Generalization of Deep Learning in Digital Pathology: Experience in Breast Cancer Metastasis Detection
Abstract
Simple Summary
Abstract
1. Introduction
Aim
2. Materials and Methods
2.1. Datasets
2.1.1. Local Linköping Data
2.1.2. CAMELYON Data
2.1.3. Data Splits
2.2. Study Design
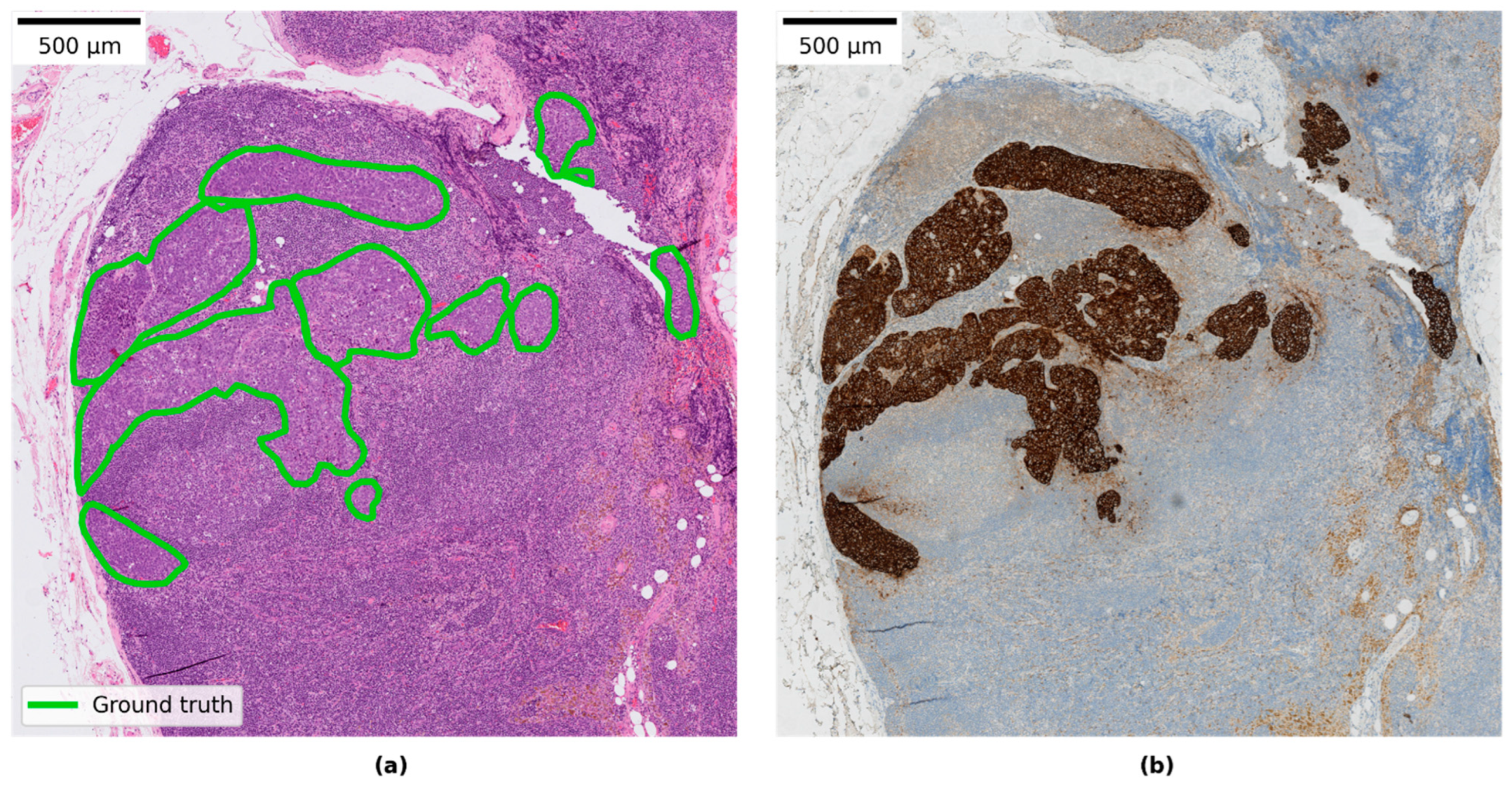
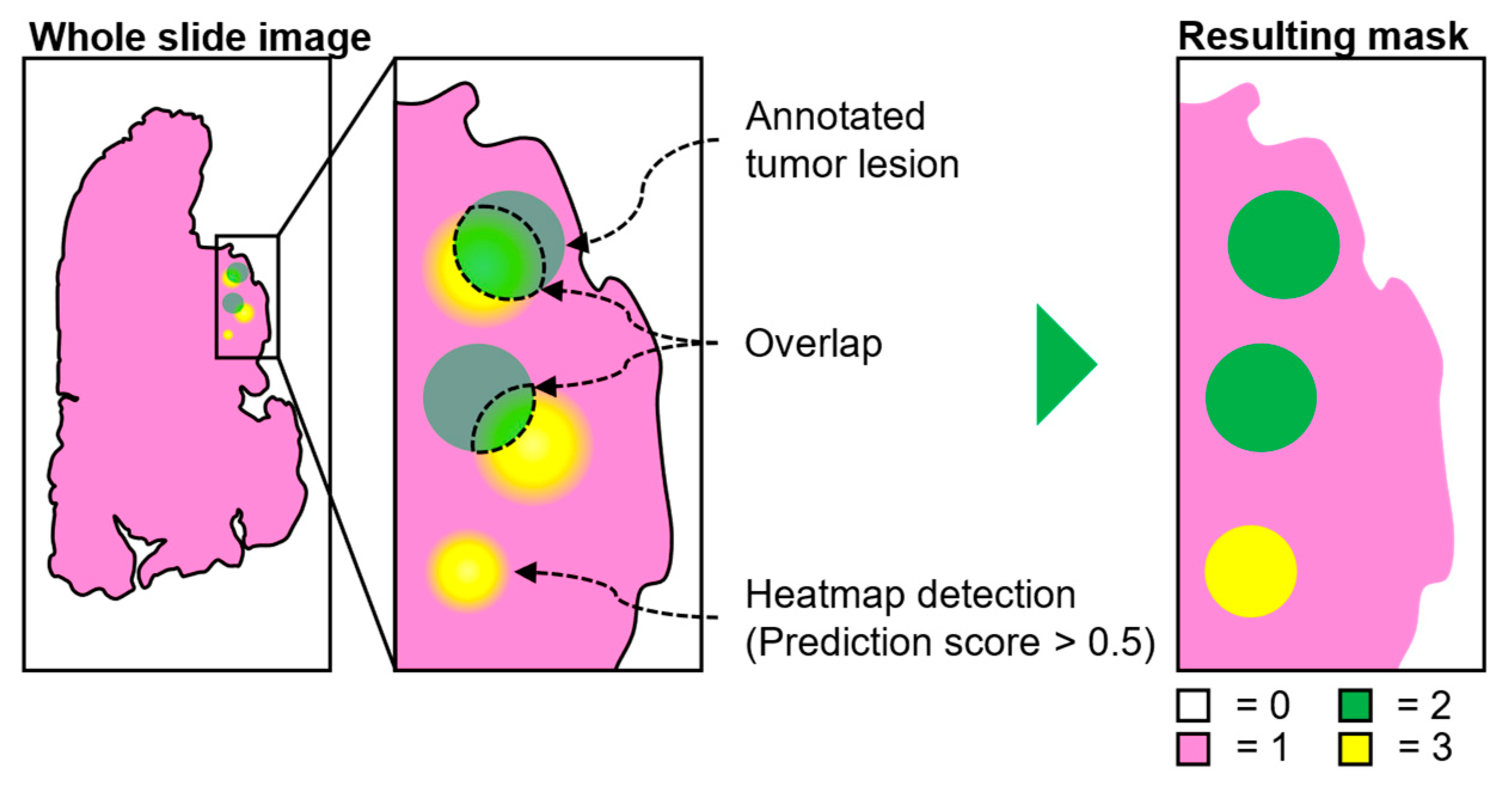
2.3. Evaluation of WSI Predictions
2.4. Metrics
2.5. Model Architecture
2.6. Model Retraining
- Transfer learning: Two modes were used when retraining the model:
- The model was retrained from scratch on available data (CAMELYON + local data)
- The base model was used as a starting point and only trained on the local data (LocalSentinel + LocalAxillary).
- Sampling rates: Two modes for patch sampling from WSI were implemented when both CAMELYON and local data were used:
- Patches were sampled uniformly from all available images.
- The patch sampling rate from LocalSentinel and LocalAxillary was increased to allow the model to learn more from the local data and combat fitting preference towards the CAMELYON data which contained the most slides.
- Hard negative mining. Increased sampling rate in negative regions that were often classified as false positives in LocalSentinel and LocalAxillary. See further description of this method below.
2.7. Hard Negative Mining
3. Results
3.1. Test of Generalisation to a New Setting: Base Model on Local Sentinel Node Data
3.2. Test of Generalisation to a New Indication: Base-Model on Local Axillary Dissection Node Data
3.3. CNN Retraining to Increase Performance on Local Data
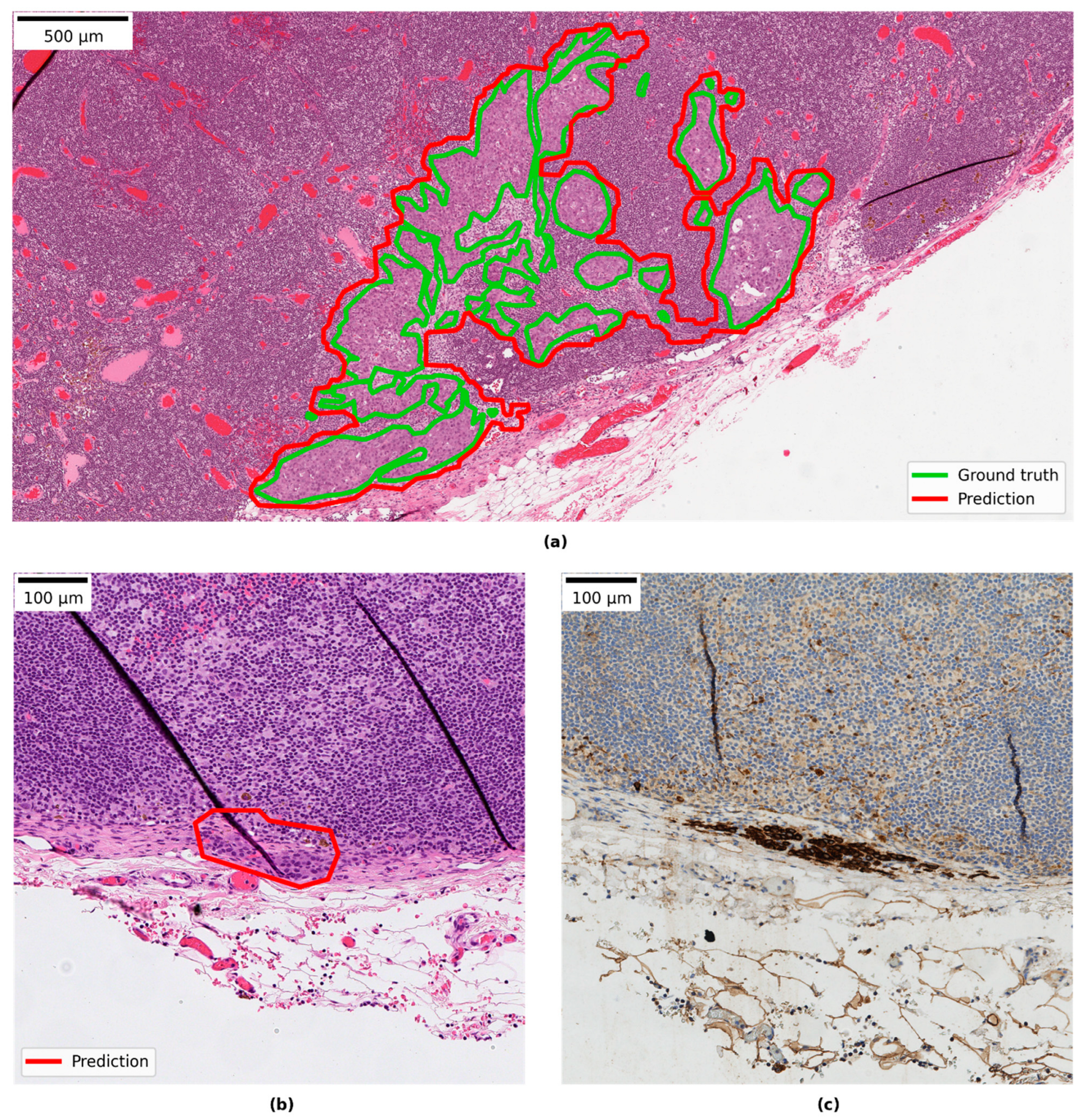
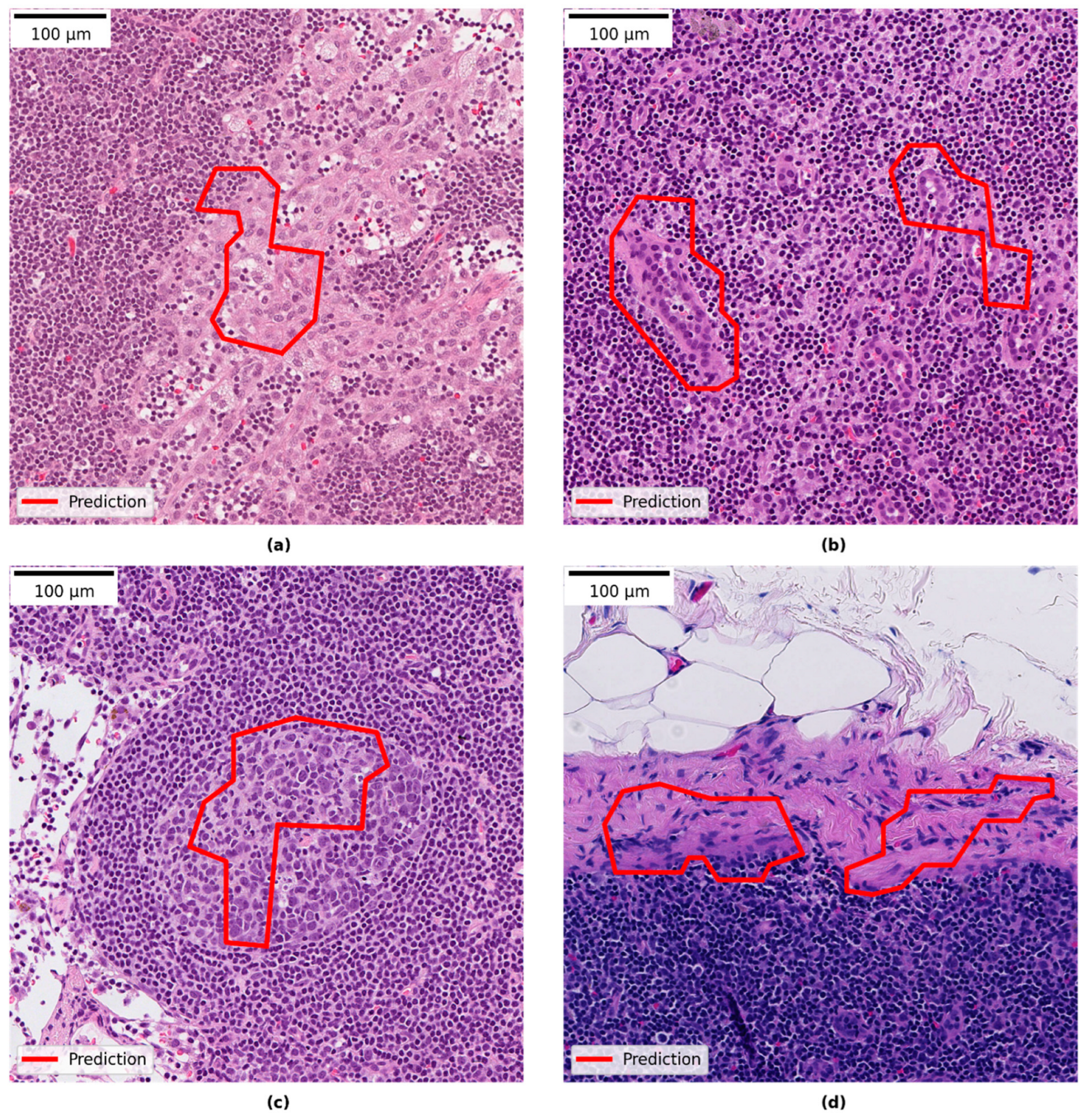
3.4. Qualitative Evaluation of Local Model Predictions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guide for Establishing a Pathology Laboratory in the Context of Cancer Control; World Health Organization: Geneva, Switzerland, 2019.
- Märkl, B.; Füzesi, L.; Huss, R.; Bauer, S.; Schaller, T. Number of pathologists in Germany: Comparison with European countries, USA, and Canada. Virchows Arch. 2021, 478, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Beckman Suurküla, M. Svensk Patologi—En Översyn och Förslag till Åtgärder; A Review of Clinical Pathology in Sweden Ordered by the Swedish Government. Översynen har Genomförts på Regeringens Uppdrag (S2011/5140/FS); Sweden. 2012. Available online: https://medlem.foreningssupport.se/foreningar/uploads/L15178/F%C3%B6reningen/Rapport_patologi_12.03.29.pdf (accessed on 5 March 2022).
- van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Ström, P.; Kartasalo, K.; Olsson, H.; Solorzano, L.; Delahunt, B.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Grignon, D.J.; Humphrey, P.A.; et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: A population-based, diagnostic study. Lancet Oncol. 2020, 21, 222–232. [Google Scholar] [CrossRef]
- Acs, B.; Rantalainen, M.; Hartman, J. Artificial intelligence as the next step towards precision pathology. J. Intern. Med. 2020, 288, 62–81. [Google Scholar] [CrossRef] [PubMed]
- Bulten, W.; Balkenhol, M.; Jean-Joël; Belinga, A.; Brilhante, A.; Çakır, A.; Egevad, L.; Eklund, M.; Farré, X.; Geronatsiou, K.; et al. Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists ISUP Pathology Imagebase Expert Panel. Mod. Pathol. 2021, 34, 660–671. [Google Scholar] [CrossRef]
- Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 5 March 2022).
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. (Eds.) TNM Classification of Malignant Tumours; Union for International Cancer Control: Oxford, UK, 2017. [Google Scholar]
- Apple, S.K. Sentinel Lymph Node in Breast Cancer: Review Article from a Pathologist’s Point of View. J. Pathol. Transl. Med. 2016, 50, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Bröstcancer Nationellt Vårdprogram Version: 4.0 (National Program for Breast Cancer in Sweden); Regionala Cancercentrum i Samverkan: Stockholm, Sweden, 2022.
- Litjens, G.; Bandi, P.; Bejnordi, B.E.; Geessink, O.; Balkenhol, M.; Bult, P.; Halilovic, A.; Hermsen, M.; van de Loo, R.; Vogels, R.; et al. 1399 H&E-stained sentinel lymph node sections of breast cancer patients: The CAMELYON dataset. GigaScience 2018, 7, giy065. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.W.M.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. J. Am. Med. Assoc. 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Steiner, D.F.; Macdonald, R.; Liu, Y.; Truszkowski, P.; Hipp, J.D.; Gammage, C.; Thng, F.; Peng, L.; Stumpe, M.C. Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. Am. J. Surg. Pathol. 2018, 42, 1636–1646. [Google Scholar] [CrossRef]
- Kleppe, A.; Skrede, O.-J.; De Raedt, S.; Liestøl, K.; Kerr, D.J.; Danielsen, H.E. Designing deep learning studies in cancer diagnostics. Nat. Rev. Cancer 2021, 21, 199–211. [Google Scholar] [CrossRef]
- Bándi, P.; Balkenhol, M.; van Dijk, M.; van Ginneken, B.; van der Laak, J.; Litjens, G. Domain adaptation strategies for cancer-independent detection of lymph node metastases. arXiv 2022, arXiv:2207.06193. [Google Scholar]
- Jarkman, S.; Lindvall, M.; Hedlund, J.; Treanor, D.; Lundstrom, C.; van der Laak, J. Axillary lymph nodes in breast cancer cases. AIDA Data Hub (AIDA Dataset Regist.) 2019. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2016; pp. 2261–2269. [Google Scholar]
- ASAP—Fluid Whole-Slide Image Viewer—Diagnostic Image Analysis Group. Available online: https://www.diagnijmegen.nl/software/asap/ (accessed on 8 May 2022).
- Bradley, A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef]
- Chakraborty, D.P. Recent developments in imaging system assessment methodology, FROC analysis and the search model. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2011, 648, S297–S301. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Efron, B.; Tibshirani, R.J. An Introduction to the Bootstrap; Chapman & Hall/CRC: Boca Raton, FL, USA, 1993. [Google Scholar]
- Bándi, P.; Geessink, O.; Manson, Q.; van Dijk, M.; Balkenhol, M.; Hermsen, M.; Ehteshami Bejnordi, B.; Lee, B.; Paeng, K.; Zhong, A.; et al. From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 Challenge. IEEE Trans. Med. Imaging 2018, 38, 550–560. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the International Conference on Computer Vision, Las Condes, Chile, 11–18 December 2015; pp. 1026–1034. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Wang, D.; Khosla, A.; Gargeya, R.; Irshad, H.; Beck, A.H. Deep Learning for Identifying Metastatic Breast Cancer. arXiv 2016, arXiv:1606.05718. [Google Scholar] [CrossRef]
- Gray, A.; Wright, A.; Jackson, P.; Hale, M.; Treanor, D. Quantification of histochemical stains using whole slide imaging: Development of a method and demonstration of its usefulness in laboratory quality control. J. Clin. Pathol. 2015, 68, 192–199. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.M.; Ciompi, F.; van der Laak, J. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef]
- Tang, H.; Sun, N.; Shen, S. Improving Generalization of Deep Learning Models for Diagnostic Pathology by Increasing Variability in Training Data: Experiments on Osteosarcoma Subtypes. J. Pathol. Inform. 2021, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Salvi, M.; Acharya, U.R.; Molinari, F.; Meiburger, K.M. The impact of pre- and post-image processing techniques on deep learning frameworks: A comprehensive review for digital pathology image analysis. Comput. Biol. Med. 2021, 128, 104129. [Google Scholar] [CrossRef] [PubMed]
- Lindvall, M.; Lundström, C.; Löwgren, J. Rapid Assisted Visual Search Supporting Digital Pathologists with Imperfect AI. In Proceedings of the 26th International Conference on Intelligent User Interfaces (IUI ’21), College Station, TX, USA, 14–17 April 2021. [Google Scholar]
- Houvenaeghel, G.; de Nonneville, A.; Cohen, M.; Chopin, N.; Coutant, C.; Reyal, F.; Mazouni, C.; Gimbergues, P.; Azuar, A.S.; Chauvet, M.P.; et al. Lack of prognostic impact of sentinel node micro-metastases in endocrine receptor-positive early breast cancer: Results from a large multicenter cohort☆. ESMO Open 2021, 6, 100151. [Google Scholar] [CrossRef]
- Viale, G.; Fusco, N. Pathology after neoadjuvant treatment—How to assess residual disease. Breast 2022, 62, S25–S28. [Google Scholar] [CrossRef] [PubMed]
- Moulin, P.; Grünberg, K.; Barale-Thomas, E.; Der Laak, J.V. IMI—Bigpicture: A Central Repository for Digital Pathology. Toxicol. Pathol. 2021, 49, 711–713. [Google Scholar] [CrossRef] [PubMed]

| Dataset | Unique Cases | Slides | Description |
|---|---|---|---|
| LocalSentinel | 161 | 161 | Retrospectively collected lymph node slides from sentinel node procedures consisting of 107 negative and 54 positive slides. One slide per unique patient case included. Cytokeratin immunohistochemically stained sections were available (AE1/AE3) and were used to aid in production of detailed ground-truth annotations. (Subset of AIDA BRLN dataset.) |
| LocalAxillary | 48 | 57 | Retrospectively collected lymph node slides from axillary dissection procedures consisting of 24 negative and 24 positive slides from unique cases. This dataset was further enriched with nine extra negative slides, overlapping with other cases, which contained only extra-nodal tissue and featured examples of fat necrosis and foreign body tissue reactions. (Subset of AIDA BRLN dataset.) |
| LocalNegativeAxillary | 24 | 259 | Retrospectively collected lymph node cases from axillary dissection procedures that were signed out as negative during initial clinical diagnostic assessment. All slides in the cases were included except for slides (n = 33) already included in LocalSentinel or LocalAxillary to avoid overlapping slides. One section per block, except in four cases 1–3 additional sections per block were presented in the clinical archive and included in this set. (Subset of AIDA BRLN dataset.) |
| CAMELYON16 | 399 | 399 | Retrospectively collected sentinel lymph node slides from two hospitals in the Netherlands. Pre-generated data splits by the CAMELYON organizers were retained in this study. Of the 160 positive slides, detailed annotations were available for 140 slides, whereas 20 slides were only partially annotated e.g., slides that contained two consecutive sections of the same tissue or slides that contained out-of-focus tumor regions, and only the delineated regions containing tumors were used in these slides during training. |
| CAMELYON17 | 200 | 1000 (344 used) | Retrospectively collected sentinel lymph node slides from five hospitals in the Netherlands. A total of 1000 slides. Only a subset of 50 positive slides contained detailed annotations, while the remaining positive slides had been given a slide label according to the TNM staging system. Of the available 50 annotated positive slides, only 34 slides contained either micro- or macro-metastases and were included in this study, the remaining 16 slides were labelled as isolated tumor cells (ITC) and were excluded. 50 randomly selected negative slides from the CAMELYON17 training set and 260 negative slides in the CAMELYON17 test set were allocated to training and testing sets, respectively, in this study. In total 344 slides from CAMELYON17 were used in this study. |
| Type | Description | Slide Label |
|---|---|---|
| Macro-metastasis | Tumor > 2 mm | Positive |
| Micro-metastasis | Tumor > 0.2 mm and ≤ 2 mm | Positive |
| Isolated tumor cells (ITC) | Tumor ≤ 0.2 mm or ≤ 200 tumor cells | Negative |
| No tumor cells | No tumor cells | Negative |
| Dataset | Summary | Training | Validation | Testing | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Neg | Pos | N | Neg | Pos | N | Neg | Pos | N | Neg | Pos | |
| CAMELYON16 | 399 | 239 | 160 | 216 | 127 | 89 | 54 | 32 | 22 | 129 | 80 | 49 (1) |
| CAMELYON17 | 344 | 310 | 34 | 55 | 40 | 15 | 19 | 10 | 9 | 270 | 260 | 10 |
| LocalSentinel | 161 | 107 | 54 | 88 | 58 | 30 | 22 | 15 | 7 | 51 | 34 | 17 (2) |
| LocalAxillary | 48 + 9 | 24 + 9 | 24 | 24 + 6 | 11 + 6 | 13 | 7 + 3 | 3 + 3 | 4 | 17 | 10 | 7 (3) |
| Total | 961 | 689 | 272 | 389 | 242 | 147 | 105 | 63 | 42 | 467 | 384 | 83 |
| LocalSentinel | LocalAxillary | CAMELYON16 | CAMELYON17 | |||||
|---|---|---|---|---|---|---|---|---|
| AUC | FROC | AUC | FROC | AUC | FROC | AUC | FROC | |
| Model | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) |
| Base | 0.929 | 0.744 | 0.898 | 0.503 | 0.969 | 0.838 | 0.997 | 0.967 |
| (0.800–0.998) | (0.566–0.912) | (0.700–1.000) | (0.201–0.911) | (0.926–0.998) | (0.757–0.913) | (0.990–1.000) | (0.886–1.000) | |
| Base | 0.992 | 0.844 | 1.000 | 0.594 | 0.976 | 0.820 | - | - |
| (only NST) | (0.966–1.000) | (0.688–1.000) | (1.000–1.000) | (0.212–1.000) | (0.936–0.999) | (0.732–0.902) | - | - |
| Local | 0.972 | 0.774 | 1.000 | 0.758 | 0.981 | 0.825 | 0.976 | 0.910 |
| (0.910–1.000) | (0.592–0.940) | (1.000–1.000) | (0.632–1.000) | (0.953–1.000) | (0.746–0.900) | (0.926–1.000) | (0.722–1.000) | |
| Local | 0.997 | 0.870 | 1.000 | 0.744 | 0.988 | 0.817 | - | - |
| (only NST) | (0.985–1.000) | (0.706–1.000) | (1.000–1.000) | (0.619–1.000) | (0.965–1.000) | (0.730–0.896) | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarkman, S.; Karlberg, M.; Pocevičiūtė, M.; Bodén, A.; Bándi, P.; Litjens, G.; Lundström, C.; Treanor, D.; van der Laak, J. Generalization of Deep Learning in Digital Pathology: Experience in Breast Cancer Metastasis Detection. Cancers 2022, 14, 5424. https://doi.org/10.3390/cancers14215424
Jarkman S, Karlberg M, Pocevičiūtė M, Bodén A, Bándi P, Litjens G, Lundström C, Treanor D, van der Laak J. Generalization of Deep Learning in Digital Pathology: Experience in Breast Cancer Metastasis Detection. Cancers. 2022; 14(21):5424. https://doi.org/10.3390/cancers14215424
Chicago/Turabian StyleJarkman, Sofia, Micael Karlberg, Milda Pocevičiūtė, Anna Bodén, Péter Bándi, Geert Litjens, Claes Lundström, Darren Treanor, and Jeroen van der Laak. 2022. "Generalization of Deep Learning in Digital Pathology: Experience in Breast Cancer Metastasis Detection" Cancers 14, no. 21: 5424. https://doi.org/10.3390/cancers14215424
APA StyleJarkman, S., Karlberg, M., Pocevičiūtė, M., Bodén, A., Bándi, P., Litjens, G., Lundström, C., Treanor, D., & van der Laak, J. (2022). Generalization of Deep Learning in Digital Pathology: Experience in Breast Cancer Metastasis Detection. Cancers, 14(21), 5424. https://doi.org/10.3390/cancers14215424






