2 is increased in human pancreatic ductal adenocarcinoma and inhibitors slow tumor growth in mice. Gastroenterology 2016, 151, 1218–1231. [Google Scholar] [CrossRef]
- Pierce, S.R.; Fang, Z.; Yin, Y.; West, L.; Asher, M.; Hao, T.; Zhang, X.; Tucker, K.; Staley, A.; Fan, Y.; et al. Targeting dopamine receptor D2 as a novel therapeutic strategy in endometrial cancer. J. Exp. Clin. Cancer Res. 2021, 40, 61. [Google Scholar] [CrossRef]
- Su, H.; Xue, Z.; Feng, Y.; Xie, Y.; Deng, B.; Yao, Y.; Tian, X.; An, Q.; Yang, L.; Yao, Q.; et al. N-arylpiperazine-containing compound (C2): An enhancer of sunitinib in the treatment of pancreatic cancer, involving D1DR activation. Toxicol. Appl. Pharmacol. 2019, 384, 114789. [Google Scholar] [CrossRef]
- Borcherding, D.C.; Tong, W.; Hugo, E.R.; Barnard, D.F.; Fox, S.; LaSance, K.; Shaughnessy, E.; Ben-Jonathan, N. Expression and therapeutic targeting of dopamine receptor-1 (D1R) in breast cancer. Oncogene 2016, 35, 3103–3113. [Google Scholar] [CrossRef]
- Yan, Y.; Pan, J.; Chen, Y.; Xing, W.; Li, Q.; Wang, D.; Zhou, X.; Xie, J.; Miao, C.; Yuan, Y.; et al. Increased dopamine and its receptor dopamine receptor D1 promote tumor growth in human hepatocellular carcinoma. Cancer Commun. 2020, 40, 694–710. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, R.; Zhang, X.; Liu, J.; Wu, H.; Li, Y.; Cui, M.; Li, T.; Song, H.; Gao, J.; et al. Dopamine improves chemotherapeutic efficacy for pancreatic cancer by regulating macrophage-derived inflammations. Cancer Immunol. Immunother. 2021, 70, 2165–2177. [Google Scholar] [CrossRef]
- Sayegh, A.I. The role of bombesin and bombesin-related peptides in the short-term control of food intake. Prog. Mol. Biol. Transl. Sci. 2013, 114, 343–370. [Google Scholar]
- Zhu, X.Z.; Ji, X.Q.; Wu, S.X.; Zou, G. Sulpiride attenuates ranatensin-M-induced antinociception. Zhongguo Yao Li Xue Bao 1991, 12, 291–293. [Google Scholar]
- Benyhe, S.; Farkas, J.; Tóth, G.; Wollemann, M. Met5-enkephalin-Arg6-Phe7, an endogenous neuropeptide, binds to multiple opioid and nonopioid sites in rat brain. J. Neurosci. Res. 1997, 48, 249–258. [Google Scholar] [CrossRef]
- Szűcs, E.; Büki, A.; Kékesi, G.; Horváth, G.; Benyhe, S. Mu-Opioid (MOP) receptor mediated G-protein signaling is impaired in specific brain regions in a rat model of schizophrenia. Neurosci. Lett. 2016, 619, 29–33. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sim, L.J.; Selley, D.E.; Childers, S.R. In vitro autoradiography of receptor-activated G proteins in rat brain by agonist-stimulated guanylyl 5’-[gamma-[35S]thio]-triphosphate binding. Proc. Natl. Acad. Sci. USA 1995, 92, 7242–7246. [Google Scholar] [CrossRef] [PubMed]
- Traynor, J.R.; Nahorski, S.R. Modulation by mu-opioid agonists of guanosine-5’-O-(3-[35S]thio)triphosphate binding to membranes from human neuroblastoma SH-SY5Y cells. Mol. Pharmacol. 1995, 47, 848–854. [Google Scholar] [PubMed]
- Mazzarino, L.; Loch-Neckel, G.; dos Santos Bubniak, L.; Ourique, F.; Otsuka, I.; Halila, S.; Pedrosa, R.C.; Santos-Silva, M.C.; Lemos-Senna, E.; Muniz, E.C.; et al. Nanoparticles made from xyloglucan-block-polycaprolactone copolymers: Safety assessment for drug delivery. Toxicol. Sci. 2015, 147, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chen, T.; Zhou, M.; Wang, L.; Su, S.; Shaw, C. Ranatensin-HL: A bombesin-related tridecapeptide from the skin secretion of the broad-folded frog, Hylarana latouchii. Molecules 2017, 22, 1110. [Google Scholar] [CrossRef] [PubMed]
- Kroog, G.S.; Jensen, R.T.; Battey, J.F. Mammalian bombesin receptors. Med. Res. Rev. 1995, 15, 389–417. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.P.; Fonkoua, L.K.; Moody, T.W. The role of gastrin and CCK receptors in pancreatic cancer and other malignancies. Int. J. Biol. Sci. 2016, 12, 283–291. [Google Scholar] [CrossRef]
- Baratto, L.; Duan, H.; Mäcke, H.; Iagaru, A. Imaging the distribution of gastrin-releasing peptide receptors in cancer. J. Nucl. Med. 2020, 61, 792–798. [Google Scholar] [CrossRef]
- Shapira, H.; Wada, E.; Battey, J.F.; Jensen, R.T.; Coy, D.H.; Kusano, K. Distinguishing bombesin receptor subtypes using the oocyte assay. Biochem. Biophys. Res. Commun. 1991, 176, 79–86. [Google Scholar] [CrossRef]
- Stahle, M.; Veit, C.; Bachfischer, U.; Schierling, K.; Skripczynski, B.; Hall, A.; Gierschik, P.; Giehl, K. Mechanisms in LPA-induced tumor cell migration: Critical role of phosphorylated ERK. J. Cell Sci. 2003, 116, 3835–3846. [Google Scholar] [CrossRef]
- Huang, H.; Wu, K.; Ma, J.; Du, Y.; Cao, C.; Nie, Y. Dopamine D2 receptor suppresses gastric cancer cell invasion and migration via inhibition of EGFR/AKT/MMP-13 pathway. Int. Immunopharmacol. 2016, 39, 113–120. [Google Scholar] [CrossRef]
- Wu, X.Y.; Zhang, C.X.; Deng, L.C.; Xiao, J.; Yuan, X.; Zhang, B.; Hou, A.B.; Scheng, Z.H.; Sun, L.; Jiang, Q.C.; et al. Overexpressed D2 dopamine receptor inhibits non-small cell lung cancer progression through inhibiting NF-kB signaling pathway. Cell. Physiol. Biochem. 2018, 48, 2258–2272. [Google Scholar] [CrossRef]
- Hoeppner, L.H.; Wang, Y.; Sharma, A.; Javeed, N.; Van Keulen, V.P.; Wang, E.; Yang, P.; Roden, A.C.; Peikert, T.; Molina, J.R.; et al. Dopamine D2 receptor agonists inhibit lung cancer progression by reducing angiogenesis and tumor infiltrating myeloid derived suppressor cells. Mol. Oncol. 2015, 9, 270–281. [Google Scholar] [CrossRef]
- Dandawate, P.; Kaushik, G.; Ghosh, C.; Standing, D.; Sayed, A.A.A.; Choudhury, S.; Subramaniam, D.; Manzardo, A.; Banerjee, T.; Santra, S.; et al. Diphenylbutylpiperidine antipsychotic drugs inhibit prolactin receptor signaling to reduce growth of pancreatic ductal adenocarcinoma in mice. Gastroenterology 2020, 158, 1433–1449. [Google Scholar] [CrossRef]
- Lee, H.; Shim, S.; Kong, J.S.; Kim, M.J.; Park, S.; Lee, S.S.; Kim, A. Overexpression of dopamine receptor D2 promotes colorectal cancer progression by activating the β-catenin/ZEB1 axis. Cancer Sci. 2021, 112, 3732–3743. [Google Scholar] [CrossRef]
- Bakadlag, R.; Jandaghi, P.; Hoheisel, J.D.; Riazalhosseini, Y. The potential of dopamine receptor D2 (DRD2) as a therapeutic target for tackling pancreatic cancer. Expert Opin. Ther. Targets 2019, 23, 65–367. [Google Scholar] [CrossRef]
- Mao, M.; Yu, T.; Hu, J.; Hu, L. Dopamine D2 receptor blocker thioridazine induces cell death in human uterine cervical carcinoma cell line SiHa. J. Obstet. Gynaecol. Res. 2015, 41, 1240–1245. [Google Scholar] [CrossRef]
- Yaseen, A.; Gull, S.; Akhtar, N.; Amin, I.; Minhas, F. HemoNet: Predicting hemolytic activity of peptides with integrated feature learning. J. Bioinform. Comput. Biol. 2021, 19, 2150021. [Google Scholar] [CrossRef]
- Liscano, Y.; Oñate-Garzón, J.; Delgado, J.P. Peptides with dual antimicrobial-anticancer activity: Strategies to overcome peptide limitations and rational design of anticancer peptides. Molecules 2020, 25, 4245. [Google Scholar] [CrossRef]
- Feder, R.; Dagan, A.; Mor, A. Structure-activity relationship study of antimicrobial dermaseptin S4 showing the consequences of peptide oligomerization on selective cytotoxicity. J. Biol. Chem. 2000, 275, 4230–4238. [Google Scholar] [CrossRef]
- Yang, Q.Z.; Wang, C.; Lang, L.; Zhou, Y.; Wang, H.; Shang, D.J. Design of potent, non-toxic anticancer peptides based on the structure of the antimicrobial peptide, temporin-1CEa. Arch. Pharm. Res. 2013, 36, 1302–1310. [Google Scholar] [CrossRef]
| IC50 (µM) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cell Line | BxPC-3 | Capan-1 | PANC-1 | NHF | ||||
| 24 h | 72 h | 24 h | 72 h | 24 h | 72 h | 24 h | 72 h | |
| Ranatensin | >60 | >60 | >60 | >60 | >60 | >60 | >60 | >60 |
| Dopamine | >60 | >60 | >60 | >60 | >60 | >60 | >60 | >60 |
| Pimozide | 21.73 ± 2.27 | 17.8 ± 1.38 | 25.32 ± 3.11 | 19.96 ± 1.79 | 15.12 ± 1.51 | 20.34 ± 2.02 | 38.71 ± 1.37 | 23.57 ± 0.92 |
| Risperidone | >60 | >60 | >60 | >60 | >60 | >60 | >60 | >60 |
| Gemcitabine | >60 | 0.2964 ± 0.15 | >60 | 0.6566 ± 0.13 | >60 | 0.2798 ± 0.031 | >60 | 1.28 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laskowska, A.K.; Szudzik, M.; Ścieżyńska, A.; Komorowski, M.; Szűcs, E.; Gombos, D.; Bączek, B.; Lipka-Miciuk, J.; Benyhe, S.; Kleczkowska, P. The Role of a Natural Amphibian Skin-Based Peptide, Ranatensin, in Pancreatic Cancers Expressing Dopamine D2 Receptors. Cancers 2022, 14, 5535. https://doi.org/10.3390/cancers14225535
Laskowska AK, Szudzik M, Ścieżyńska A, Komorowski M, Szűcs E, Gombos D, Bączek B, Lipka-Miciuk J, Benyhe S, Kleczkowska P. The Role of a Natural Amphibian Skin-Based Peptide, Ranatensin, in Pancreatic Cancers Expressing Dopamine D2 Receptors. Cancers. 2022; 14(22):5535. https://doi.org/10.3390/cancers14225535
Chicago/Turabian StyleLaskowska, Anna K., Mateusz Szudzik, Aneta Ścieżyńska, Michał Komorowski, Edina Szűcs, Dávid Gombos, Bartłomiej Bączek, Jowita Lipka-Miciuk, Sandor Benyhe, and Patrycja Kleczkowska. 2022. "The Role of a Natural Amphibian Skin-Based Peptide, Ranatensin, in Pancreatic Cancers Expressing Dopamine D2 Receptors" Cancers 14, no. 22: 5535. https://doi.org/10.3390/cancers14225535
APA StyleLaskowska, A. K., Szudzik, M., Ścieżyńska, A., Komorowski, M., Szűcs, E., Gombos, D., Bączek, B., Lipka-Miciuk, J., Benyhe, S., & Kleczkowska, P. (2022). The Role of a Natural Amphibian Skin-Based Peptide, Ranatensin, in Pancreatic Cancers Expressing Dopamine D2 Receptors. Cancers, 14(22), 5535. https://doi.org/10.3390/cancers14225535







