Identification of Factors Driving Doxorubicin-Resistant Ewing Tumor Cells to Survival
Abstract
Simple Summary
Abstract
1. Background
2. Materials and Methods
2.1. Materials
2.2. Primary Tissue Samples
2.3. Cell Cultures
2.4. cDNA Library Preparation and Single-Cell RNA-Seq
2.5. Proteomic Data Generation and Analysis
2.6. Data Source, Data Processing, and Data Distribution
2.7. Functional GO Enrichment Analysis
2.8. Experimental Validation of Fold-Change Values Using Semiquantitative RT-PCR Analysis
2.9. Statistical Analyses
2.10. Availability of Data and Materials
3. Results
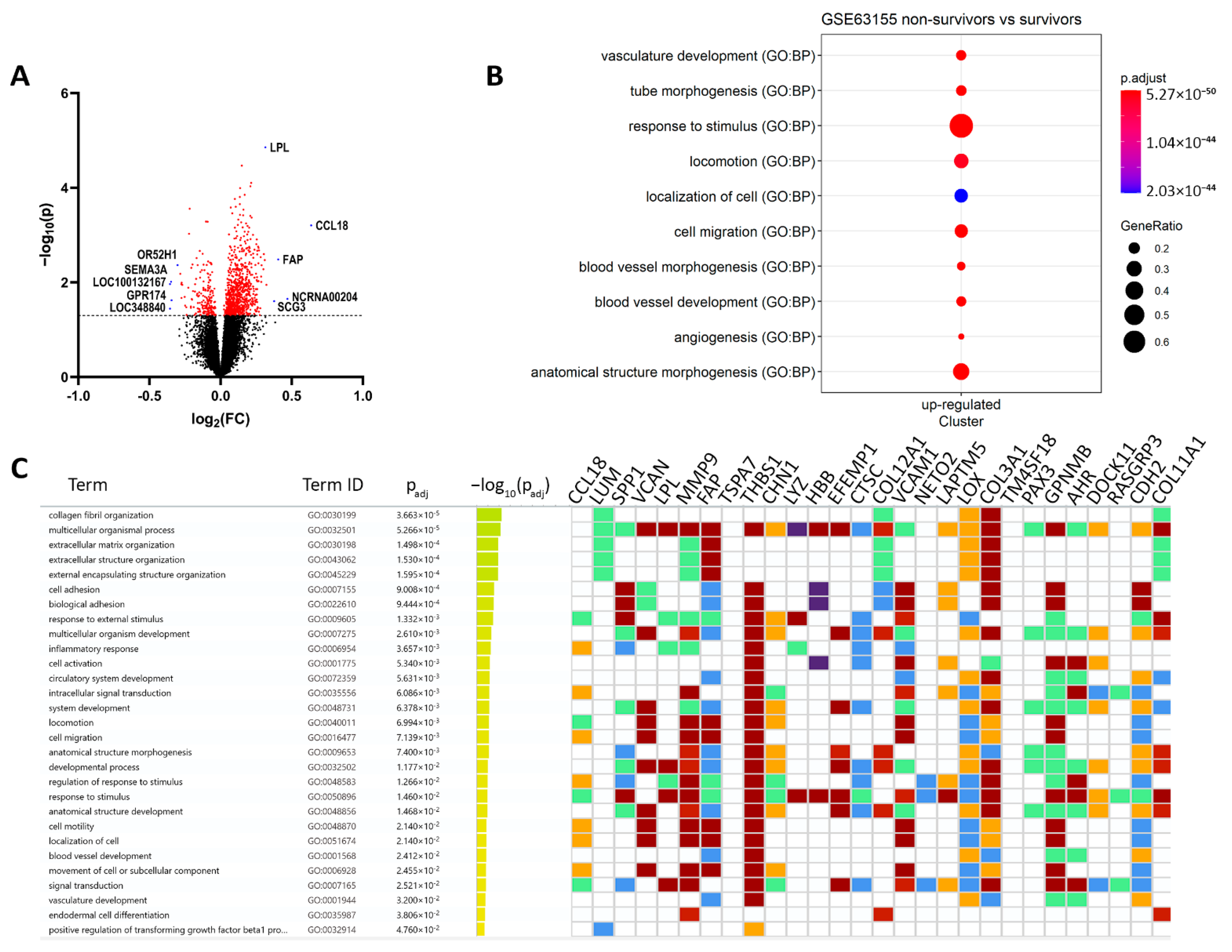
3.1. Hypoxia and Metastatic Traits Distinguish Tumor Cells from Non-Neoplastic Cells
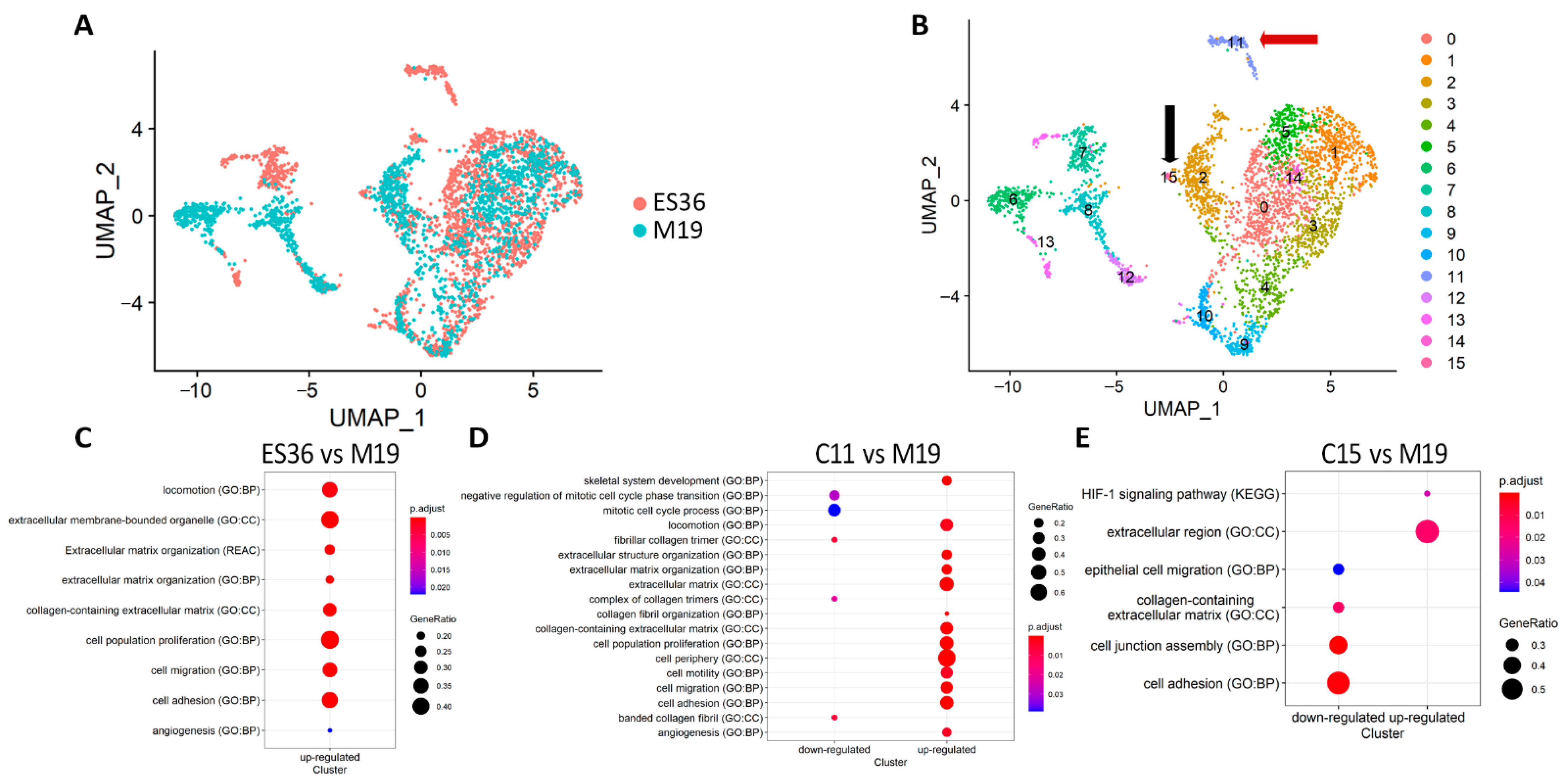
3.2. Embryonic Fibroblasts (M19) and ES (ES36) Cells Are Genetically Close
3.3. Proteomic Profiling of DOX-Treated ES36 Cells Provides a Unique Signature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koelsche, C.; Kriegsmann, M.; Kommoss, F.K.F.; Stichel, D.; Kriegsmann, K.; Vokuhl, C.; Grunewald, T.G.P.; Romero-Perez, L.; Kirchner, T.; de Alava, E.; et al. DNA methylation profiling distinguishes Ewing-like sarcoma with EWSR1-NFATc2 fusion from Ewing sarcoma. J. Cancer Res. Clin. Oncol. 2019, 145, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Sankar, S.; Lessnick, S.L. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011, 204, 351–365. [Google Scholar] [CrossRef]
- Shulman, D.S.; Whittle, S.B.; Surdez, D.; Bailey, K.M.; de Alava, E.; Yustein, J.T.; Shlien, A.; Hayashi, M.; Bishop, A.J.R.; Crompton, B.D.; et al. An international working group consensus report for the prioritization of molecular biomarkers for Ewing sarcoma. NPJ Precis. Oncol. 2022, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- Miser, J.S.; Goldsby, R.E.; Chen, Z.; Krailo, M.D.; Tarbell, N.J.; Link, M.P.; Fryer, C.J.; Pritchard, D.J.; Gebhardt, M.C.; Dickman, P.S.; et al. Treatment of metastatic Ewing sarcoma/primitive neuroectodermal tumor of bone: Evaluation of increasing the dose intensity of chemotherapy—A report from the Children’s Oncology Group. Pediatr. Blood Cancer 2007, 49, 894–900. [Google Scholar] [CrossRef]
- Leavey, P.J.; Mascarenhas, L.; Marina, N.; Chen, Z.; Krailo, M.; Miser, J.; Brown, K.; Tarbell, N.; Bernstein, M.L.; Granowetter, L.; et al. Prognostic factors for patients with Ewing sarcoma (EWS) at first recurrence following multi-modality therapy: A report from the Children’s Oncology Group. Pediatr. Blood Cancer 2008, 51, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hong, T.; Liu, W.; Dong, S.; Wang, H.; Tang, Z.R.; Li, W.; Wang, B.; Hu, Z.; Liu, Q.; et al. Development of a Machine Learning-Based Predictive Model for Lung Metastasis in Patients With Ewing Sarcoma. Front. Med. 2022, 9, 807382. [Google Scholar] [CrossRef]
- Margulies, B.S.; DeBoyace, S.D.; Damron, T.A.; Allen, M.J. Ewing’s sarcoma of bone tumor cells produces MCSF that stimulates monocyte proliferation in a novel mouse model of Ewing’s sarcoma of bone. Bone 2015, 79, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.; Knowles, H.J.; Athanasou, N.A. Ewing sarcoma cells express RANKL and support osteoclastogenesis. J. Pathol. 2011, 225, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Neve, A.; Cantatore, F.P.; Maruotti, N.; Corrado, A.; Ribatti, D. Extracellular matrix modulates angiogenesis in physiological and pathological conditions. BioMed Res. Int. 2014, 2014, 756078. [Google Scholar] [CrossRef] [PubMed]
- Perut, F.; Carta, F.; Bonuccelli, G.; Grisendi, G.; Di Pompo, G.; Avnet, S.; Sbrana, F.V.; Hosogi, S.; Dominici, M.; Kusuzaki, K.; et al. Carbonic anhydrase IX inhibition is an effective strategy for osteosarcoma treatment. Expert Opin. Ther. Targets 2015, 19, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- Hatano, M.; Matsumoto, Y.; Fukushi, J.; Matsunobu, T.; Endo, M.; Okada, S.; Iura, K.; Kamura, S.; Fujiwara, T.; Iida, K.; et al. Cadherin-11 regulates the metastasis of Ewing sarcoma cells to bone. Clin. Exp. Metastasis 2015, 32, 579–591. [Google Scholar] [CrossRef]
- Supuran, C.T.; Alterio, V.; Di Fiore, A.; D’Ambrosio, K.; Carta, F.; Monti, S.M.; De Simone, G. Inhibition of carbonic anhydrase IX targets primary tumors, metastases, and cancer stem cells: Three for the price of one. Med. Res. Rev. 2018, 38, 1799–1836. [Google Scholar] [CrossRef] [PubMed]
- Vieira, G.M.; Roberto, G.M.; Lira, R.C.; Engel, E.E.; Tone, L.G.; Brassesco, M.S. Prognostic value and functional role of ROCK2 in pediatric Ewing sarcoma. Oncol. Lett. 2018, 15, 2296–2304. [Google Scholar] [CrossRef]
- Hetland, T.E.; Nymoen, D.A.; Emilsen, E.; Kaern, J.; Trope, C.G.; Florenes, V.A.; Davidson, B. MGST1 expression in serous ovarian carcinoma differs at various anatomic sites, but is unrelated to chemoresistance or survival. Gynecol. Oncol. 2012, 126, 460–465. [Google Scholar] [CrossRef] [PubMed]
- van Kuijk, S.J.; Yaromina, A.; Houben, R.; Niemans, R.; Lambin, P.; Dubois, L.J. Prognostic Significance of Carbonic Anhydrase IX Expression in Cancer Patients: A Meta-Analysis. Front. Oncol. 2016, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.K.; Liu, J.; Chen, C.; Yang, K.Y.; Deng, Y.T.; Jiang, Y. Locally advanced malignant solitary fibrous tumour successfully treated with conversion chemotherapy, operation and postoperative radiotherapy: A case report. J. Int. Med. Res. 2021, 49, 300060521996940. [Google Scholar] [CrossRef]
- Ulanet, D.B.; Ludwig, D.L.; Kahn, C.R.; Hanahan, D. Insulin receptor functionally enhances multistage tumor progression and conveys intrinsic resistance to IGF-1R targeted therapy. Proc. Natl. Acad. Sci. USA 2010, 107, 10791–10798. [Google Scholar] [CrossRef] [PubMed]
- Scotlandi, K.; Remondini, D.; Castellani, G.; Manara, M.C.; Nardi, F.; Cantiani, L.; Francesconi, M.; Mercuri, M.; Caccuri, A.M.; Serra, M.; et al. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J. Clin. Oncol. 2009, 27, 2209–2216. [Google Scholar] [CrossRef]
- Yarovinsky, T.O.; Gorlina, N.K.; Cheredeev, A.N.; Kozlov, I.G.; Zorin, N.A.; Zorina, R.M. Alpha2-Macroglobulin Modulates Interactions between Lymphocytes and Fibroblasts. Russ. J. Immunol. 2001, 6, 1–8. [Google Scholar]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Volchenboum, S.L.; Andrade, J.; Huang, L.; Barkauskas, D.A.; Krailo, M.; Womer, R.B.; Ranft, A.; Potratz, J.; Dirksen, U.; Triche, T.J.; et al. Gene Expression Profiling of Ewing Sarcoma Tumors Reveals the Prognostic Importance of Tumor-Stromal Interactions: A Report from the Children’s Oncology Group. J. Pathol. Clin. Res. 2015, 1, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.P.; Ding, S.Y.; Sun, Y.Y.; Li, Y.H.; Fu, W.N. MYCT1 Inhibits the Adhesion and Migration of Laryngeal Cancer Cells Potentially Through Repressing Collagen VI. Front. Oncol. 2020, 10, 564733. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, Z.; Zou, M.; Wan, R.; Wu, T.; Luo, Y.; Wu, G.; Wang, W.; Liu, T. Prognosis and Immune Infiltration of Chromobox Family Genes in Sarcoma. Front. Oncol. 2021, 11, 657595. [Google Scholar] [CrossRef]
- Nakatani, F.; Ferracin, M.; Manara, M.C.; Ventura, S.; Del Monaco, V.; Ferrari, S.; Alberghini, M.; Grilli, A.; Knuutila, S.; Schaefer, K.L.; et al. miR-34a predicts survival of Ewing’s sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J. Pathol. 2012, 226, 796–805. [Google Scholar] [CrossRef]
- Krueger, S.; Kellner, U.; Buehling, F.; Roessner, A. Cathepsin L antisense oligonucleotides in a human osteosarcoma cell line: Effects on the invasive phenotype. Cancer Gene Ther. 2001, 8, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Yeh, L.T.; Lin, C.W.; Lu, K.H.; Hsieh, Y.H.; Yeh, C.B.; Yang, S.F.; Yang, J.S. Niclosamide Suppresses Migration and Invasion of Human Osteosarcoma Cells by Repressing TGFBI Expression via the ERK Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 484. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Mu, T. LncRNA LINC00958 promotes tumor progression through miR-4306/CEMIP axis in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 3182–3199. [Google Scholar] [CrossRef]
- Park, I.C.; Lee, S.Y.; Jeon, D.G.; Lee, J.S.; Hwang, C.S.; Hwang, B.G.; Lee, S.H.; Hong, W.S.; Hong, S.I. Enhanced expression of cathepsin L in metastatic bone tumors. J. Korean Med. Sci. 1996, 11, 144–148. [Google Scholar] [CrossRef]
- Wiggers, F.; Wohl, S.; Dubovetskyi, A.; Rosenblum, G.; Zheng, W.; Hofmann, H. Diffusion of a disordered protein on its folded ligand. Proc. Natl. Acad. Sci. USA 2021, 118, e2106690118. [Google Scholar] [CrossRef]
- Tang, M.; Liu, P.; Wu, X.; Gong, J.; Weng, J.; Gao, G.; Liu, Y.; Gan, L. COL3A1 and Its Related Molecules as Potential Biomarkers in the Development of Human Ewing’s Sarcoma. BioMed Res. Int. 2021, 2021, 7453500. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, J.; Liu, W.; Ding, L.; Cheng, D.; Xiao, H. Identification of Common Oncogenic Genes and Pathways Both in Osteosarcoma and Ewing’s Sarcoma Using Bioinformatics Analysis. J. Immunol. Res. 2022, 2022, 3655908. [Google Scholar] [CrossRef] [PubMed]
- Fellenberg, J.; Bernd, L.; Delling, G.; Witte, D.; Zahlten-Hinguranage, A. Prognostic significance of drug-regulated genes in high-grade osteosarcoma. Mod. Pathol. 2007, 20, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.H.; Fu, L.; Chen, J.; Wei, F.; Shi, W.X. Decreased expression of ferritin light chain in osteosarcoma and its correlation with epithelial-mesenchymal transition. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2580–2587. [Google Scholar] [CrossRef]
- Mao, M.; Wang, W. SerpinE2 promotes multiple cell proliferation and drug resistance in osteosarcoma. Mol. Med. Rep. 2016, 14, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Brooks, M.; Mo, Q.; Krasnow, R.; Ho, P.L.; Lee, Y.C.; Xiao, J.; Kurtova, A.; Lerner, S.; Godoy, G.; Jian, W.; et al. Positive association of collagen type I with non-muscle invasive bladder cancer progression. Oncotarget 2016, 7, 82609–82619. [Google Scholar] [CrossRef]
- Yang, L.; Jing, J.; Sun, L.; Yue, Y. Exploring prognostic genes in ovarian cancer stage-related coexpression network modules. Medicine 2018, 97, e11895. [Google Scholar] [CrossRef]
- Lovitt, C.J.; Shelper, T.B.; Avery, V.M. Doxorubicin resistance in breast cancer cells is mediated by extracellular matrix proteins. BMC Cancer 2018, 18, 41. [Google Scholar] [CrossRef]
- Guzel Tanoglu, E.; Ozturk, S. miR-145 suppresses epithelial-mesenchymal transition by targeting stem cells in Ewing sarcoma cells. Bratisl. Lek. Listy 2021, 122, 71–77. [Google Scholar] [CrossRef]
- Wiles, E.T.; Bell, R.; Thomas, D.; Beckerle, M.; Lessnick, S.L. ZEB2 Represses the Epithelial Phenotype and Facilitates Metastasis in Ewing Sarcoma. Genes Cancer 2013, 4, 486–500. [Google Scholar] [CrossRef]
- Garofalo, C.; Manara, M.C.; Nicoletti, G.; Marino, M.T.; Lollini, P.L.; Astolfi, A.; Pandini, G.; Lopez-Guerrero, J.A.; Schaefer, K.L.; Belfiore, A.; et al. Efficacy of and resistance to anti-IGF-1R therapies in Ewing’s sarcoma is dependent on insulin receptor signaling. Oncogene 2011, 30, 2730–2740. [Google Scholar] [CrossRef]
- Hong, S.H.; Tilan, J.U.; Galli, S.; Izycka-Swieszewska, E.; Polk, T.; Horton, M.; Mahajan, A.; Christian, D.; Jenkins, S.; Acree, R.; et al. High neuropeptide Y release associates with Ewing sarcoma bone dissemination—In vivo model of site-specific metastases. Oncotarget 2015, 6, 7151–7165. [Google Scholar] [CrossRef] [PubMed]
- Sainz-Jaspeado, M.; Lagares-Tena, L.; Lasheras, J.; Navid, F.; Rodriguez-Galindo, C.; Mateo-Lozano, S.; Notario, V.; Sanjuan, X.; Garcia Del Muro, X.; Fabra, A.; et al. Caveolin-1 modulates the ability of Ewing’s sarcoma to metastasize. Mol. Cancer Res. 2010, 8, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Dominguez, D.J.; Hajji, N.; Lopez-Alemany, R.; Sanchez-Molina, S.; Figuerola-Bou, E.; Moron Civanto, F.J.; Rello-Varona, S.; Andres-Leon, E.; Benito, A.; Keun, H.C.; et al. Selective histone methyltransferase G9a inhibition reduces metastatic development of Ewing sarcoma through the epigenetic regulation of NEU1. Oncogene 2022, 41, 2638–2650. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, Y.; Gao, H.; Peng, T.; Shi, H.; Tang, Y.; Li, H.; Chen, L.; Hu, K.; Han, A. A novel HDGF-ALCAM axis promotes the metastasis of Ewing sarcoma via regulating the GTPases signaling pathway. Oncogene 2021, 40, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Alemany, R.; Tirado, O.M. Metastasis Assessment in Ewing Sarcoma Using Orthotopic Xenografts. Methods Mol. Biol. 2021, 2226, 201–213. [Google Scholar] [CrossRef]
- Choo, S.; Wang, P.; Newbury, R.; Roberts, W.; Yang, J. Reactivation of TWIST1 contributes to Ewing sarcoma metastasis. Pediatr. Blood Cancer 2018, 65, e26721. [Google Scholar] [CrossRef]
- Cerra, C.; Harris, M.A.; Hawkins, C.J. Establishment and Characterisation of Metastatic Extraskeletal Ewing Sarcoma Mouse Models. In Vivo 2021, 35, 3097–3106. [Google Scholar] [CrossRef]
- Grossman, T.R.; Gamliel, A.; Wessells, R.J.; Taghli-Lamallem, O.; Jepsen, K.; Ocorr, K.; Korenberg, J.R.; Peterson, K.L.; Rosenfeld, M.G.; Bodmer, R.; et al. Over-expression of DSCAM and COL6A2 cooperatively generates congenital heart defects. PLoS Genet. 2011, 7, e1002344. [Google Scholar] [CrossRef]
- McIlwain, C.C.; Townsend, D.M.; Tew, K.D. Glutathione S-transferase polymorphisms: Cancer incidence and therapy. Oncogene 2006, 25, 1639–1648. [Google Scholar] [CrossRef]
- Drozd, E.; Krzyszton-Russjan, J.; Marczewska, J.; Drozd, J.; Bubko, I.; Bielak, M.; Lubelska, K.; Wiktorska, K.; Chilmonczyk, Z.; Anuszewska, E.; et al. Up-regulation of glutathione-related genes, enzyme activities and transport proteins in human cervical cancer cells treated with doxorubicin. Biomed. Pharmacother. 2016, 83, 397–406. [Google Scholar] [CrossRef]


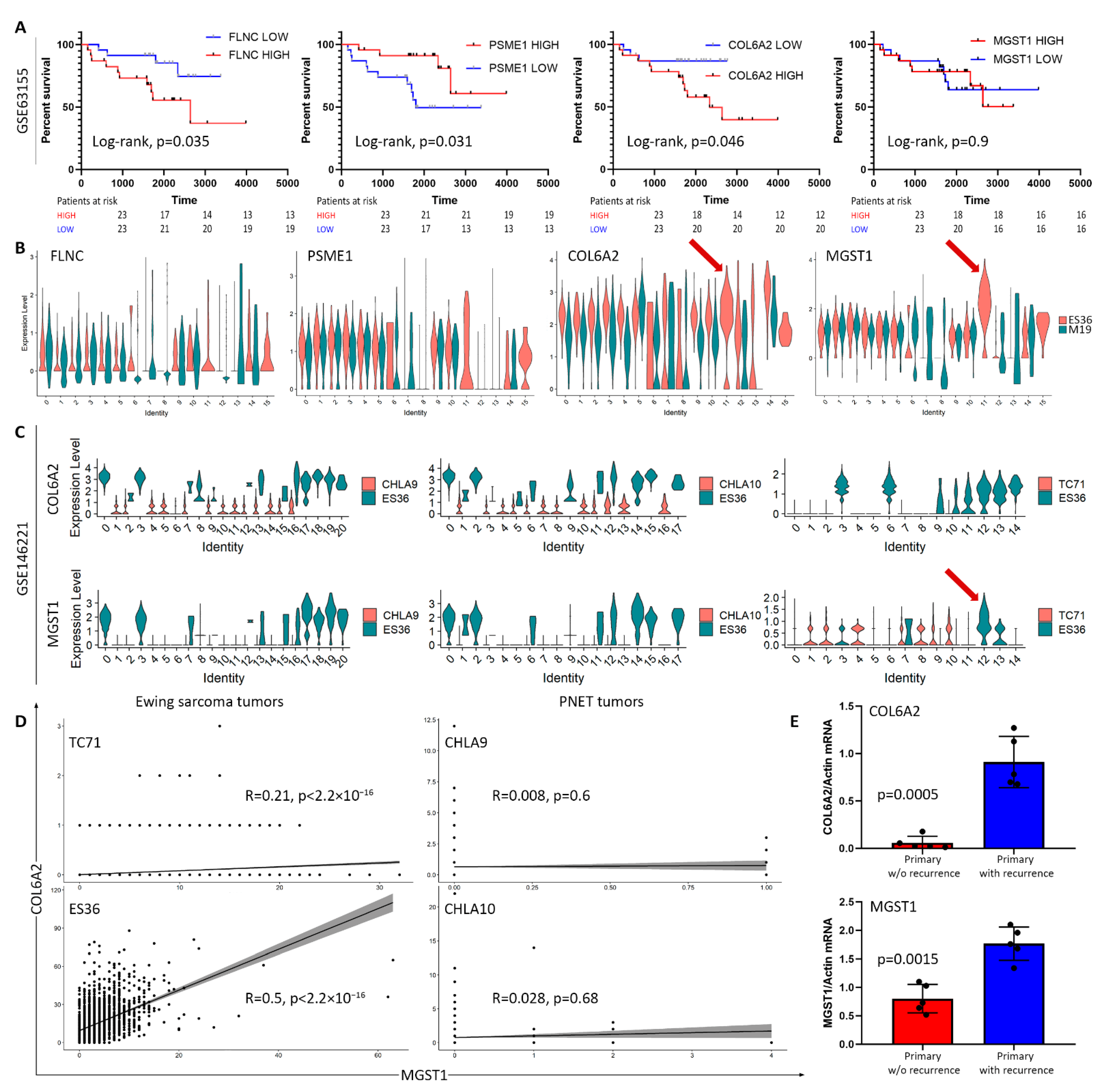
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Beta | HR (95% CI) | p.Value | Beta | HR (95% CI) | p.Value | |
| Sex | −0.47 | 0.62 (0.21–1.9) | 0.4 | −2.358 | 0.095 (0.005–1.84) | 0.120 |
| Age_at_enrollment_days | 0.00033 | 1 (1–1) | 0.07 | −0.004 | 0.996 (0.98–1.0) | 0.465 |
| Age | 0.11 | 1.1 (0.99–1.3) | 0.075 | 1.637 | 5.14 (0.085–309.385) | 0.433 |
| Primary_tumor_site | −0.28 | 0.76 (0.35–1.6) | 0.47 | 1.502 | 4.492 (0.377–53.53) | 0.235 |
| Efs_event | 2 | 7.7 (3.4–17) | 9.5e-07 | 3.941 | 51.476 (3.736–709.32) | 0.003 |
| Tumor_content | 0.77 | 2.2 (0.57–8.2) | 0.26 | −0.562 | 0.57 (0.088–3.676) | 0.554 |
| Description | ID | p Value | Count/Gene ID | |
|---|---|---|---|---|
| Downregulated | locomotion | GO:0040011 | 1.14 × 10−26 | CCL18/VCAN/MMP9/FAP/THBS1/CHN1/VCAM1/LOX/COL3A1/GPNMB/CDH2/SULF1/GJA1/ACTA2/TGFBR2/SMOC2/CCL21/MMP2/DCLK1/AKT3/CD200/MAP1B/RND3/KDR/MCC/LEF1/SGK1/SDC2/ENPP2/PECAM1/KITLG/CCR1/ADAMTS1/ITGA6/EPS8/ADAMTS9/EFNB2/CD34/THY1/SEMA6A/LYN/ALX1/S100A8/ITGA9/MTUS1/NRP1/PLVAP/PRKD1/ETS1/LYVE1/CDH5/PLXNC1/PODXL/ITGA2/FCER1G/TLR4/TEK/NRP2/PDGFRB/LAMB1/ELMO1/EMP2/MCTP1/FLT1/DOCK2/PTPRC/XBP1/CHL1/SATB2/PLXNA2/DLC1 |
| angiogenesis | GO:0001525 | 4.39 × 10−26 | FAP/THBS1/GPNMB/SULF1/THBS2/PTPRB/TGFBR2/SMOC2/MMP2/CALCRL/CYBB/ADAM12/AKT3/ANPEP/KDR/LEF1/ENPP2/COL15A1/CEMIP2/TGFBI/APLNR/ADAMTS1/ADAMTS9/EFNB2/CD34/THY1/SEMA6A/SAT1/NRP1/PRKD1/ETS1/COL4A1/CDH5/EMCN/EPAS1/TEK/COL4A2/NRP2/PDGFRB/EMP2/FLT1/XBP1 | |
| positive regulation of cell migration | GO:0030335 | 2.52 × 10−19 | MMP9/THBS1/GPNMB/ACTA2/TGFBR2/SMOC2/CCL21/MMP2/AKT3/KDR/LEF1/ENPP2/PECAM1/KITLG/CCR1/ADAMTS1/ITGA6/THY1/SEMA6A/LYN/NRP1/PLVAP/PRKD1/ETS1/LYVE1/CDH5/PODXL/ITGA2/TLR4/TEK/NRP2/PDGFRB/LAMB1/FLT1/PTPRC/XBP1 | |
| cell differentiation | GO:0030154 | 6.24 × 10−16 | SPP1/VCAN/LPL/MMP9/CHN1/EFEMP1/COL12A1/VCAM1/LOX/COL3A1/DOCK11/CDH2/COL11A1/KRT10/SULF1/STEAP4/GJA1/A2M/ACTA2/TGFBR2/CCL21/MMP2/CD53/GPM6B/DCLK1/ADAM12/PLEK/MAP1B/ANPEP/KDR/ESRP1/LEF1/SGK1/SDC2/PECAM1/COL15A1/KITLG/CCR1/TGFBI/APLNR/ITGA6/MAP2/ADAMTS9/MTSS1/EFNB2/CD34/CD36/THY1/SEMA6A/LYN/HEY2/RAI14/ALX1/S100A8/RPS6KA2/SPRY4/NRP1/PRKD1/TMEM119/ETS1/PRICKLE1/COL4A1/CDH5/PLXNC1/PODXL/ITGA2/SLC6A6/EPAS1/FCER1G/TLR4/TEK/COL4A2/FRMD6/NRP2/PDGFRB/LAMB1/EMP2/FLT1/FARP1/DOCK2/PTPRC/XBP1/CHL1/RPS6KA3/SATB2/PLXNA2/TAGLN/BHLHE40 | |
| extracellular matrix organization | GO:0030198 | 8.07 × 10−14 | LUM/MMP9/FAP/COL12A1/LOX/COL3A1/COL11A1/SULF1/SMOC2/MMP2/GPM6B/COL14A1/MMP16/COL15A1/TGFBI/ADAMTS1/ITGA6/ADAMTS9/ITGA9/FBLN5/COL4A1/ITGA2/COL4A2/LAMB1/NID2 | |
| cell-substrate adhesion | GO:0031589 | 7.69 × 10−11 | THBS1/VCAM1/COL3A1/EDIL3/CCL21/GPM6B/VWF/KDR/ITGA6/ADAMTS9/CD34/CD36/THY1/SPRY4/NRP1/FBLN5/LYVE1/ITGA2/TEK/LAMB1/EMP2/NID2/DLC1 | |
| chemotaxis | GO:0006935 | 7.74 × 10−11 | CCL18/THBS1/CHN1/VCAM1/LOX/GPNMB/SMOC2/CCL21/KDR/LEF1/ENPP2/CCR1/EFNB2/SEMA6A/LYN/S100A8/ITGA9/MTUS1/NRP1/PRKD1/PLXNC1/ITGA2/FCER1G/NRP2/PDGFRB/FLT1/DOCK2/CHL1/PLXNA2 | |
| cell-matrix adhesion | GO:0007160 | 1.55 × 10−9 | THBS1/VCAM1/COL3A1/CCL21/GPM6B/KDR/ADAMTS9/CD34/CD36/THY1/NRP1/FBLN5/LYVE1/ITGA2/TEK/EMP2/NID2/DLC1 | |
| cell-substrate junction assembly | GO:0007044 | 0.0002 | THBS1/GPM6B/KDR/ITGA6/THY1/NRP1/ITGA2/TEK/DLC1 | |
| collagen-containing extracellular matrix | GO:0062023 | 3.43 × 10−21 | LUM/VCAN/MMP9/THBS1/EFEMP1/COL12A1/CTSC/COL3A1/CDH2/COL11A1/EDIL3/SULF1/A2M/THBS2/SMOC2/MMP2/VWF/COL14A1/SDC2/SPON1/PCOLCE/COL15A1/TGFBI/ADAMTS1/SPARCL/ADAMTS9/S100A8/BGN/HAPLN1/FBLN5/COL4A1/COL4A2/LAMB1/NID2 | |
| extracellular exosome | GO:0070062 | 6.20 × 10−10 | LUM/SPP1/MMP9/THBS1/LYZ/HBB/EFEMP1/COL12A1/CTSC/VCAM1/PPIC/EDIL3/KRT10/STEAP4/A2M/ACTA2/CD53/VWF/ANPEP/MAN1A1/PECAM1/PCOLCE/COL15A1/CEMIP2/TGFBI/EPS8/PRSS23/THY1/LYN/S100A8/CD14/BGN/PLVAP/MYO1B/ENTPD1/FBLN5/LYVE1/RFTN1/PODXL/NT5E/FCGR3A/COL4A2/LAMB1/DOCK2/RAB27B/PTPRC/MARCKS/CHL1/NID2/TNFSF10/PYGL | |
| collagen trimer | GO:0005581 | 1.90 × 10−6 | LUM/COL12A1/LOX/COL3A1/COL11A1/COL14A1/COL15A1/CD36/COL4A1/COL4A2 | |
| collagen binding | GO:0005518 | 1.53 × 10−8 | LUM/MMP9/THBS1/LOX/VWF/COL14A1/PCOLCE/TGFBI/SPARCL1/ITGA2/NID2 | |
| glycosaminoglycan binding | GO:0005539 | 3.92 × 10−8 | VCAN/LPL/THBS1/GPNMB/COL11A1/SULF1/THBS2/TGFBR2/SMOC2/PCOLCE/ADAMTS1/NRP1/BGN/HAPLN1/LYVE1/NRP2/PTPRC | |
| Degradation of the extracellular matrix | REAC:R-HSA-1474228 | 6.00 × 10−8 | SPP1/MMP9/COL12A1/COL3A1/COL11A1/A2M/MMP2/COL14A1/MMP16/COL15A1/ADAMTS1/ADAMTS9/COL4A1/COL4A2/LAMB1 | |
| Collagen formation | REAC:R-HSA-1474290 | 4.10 × 10−6 | MMP9/COL12A1/LOX/COL3A1/COL11A1/COL14A1/PCOLCE/COL15A1/ITGA6/COL4A1/COL4A2 | |
| Collagen degradation | REAC:R-HSA-1442490 | 2.99 × 10−5 | MMP9/COL12A1/COL3A1/COL11A1/MMP2/COL14A1/COL15A1/COL4A1/COL4A2 | |
| Collagen chain trimerization | REAC:R-HSA-8948216 | 0.0004 | COL12A1/COL3A1/COL11A1/COL14A1/COL15A1/COL4A1/COL4A2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yakushov, S.; Menyailo, M.; Denisov, E.; Karlina, I.; Zainullina, V.; Kirgizov, K.; Romantsova, O.; Timashev, P.; Ulasov, I. Identification of Factors Driving Doxorubicin-Resistant Ewing Tumor Cells to Survival. Cancers 2022, 14, 5498. https://doi.org/10.3390/cancers14225498
Yakushov S, Menyailo M, Denisov E, Karlina I, Zainullina V, Kirgizov K, Romantsova O, Timashev P, Ulasov I. Identification of Factors Driving Doxorubicin-Resistant Ewing Tumor Cells to Survival. Cancers. 2022; 14(22):5498. https://doi.org/10.3390/cancers14225498
Chicago/Turabian StyleYakushov, Semyon, Maxim Menyailo, Evgeny Denisov, Irina Karlina, Viktoria Zainullina, Kirill Kirgizov, Olga Romantsova, Peter Timashev, and Ilya Ulasov. 2022. "Identification of Factors Driving Doxorubicin-Resistant Ewing Tumor Cells to Survival" Cancers 14, no. 22: 5498. https://doi.org/10.3390/cancers14225498
APA StyleYakushov, S., Menyailo, M., Denisov, E., Karlina, I., Zainullina, V., Kirgizov, K., Romantsova, O., Timashev, P., & Ulasov, I. (2022). Identification of Factors Driving Doxorubicin-Resistant Ewing Tumor Cells to Survival. Cancers, 14(22), 5498. https://doi.org/10.3390/cancers14225498









