Effect of Collagen Matrix on Doxorubicin Distribution and Cancer Cells’ Response to Treatment in 3D Tumor Model
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Cell Cultures
2.2. Collagen-Based 3D-Model
2.3. Treatment with Doxorubicin
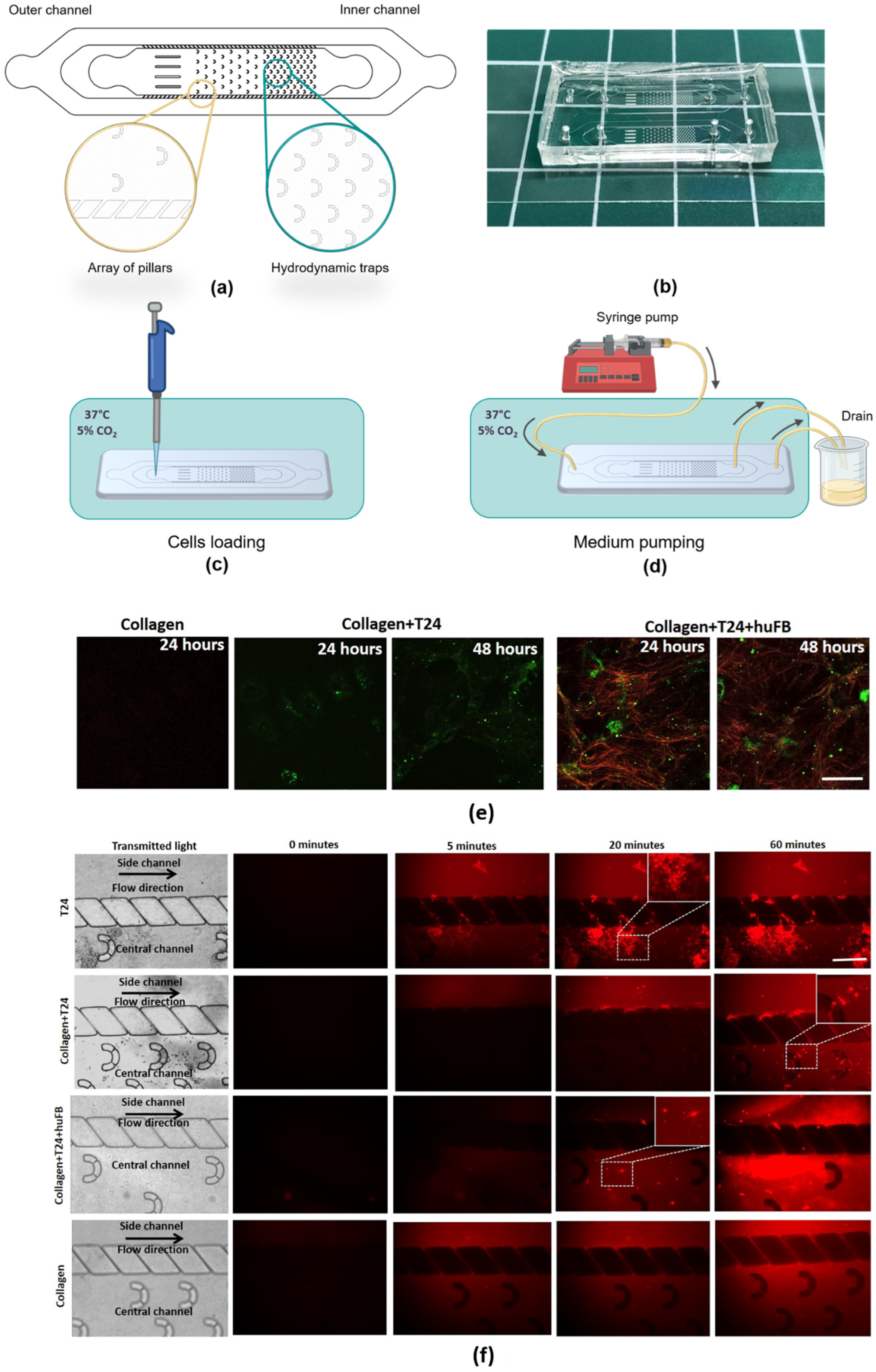
2.4. Design of the Microfluidic Chip (MFC)
2.5. Experimental Setup with MFCs for Drug Delivery Analysis
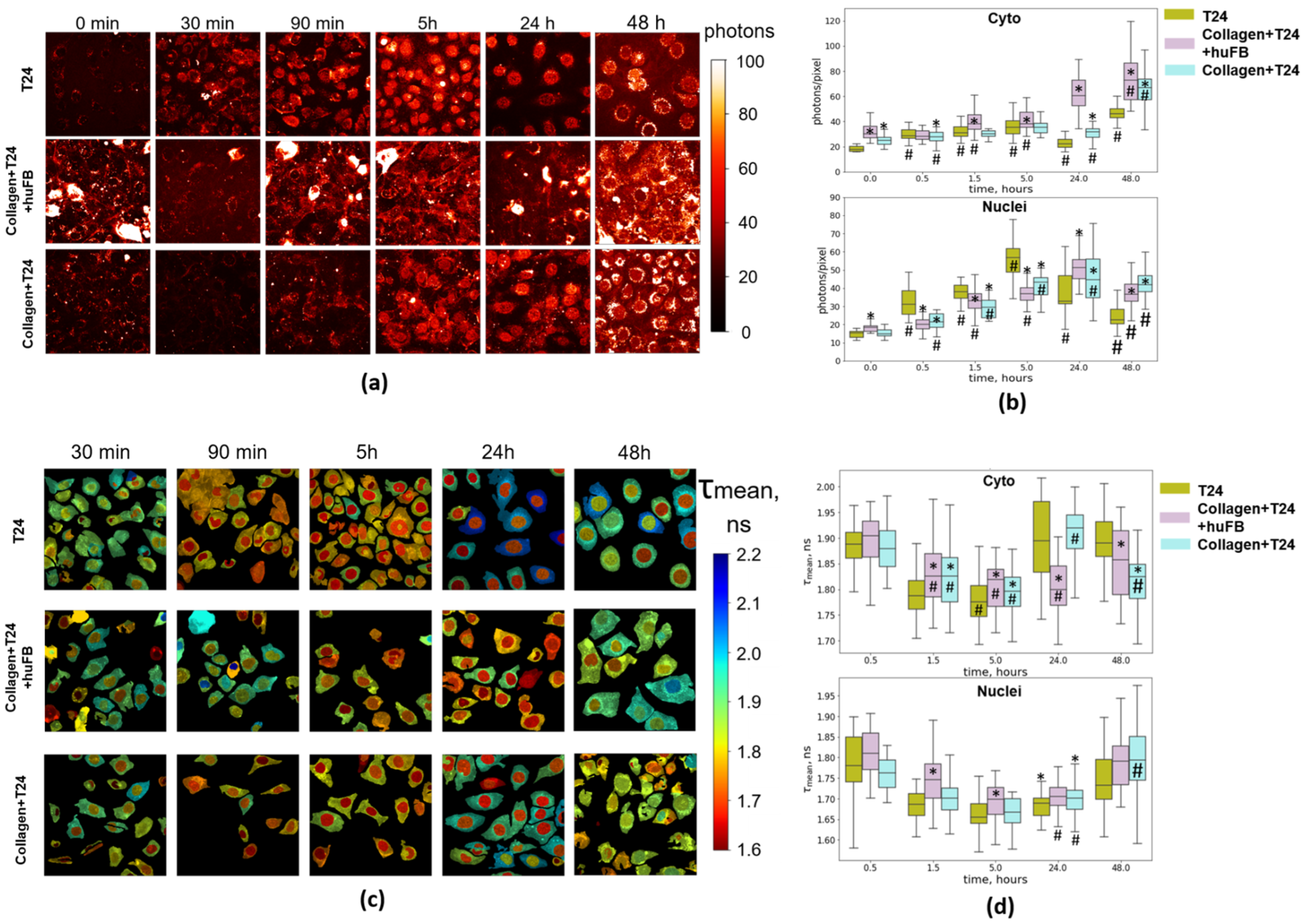
2.6. Multiphoton Microscopy for NAD(P)H and DOX Imaging
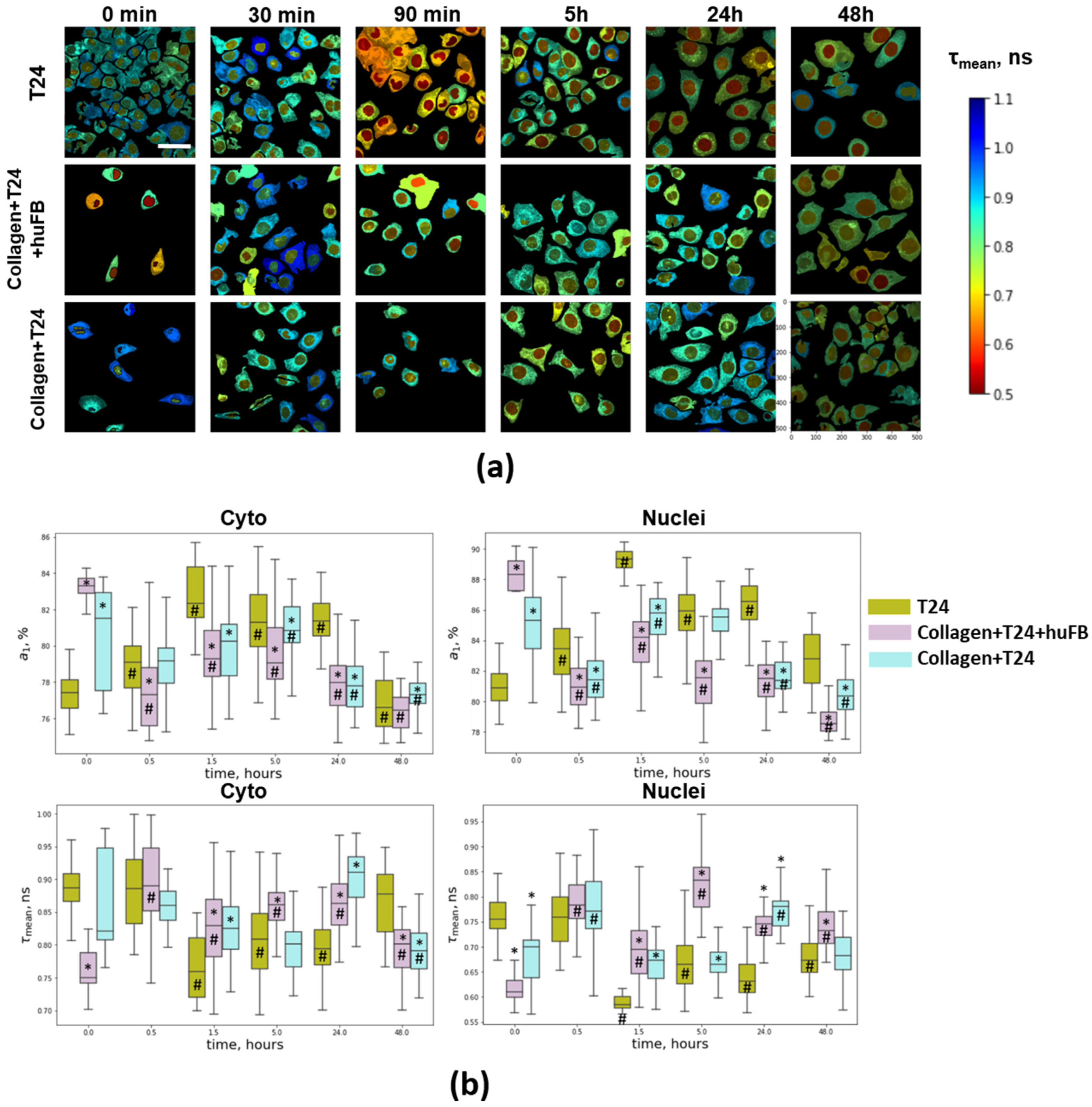
2.7. Single-Cell FLIM Analysis
2.8. Second Harmonic Generation (SHG) Microscopy
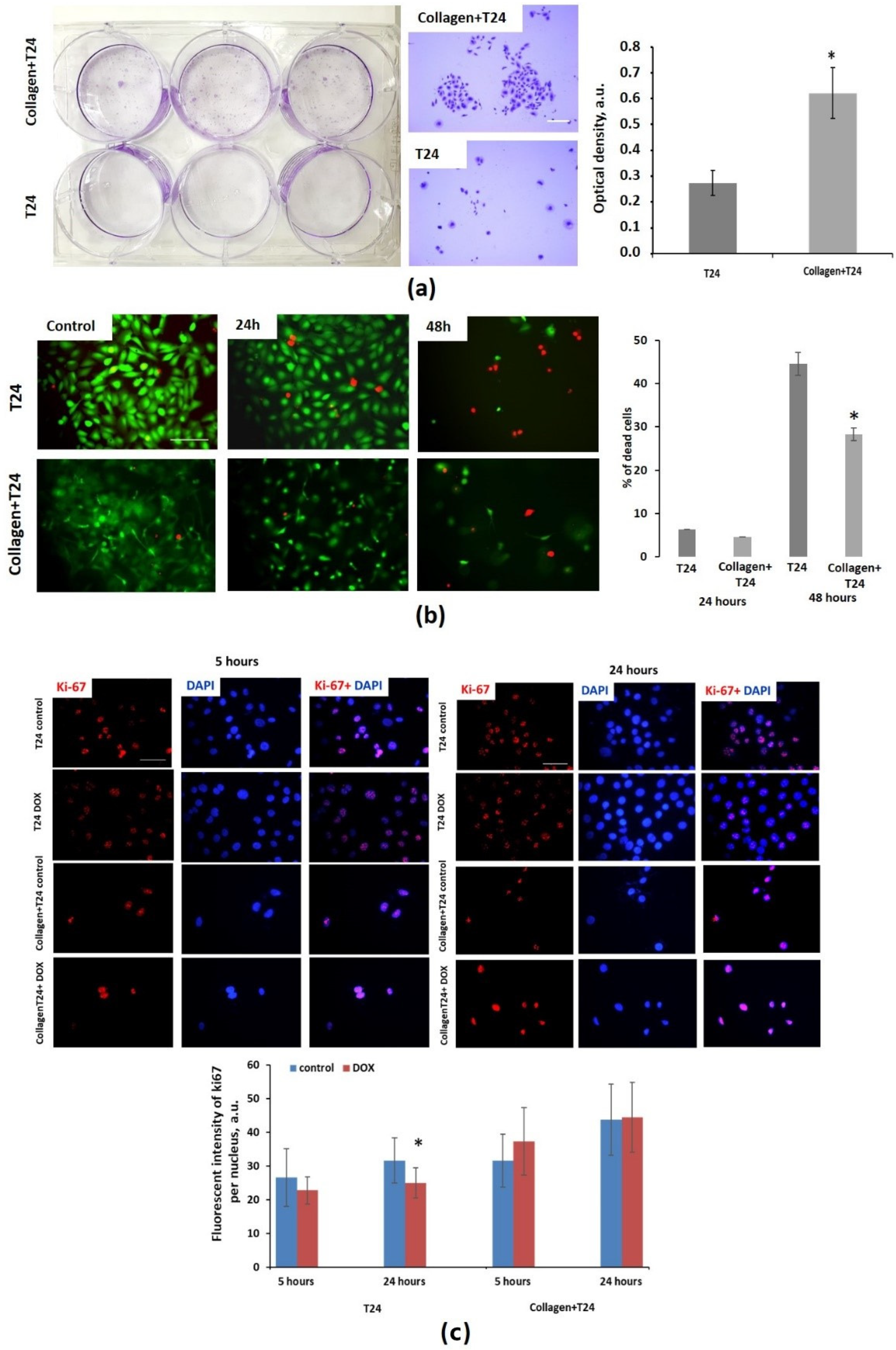
2.9. Colony-Forming Assay
2.10. MTT Assay
2.11. Live/Dead Cell Assay
2.12. Immunocytochemistry for ki-67
2.13. Assessment of Mitochondrial Potential
2.14. Isolation of RNA and Quantitative RT-PCR Analysis
2.15. Statistical Analysis
3. Results
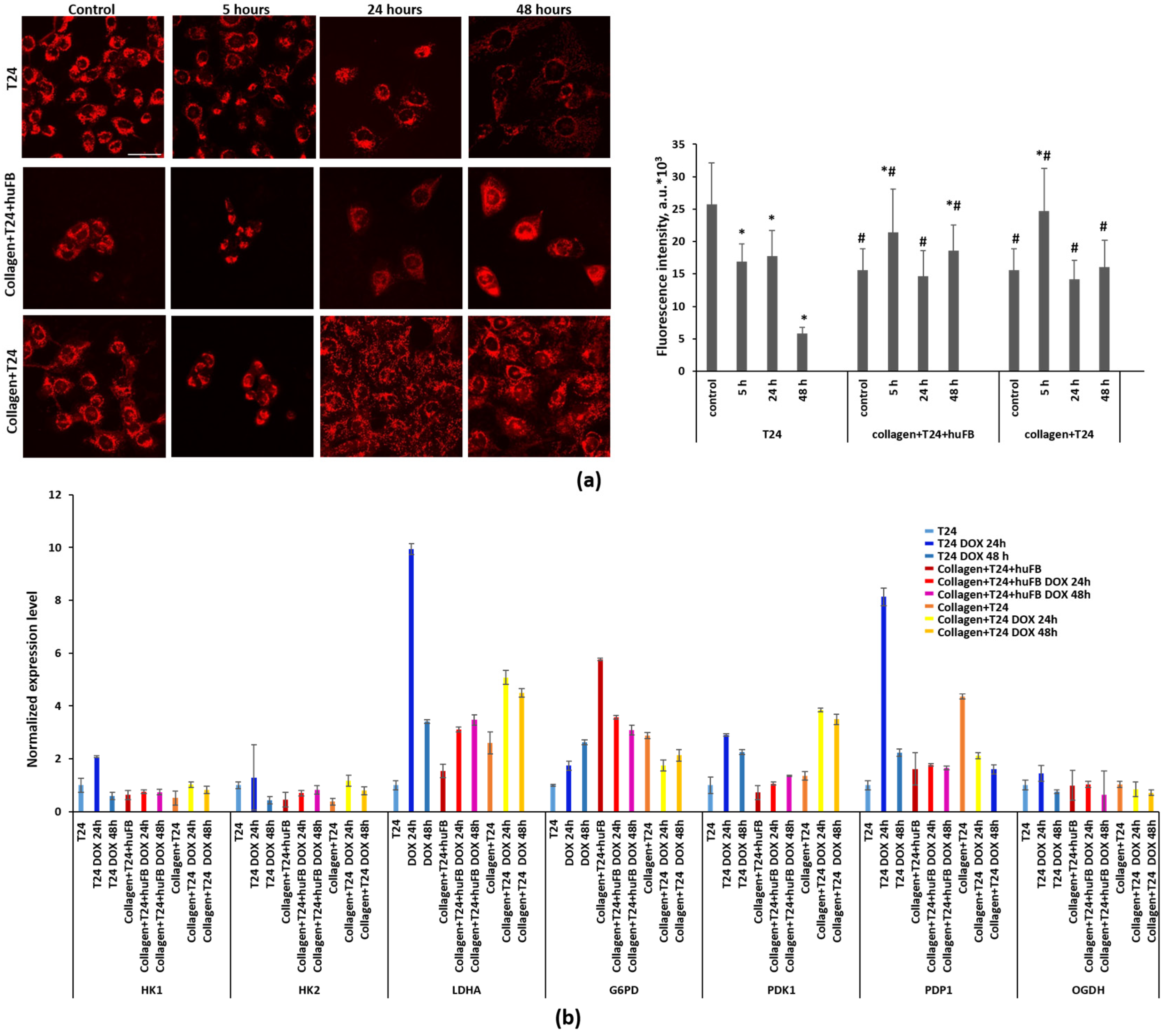
3.1. Effect of Collagen on DOX Delivery to Cancer Cells in MFC
3.2. Effect of Collagen on Cytotoxicity of Doxorubicin
3.3. Analysis of Doxorubicin Distribution by Multiphoton Microscopy
3.4. NAD(P)H FLIM of T24 Cells under Doxorubicin Exposure
3.5. Metabolic Activity of T24 Cells after Treatment with Doxorubicin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Minchinton, A.I.; Tannock, I.F. Drug penetration in solid tumours. Nature reviews. Cancer 2006, 6, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Hastings, J.F.; Skhinas, J.N.; Fey, D.; Croucher, D.R.; Cox, T.R. The extracellular matrix as a key regulator of intracellular signalling networks. Br. J. Pharmacol. 2019, 176, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Poltavets, V.; Kochetkova, M.; Pitson, S.M.; Samuel, M.S. The Role of the Extracellular Matrix and Its Molecular and Cellular Regulators in Cancer Cell Plasticity. Front. Oncol. 2018, 8, 431. [Google Scholar] [CrossRef]
- Dewhirst, M.W.; Secomb, T.W. Transport of drugs from blood vessels to tumour tissue. Nature reviews. Cancer 2017, 17, 738–750. [Google Scholar] [CrossRef]
- Rossow, L.; Veitl, S.; Vorlová, S.; Wax, J.K.; Kuhn, A.E.; Maltzahn, V.; Upcin, B.; Karl, F.; Hoffmann, H.; Gätzner, S.; et al. LOX-catalyzed collagen stabilization is a proximal cause for intrinsic resistance to chemotherapy. Oncogene 2018, 37, 4921–4940. [Google Scholar] [CrossRef] [PubMed]
- Jacobetz, M.A.; Chan, D.S.; Neesse, A.; Bapiro, T.E.; Cook, N.; Frese, K.K.; Feig, C.; Nakagawa, T.; Caldwell, M.E.; Zecchini, H.I.; et al. Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut 2013, 62, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Eikenes, L.; Tari, M.; Tufto, I.; Bruland, O.S.; de Lange Davies, C. Hyaluronidase induces a transcapillary pressure gradient and improves the distribution and uptake of liposomal doxorubicin (Caelyx) in human osteosarcoma xenografts. Br. J. Cancer 2005, 93, 81–88. [Google Scholar] [CrossRef]
- Wang, X.; Luo, J.; He, L.; Cheng, X.; Yan, G.; Wang, J.; Tang, R. Hybrid pH-sensitive nanogels surface-functionalized with collagenase for enhanced tumor penetration. J. Colloid Interface Sci. 2018, 525, 269–281. [Google Scholar] [CrossRef]
- Johnson-Arbor, K.; Dubey, R. Doxorubicin. In StatPearls; StatPearls Publishing Copyright © 2022; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, S.J.; Kim, B.J.; Rah, S.Y.; Chung, S.M.; Im, M.J.; Kim, U.H. Doxorubicin-induced reactive oxygen species generation and intracellular Ca2+ increase are reciprocally modulated in rat cardiomyocytes. Exp. Mol. Med. 2006, 38, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Taymaz-Nikerel, H.; Karabekmez, M.E.; Eraslan, S.; Kırdar, B. Doxorubicin induces an extensive transcriptional and metabolic rewiring in yeast cells. Sci. Rep. 2018, 8, 13672. [Google Scholar] [CrossRef] [PubMed]
- Achkar, I.W.; Kader, S.; Dib, S.S.; Junejo, K.; Al-Bader, S.B.; Hayat, S.; Bhagwat, A.M.; Rousset, X.; Wang, Y.; Viallet, J.; et al. Metabolic Signatures of Tumor Responses to Doxorubicin Elucidated by Metabolic Profiling in Ovo. Metabolites 2020, 10, 268. [Google Scholar] [CrossRef] [PubMed]
- Marina, V.; Shirmanova, V.; Shcheslavskiy, I.; Lukina, M.M.; Becker, W.; Zagaynova, E.V. Exploring Tumor Metabolism with Time-Resolved Fluo-Rescence Methods: From Single Cells to a Whole Tumor // Chapter 3 in Multimodal Optical Diagnostics of Cancer; Tuchin, V., Popp, J., Zakharov, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 133–155. ISBN 978-3-030-44594-2. [Google Scholar]
- Kalinina, S.; Freymueller, C.; Naskar, N.; von Einem, B.; Reess, K.; Sroka, R.; Rueck, A. Bioenergetic Alterations of Metabolic Redox Coenzymes as NADH, FAD and FMN by Means of Fluorescence Lifetime Imaging Techniques. Int. J. Mol. Sci. 2021, 22, 5952. [Google Scholar] [CrossRef]
- Ke, X.Y.; Lin Ng, V.W.; Gao, S.J.; Tong, Y.W.; Hedrick, J.L.; Yang, Y.Y. Co-delivery of thioridazine and doxorubicin using polymeric micelles for targeting both cancer cells and cancer stem cells. Biomaterials 2014, 35, 1096–1108. [Google Scholar] [CrossRef]
- Xiong, H.; Du, S.; Ni, J.; Zhou, J.; Yao, J. Mitochondria and nuclei dual-targeted heterogeneous hydroxyapatite nanoparticles for enhancing therapeutic efficacy of doxorubicin. Biomaterials 2016, 94, 70–83. [Google Scholar] [CrossRef]
- Alam, S.R.; Wallrabe, H.; Svindrych, Z.; Chaudhary, A.K.; Christopher, K.G.; Chandra, D.; Periasamy, A. Investigation of Mitochondrial Metabolic Response to Doxorubicin in Prostate Cancer Cells: An NADH, FAD and Tryptophan FLIM Assay. Sci. Rep. 2017, 7, 10451. [Google Scholar] [CrossRef]
- Wallrabe, H.; Svindrych, Z.; Alam, S.R.; Siller, K.H.; Wang, T.; Kashatus, D.; Hu, S.; Periasamy, A. Segmented cell analyses to measure redox states of autofluorescent NAD(P)H, FAD & Trp in cancer cells by FLIM. Sci. Rep. 2018, 8, 79. [Google Scholar] [CrossRef]
- Druzhkova, I.; Shirmanova, M.; Ignatova, N.; Dudenkova, V.; Lukina, M.; Zagaynova, E.; Safina, D.; Kostrov, S.; Didych, D.; Kuzmich, A.; et al. Expression of EMT-Related Genes in Hybrid E/M Colorectal Cancer Cells Determines Fibroblast Activation and Collagen Remodeling. Int. J. Mol. Sci. 2020, 21, 8119. [Google Scholar] [CrossRef]
- Druzhkova, I.N.; Shirmanova, M.V.; Lukina, M.M.; Dudenkova, V.V.; Mishina, N.M.; Zagaynova, E.V. The metabolic interaction of cancer cells and fibroblasts—Coupling between NAD(P)H and FAD, intracellular pH and hydrogen peroxide. Cell Cycle 2016, 15, 1257–1266. [Google Scholar] [CrossRef]
- Shirshin, E.A.; Shirmanova, M.V.; Gayer, A.V.; Lukina, M.M.; Nikonova, E.E.; Yakimov, B.P.; Budylin, G.S.; Dudenkova, V.V.; Ignatova, N.I.; Komarov, D.V.; et al. Label-free sensing of cells with fluorescence lifetime imaging: The quest for metabolic heterogeneity. Proc. Natl. Acad. Sci. USA 2022, 119, e2118241119. [Google Scholar] [CrossRef] [PubMed]
- Carlson, M.; Watson, A.L.; Anderson, L.; Largaespada, D.A.; Provenzano, P.P. Multiphoton fluorescence lifetime imaging of chemotherapy distribution in solid tumors. J. Biomed. Opt. 2017, 22, 116010. [Google Scholar] [CrossRef]
- Freund, I.; Deutsch, M. Second-harmonic microscopy of biological tissue. Opt. Lett. 1986, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.T.; Wu, C.Y.; Chung, C.Y.; Hwu, Y.; Cheng, S.H.; Mou, C.Y.; Lo, L.W. Probing the dynamics of doxorubicin-DNA intercalation during the initial activation of apoptosis by fluorescence lifetime imaging microscopy (FLIM). PLoS ONE 2012, 7, e44947. [Google Scholar] [CrossRef]
- Heikal, A.A. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark. Med. 2010, 4, 241–263. [Google Scholar] [CrossRef]
- Skala, M.C.; Riching, K.M.; Bird, D.K.; Gendron-Fitzpatrick, A.; Eickhoff, J.; Eliceiri, K.W.; Keely, P.J.; Ramanujam, N. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J. Biomed. Opt. 2007, 12, 024014. [Google Scholar] [CrossRef]
- Lovitt, C.J.; Shelper, T.B.; Avery, V.M. Doxorubicin resistance in breast cancer cells is mediated by extracellular matrix proteins. BMC Cancer 2018, 18, 41. [Google Scholar] [CrossRef]
- Li, X.; Shepard, H.M.; Cowell, J.A.; Zhao, C.; Osgood, R.J.; Rosengren, S.; Blouw, B.; Garrovillo, S.A.; Pagel, M.D.; Whatcott, C.J.; et al. Parallel Accumulation of Tumor Hyaluronan, Collagen, and Other Drivers of Tumor Progression. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 4798–4807. [Google Scholar] [CrossRef]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef]
- Al-Abd, A.M.; Lee, J.H.; Kim, S.Y.; Kun, N.; Kuh, H.J. Novel application of multicellular layers culture for in situ evaluation of cytotoxicity and penetration of paclitaxel. Cancer Sci. 2008, 99, 423–431. [Google Scholar] [CrossRef]
- Regnault, C.; Dheeman, D.S.; Hochstetter, A. Microfluidic Devices for Drug Assays. High-Throughput 2018, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Na, K.; Song, S.-C.; Lee, J.; Kuh, H.-J. The distribution and retention of paclitaxel and doxorubicin in multicellular layer cultures. Oncol. Rep. 2012, 27, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Van Pham, P. Cancer Biology and Advances in Treatment; Springer International Publishing: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Changenet-Barret, P.; Gustavsson, T.; Markovitsi, D.; Manet, I.; Monti, S. Unravelling molecular mechanisms in the fluorescence spectra of doxorubicin in aqueous solution by femtosecond fluorescence spectroscopy. Phys. Chem. Chem. Phys. 2013, 15, 2937–2944. [Google Scholar] [CrossRef] [PubMed]
- Sparks, H.; Kondo, H.; Hooper, S.; Munro, I.; Kennedy, G.; Dunsby, C.; French, P.; Sahai, E. Heterogeneity in tumor chromatin-doxorubicin binding revealed by in vivo fluorescence lifetime imaging confocal endomicroscopy. Nat. Commun. 2018, 9, 2662. [Google Scholar] [CrossRef] [PubMed]
- Wallace, K.B.; Sardão, V.A.; Oliveira, P.J. Mitochondrial Determinants of Doxorubicin-Induced Cardiomyopathy. Circ. Res. 2020, 126, 926–941. [Google Scholar] [CrossRef]
- Abdullah, C.S.; Alam, S.; Aishwarya, R.; Miriyala, S.; Bhuiyan, M.A.N.; Panchatcharam, M.; Pattillo, C.B.; Orr, A.W.; Sadoshima, J.; Hill, J.A.; et al. Doxorubicin-induced cardiomyopathy associated with inhibition of autophagic degradation process and defects in mitochondrial respiration. Sci. Rep. 2019, 9, 2002. [Google Scholar] [CrossRef]
- Aryal, B.; Rao, V.A. Deficiency in Cardiolipin Reduces Doxorubicin-Induced Oxidative Stress and Mitochondrial Damage in Human B-Lymphocytes. PLoS ONE 2016, 11, e0158376. [Google Scholar] [CrossRef]
- Santos, S.M.; Hartman, J.L. A yeast phenomic model for the influence of Warburg metabolism on genetic buffering of doxorubicin. Cancer Metab. 2019, 7, 9. [Google Scholar] [CrossRef]
- Dornfeld, K.; Bjork, J.; Folkert, G.; Skildum, A.; Wallace, K.B. Mitochondrial activities play a pivotal role in regulating cell cycle in response to doxorubicin. Cell Cycle 2021, 20, 1067–1079. [Google Scholar] [CrossRef]
- Lukina, M.M.; Dudenkova, V.V.; Ignatova, N.I.; Druzhkova, I.N.; Shimolina, L.E.; Zagaynova, E.V.; Shirmanova, M.V. Metabolic cofactors NAD(P)H and FAD as potential indicators of cancer cell response to chemotherapy with paclitaxel. Biochimica et biophysica acta. Gen. Subj. 2018, 1862, 1693–1700. [Google Scholar] [CrossRef]
- Lukina, M.M.; Dudenkova, V.V.; Shimolina, L.E.; Snopova, L.B.; Zagaynova, E.V.; Shirmanova, M.V. In vivo metabolic and SHG imaging for monitoring of tumor response to chemotherapy. Cytometry Part A J. Int. Soc. Anal. Cytol. 2019, 95, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Shirmanova, M.V.; Druzhkova, I.N.; Lukina, M.M.; Dudenkova, V.V.; Ignatova, N.I.; Snopova, L.B.; Shcheslavskiy, V.I.; Belousov, V.V.; Zagaynova, E.V. Chemotherapy with cisplatin: Insights into intracellular pH and metabolic landscape of cancer cells in vitro and in vivo. Sci. Rep. 2017, 7, 8911. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, P.M.; Hilpert, D.; Niederschweiberer, M.; Neuhauser, L.; Kalinina, S.; Calzia, E.; Rueck, A.; von Einem, B.; von Arnim, C.A.F. Mitochondrial matrix pH as a decisive factor in neurometabolic imaging. Neurophotonics 2017, 4, 045004. [Google Scholar] [CrossRef][Green Version]
- Shimolina, L.; Potekhina, E.; Druzhkova, I.; Lukina, M.; Dudenkova, V.; Belousov, V.; Shcheslavskiy, V.; Zagaynova, E.; Shirmanova, M. Fluorescence lifetime-based pH mapping of tumors in vivo using genetically encoded sensor SypHerRed. Biophys. J. 2022, 121, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Bean, J.F.; Qiu, Y.Y.; Yu, S.; Clark, S.; Chu, F.; Madonna, M.B. Glycolysis inhibition and its effect in doxorubicin resistance in neuroblastoma. J. Pediatric Surg. 2014, 49, 981–984, discussion 984. [Google Scholar] [CrossRef] [PubMed]
- Yadav, N.; Kumar, S.; Marlowe, T.; Chaudhary, A.K.; Kumar, R.; Wang, J.; O’Malley, J.; Boland, P.M.; Jayanthi, S.; Kumar, T.K.; et al. Oxidative phosphorylation-dependent regulation of cancer cell apoptosis in response to anticancer agents. Cell Death Dis. 2015, 6, e1969. [Google Scholar] [CrossRef] [PubMed]
- Kule, C.; Ondrejickova, O.; Verner, K. Doxorubicin, daunorubicin, and mitoxantrone cytotoxicity in yeast. Mol. Pharmacol. 1994, 46, 1234–1240. [Google Scholar]
- Micallef, I.; Baron, B. Doxorubicin: An Overview of the Anti-Cancer and Chemoresistance Mechanisms. Ann. Clin. Toxicol. 2020, 3, 1031. [Google Scholar]
- Bertero, T.; Oldham, W.M.; Grasset, E.M.; Bourget, I.; Boulter, E.; Pisano, S.; Hofman, P.; Bellvert, F.; Meneguzzi, G.; Bulavin, D.V.; et al. Tumor-Stroma Mechanics Coordinate Amino Acid Availability to Sustain Tumor Growth and Malignancy. Cell Metab. 2019, 29, 124–140.e110. [Google Scholar] [CrossRef]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B. The intermediary metabolism of the prostate: A key to understanding the pathogenesis and progression of prostate malignancy. Oncology 2000, 59, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Vayalil, P.K. Mitochondrial oncobioenergetics of prostate tumorigenesis (Review). Oncol. Lett. 2019, 18, 4367–4376. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Pouli, D.; Alonzo, C.A.; Varone, A.; Karaliota, S.; Quinn, K.P.; Münger, K.; Karalis, K.P.; Georgakoudi, I. Mapping metabolic changes by noninvasive, multiparametric, high-resolution imaging using endogenous contrast. Sci. Adv. 2018, 4, eaap9302. [Google Scholar] [CrossRef] [PubMed]
- Paredes, F.; Williams, H.C.; San Martin, A. Metabolic adaptation in hypoxia and cancer. Cancer Lett. 2021, 502, 133–142. [Google Scholar] [CrossRef]
- Yao, C.-H.; Wang, R.; Wang, Y.; Kung, C.-P.; Weber, J.D.; Patti, G.J. Mitochondrial fusion supports increased oxidative phosphorylation during cell proliferation. eLife 2019, 8, e41351. [Google Scholar] [CrossRef]
- Meyer, J.N.; Leuthner, T.C.; Luz, A.L. Mitochondrial fusion, fission, and mitochondrial toxicity. Toxicology 2017, 391, 42–53. [Google Scholar] [CrossRef]
| Primer Target | Description | Primer Sequence (5′→3′) |
|---|---|---|
| HK1 | Hexokinase-1 | Forward Primer-CACCTGTGAGGTTGGACTCA Reverse Primer-CCACCATCTCCACGTTCTTC |
| HK2 | Hexokinase-2 | Forward Primer-GAGTTTGACCTGGATGTGGTTGC Reverse Primer-CCTCCATGTAGCAGGCATTGCT |
| PDK1 | Pyruvate dehydrogenase kinase | Forward Primer-CCGCTCTCCATGAAGCAGTT Reverse Primer-TTGCCGCAGAAACATAAATGAG |
| LDHA | Lactate dehydrogenase A | Forward Primer-AGCCCGATTCCGTTACCT Reverse Primer-CACCAGCAACATTCATTCCA |
| G6PD | Glucose-6-phosphate dehydrogenase | Forward Primer-CTGTTCCGTGAGGACCAGATCT Reverse Primer-TGAAGGTGAGGATAACGCAGGC |
| OGDH | 2-Oxoglutarate dehydrogenase | Forward Primer-GAGGCTGTCATGTACGTGTGCA Reverse Primer-TACATGAGCGGCTGCGTGAACA |
| ABL1 | Tyrosine-protein kinase | Forward Primer-CCAGGTGTATGAGCTGCTAGAG Reverse Primer-GTCAGAGGGATTCCACTGCCAA |
| SDHA1 | Succinate dehydrogenase Complex flavoprotein Subunite A | Forward Primer-GAGATGTGGTGTCTCGGTCCAT Reverse Primer-GCTGTCTCTGAAATGCCAGGCA |
| PDP1 | Pyruvate dehydrogenase Phosphatase catalytic Subunit 1 | Forward Primer-TTCTGGAGCCACTGCTTGTGTG Reverse Primer-ACAGCGTGACTGCTGACCATGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Druzhkova, I.; Nikonova, E.; Ignatova, N.; Koryakina, I.; Zyuzin, M.; Mozherov, A.; Kozlov, D.; Krylov, D.; Kuznetsova, D.; Lisitsa, U.; et al. Effect of Collagen Matrix on Doxorubicin Distribution and Cancer Cells’ Response to Treatment in 3D Tumor Model. Cancers 2022, 14, 5487. https://doi.org/10.3390/cancers14225487
Druzhkova I, Nikonova E, Ignatova N, Koryakina I, Zyuzin M, Mozherov A, Kozlov D, Krylov D, Kuznetsova D, Lisitsa U, et al. Effect of Collagen Matrix on Doxorubicin Distribution and Cancer Cells’ Response to Treatment in 3D Tumor Model. Cancers. 2022; 14(22):5487. https://doi.org/10.3390/cancers14225487
Chicago/Turabian StyleDruzhkova, Irina, Elena Nikonova, Nadezhda Ignatova, Irina Koryakina, Mikhail Zyuzin, Artem Mozherov, Dmitriy Kozlov, Dmitry Krylov, Daria Kuznetsova, Uliyana Lisitsa, and et al. 2022. "Effect of Collagen Matrix on Doxorubicin Distribution and Cancer Cells’ Response to Treatment in 3D Tumor Model" Cancers 14, no. 22: 5487. https://doi.org/10.3390/cancers14225487
APA StyleDruzhkova, I., Nikonova, E., Ignatova, N., Koryakina, I., Zyuzin, M., Mozherov, A., Kozlov, D., Krylov, D., Kuznetsova, D., Lisitsa, U., Shcheslavskiy, V., Shirshin, E. A., Zagaynova, E., & Shirmanova, M. (2022). Effect of Collagen Matrix on Doxorubicin Distribution and Cancer Cells’ Response to Treatment in 3D Tumor Model. Cancers, 14(22), 5487. https://doi.org/10.3390/cancers14225487







