Liquid Biopsy in Cancer: Focus on Lymphoproliferative Disorders
Abstract
Simple Summary
Abstract
1. Introduction
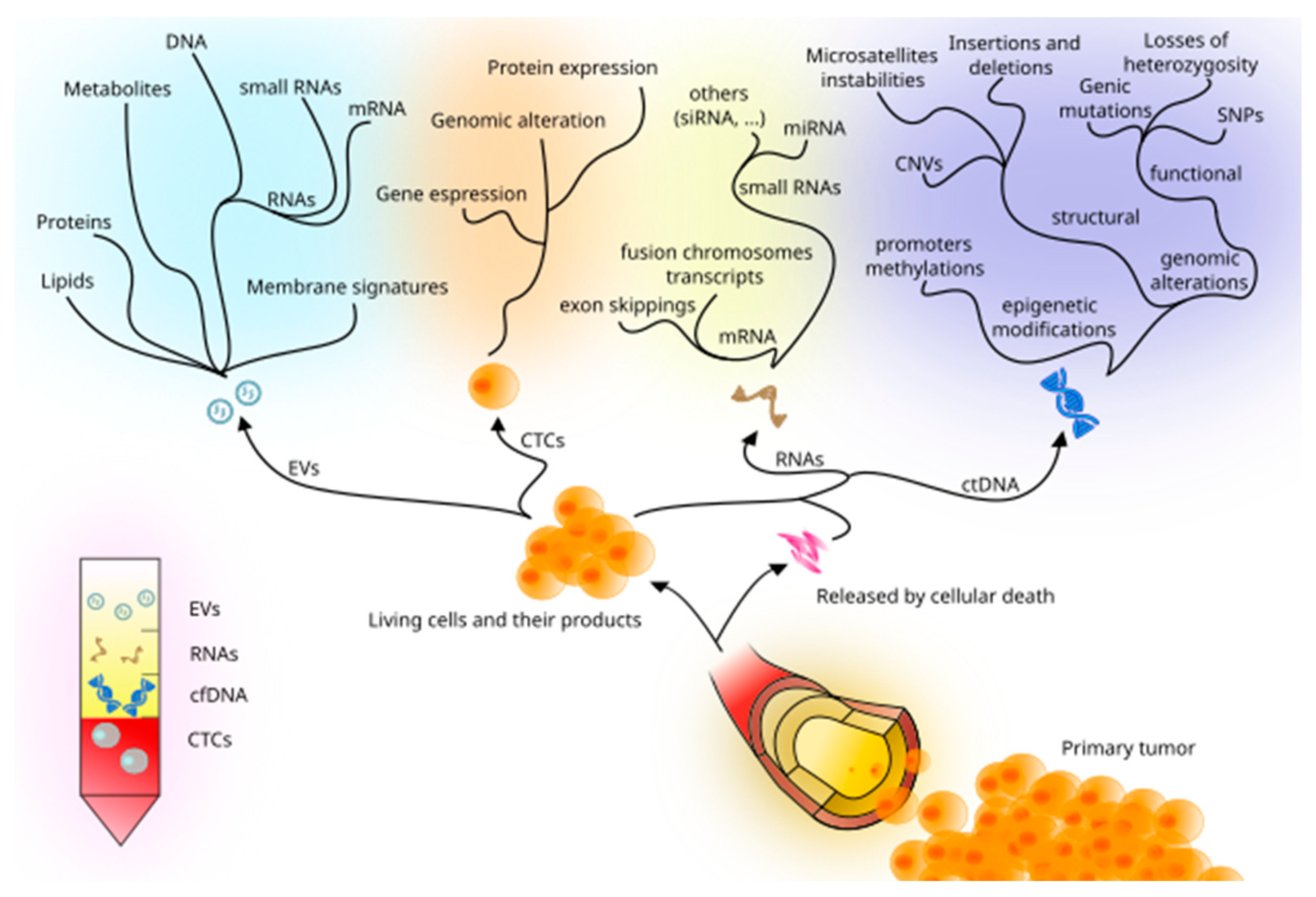
2. Liquid Biopsy: New Techniques and Biomarkers
2.1. cfDNA, ctDNA, and Circulating RNA
2.2. CTCs
2.3. Methylation Markers
2.4. Extracellular Vesicles
3. Liquid Biopsy in Lymphoproliferative Diseases
3.1. Diagnosis and Prognosis
3.2. Follow-Up: MRD and Relapse Settings
3.3. Drug Sensitivity and Resistance
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [PubMed]
- Togneri, F.S.; Ward, D.G.; Foster, J.M.; Devall, A.J.; Wojtowicz, P.; Alyas, S.; Vasques, F.R.; Oumie, A.; James, N.D.; Cheng, K.K.; et al. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur. J. Hum. Genet. 2016, 24, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Kaczor-Urbanowicz, K.E.; Wei, F.; Rao, S.L.; Kim, J.; Shin, H.; Cheng, J.; Tu, M.; Wong, D.T.W.; Kim, Y. Clinical validity of saliva and novel technology for cancer detection. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Imperiale, T.F.; Ransohoff, D.F.; Itzkowitz, S.H. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014, 371, 187–188. [Google Scholar] [CrossRef] [PubMed]
- De Mattos-Arruda, L.; Mayor, R.; Ng, C.K.Y.; Weigelt, B.; Martinez-Ricarte, F.; Torrejon, D.; Oliveira, M.; Arias, A.; Raventos, C.; Tang, J.; et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 2015, 6, 8839. [Google Scholar] [CrossRef]
- Malapelle, U.; Sirera, R.; Jantus-Lewintre, E.; Reclusa, P.; Calabuig-Farinas, S.; Blasco, A.; Pisapia, P.; Rolfo, C.; Camps, C. Profile of the Roche cobas(R) EGFR mutation test v2 for non-small cell lung cancer. Expert. Rev. Mol. Diagn. 2017, 17, 209–215. [Google Scholar] [CrossRef]
- FDA. FDA Approves First Liquid Biopsy Next-Generation Sequencing Companion Diagnostic Test; FDA: Silver Spring, MD, USA, 2020.
- FDA. FDA Approves Liquid Biopsy NGS Companion Diagnostic Test for Multiple Cancers and Biomarkers; FDA: Silver Spring, MD, USA, 2020.
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180. [Google Scholar] [CrossRef]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef]
- Markou, A.; Tzanikou, E.; Lianidou, E. The potential of liquid biopsy in the management of cancer patients. Semin. Cancer Biol. 2022, 84, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Borghesi, M.; Ahmed, H.; Nam, R.; Schaeffer, E.; Schiavina, R.; Taneja, S.; Weidner, W.; Loeb, S. Complications After Systematic, Random, and Image-guided Prostate Biopsy. Eur. Urol. 2017, 71, 353–365. [Google Scholar] [CrossRef]
- Lu, M.; Shi, B.; Wang, J.; Cao, Q.; Cui, Q. TAM: A method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinform. 2010, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Lucci, A.; Ignatiadis, M.; Jeffrey, S.S. Cell-free circulating tumor DNA profiling in cancer management. Trends Mol. Med. 2021, 27, 1014–1015. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Pascher, A.; Juratli, M.A.; Sporn, J.C. The Potential of Aptamer-Mediated Liquid Biopsy for Early Detection of Cancer. Int. J. Mol. Sci. 2021, 22, 5601. [Google Scholar] [CrossRef]
- Sacher, A.G.; Paweletz, C.; Dahlberg, S.E.; Alden, R.S.; O’Connell, A.; Feeney, N.; Mach, S.L.; Janne, P.A.; Oxnard, G.R. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016, 2, 1014–1022. [Google Scholar] [CrossRef]
- Valihrach, L.; Androvic, P.; Kubista, M. Circulating miRNA analysis for cancer diagnostics and therapy. Mol. Aspects Med. 2020, 72, 100825. [Google Scholar] [CrossRef] [PubMed]
- Tural-Kara, T.; Ozdemir, H.; Fitoz, S.; Ciftci, E.; Yalcinkaya, F. Bone marrow aspiration complications: Iliopsoas abscess and sacroiliac osteomyelitis. Turk J. Pediatr. 2016, 58, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Abhishek, K.; Khunger, N. Complications of skin biopsy. J. Cutan. Aesthet. Surg. 2015, 8, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Osumi, H.; Shinozaki, E.; Yamaguchi, K.; Zembutsu, H. Clinical utility of circulating tumor DNA for colorectal cancer. Cancer Sci. 2019, 110, 1148–1155. [Google Scholar] [CrossRef]
- Maisano, D.; Mimmi, S.; Dattilo, V.; Marino, F.; Gentile, M.; Vecchio, E.; Fiume, G.; Nistico, N.; Aloisio, A.; de Santo, M.P.; et al. A novel phage display based platform for exosome diversity characterization. Nanoscale 2022, 14, 2998–3003. [Google Scholar] [CrossRef] [PubMed]
- Plattner, B.W.; Chen, P.; Cross, R.; Leavitt, M.A.; Killen, P.D.; Heung, M. Complications and adequacy of transplant kidney biopsies: A comparison of techniques. J. Vasc. Access 2018, 19, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, H.; Chau, A.; Jeng, Z.; Gardner, G.; Yoo, R.; Zhang, W.; Hernandez, J. Percutaneous Lung Biopsy in Immunocompromised Pediatric Patients. J. Vasc. Interv. Radiol. 2020, 31, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.B.; Siu, L.L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef]
- Rolfo, C.; Cardona, A.F.; Cristofanilli, M.; Paz-Ares, L.; Diaz Mochon, J.J.; Duran, I.; Raez, L.E.; Russo, A.; Lorente, J.A.; Malapelle, U.; et al. Challenges and opportunities of cfDNA analysis implementation in clinical practice: Perspective of the International Society of Liquid Biopsy (ISLB). Crit. Rev. Oncol. Hematol. 2020, 151, 102978. [Google Scholar] [CrossRef]
- Reckamp, K.L.; Melnikova, V.O.; Karlovich, C.; Sequist, L.V.; Camidge, D.R.; Wakelee, H.; Perol, M.; Oxnard, G.R.; Kosco, K.; Croucher, P.; et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J. Thorac. Oncol. 2016, 11, 1690–1700. [Google Scholar] [CrossRef]
- Villatoro, S.; Mayo-de-Las-Casas, C.; Jordana-Ariza, N.; Viteri-Ramirez, S.; Garzon-Ibanez, M.; Moya-Horno, I.; Garcia-Pelaez, B.; Gonzalez-Cao, M.; Malapelle, U.; Balada-Bel, A.; et al. Prospective detection of mutations in cerebrospinal fluid, pleural effusion, and ascites of advanced cancer patients to guide treatment decisions. Mol. Oncol. 2019, 13, 2633–2645. [Google Scholar] [CrossRef]
- Zheng, M.M.; Li, Y.S.; Tu, H.Y.; Jiang, B.Y.; Yang, J.J.; Zhou, Q.; Xu, C.R.; Yang, X.R.; Wu, Y.L. Genotyping of Cerebrospinal Fluid Associated With Osimertinib Response and Resistance for Leptomeningeal Metastases in EGFR-Mutated NSCLC. J. Thorac. Oncol. 2021, 16, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Jiang, P.; Chan, K.C.A.; Lo, Y.M.D. Liver-derived cell-free nucleic acids in plasma: Biology and applications in liquid biopsies. J. Hepatol. 2019, 71, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Bauml, J.; Levy, B. Clonal Hematopoiesis: A New Layer in the Liquid Biopsy Story in Lung Cancer. Clin. Cancer Res. 2018, 24, 4352–4354. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ren, J.; Luo, N.; Guo, H.; Zheng, Y.; Li, J.; Tang, F.; Wen, L.; Peng, J. Comprehensive DNA methylation analysis of tissue of origin of plasma cell-free DNA by methylated CpG tandem amplification and sequencing (MCTA-Seq). Clin. Epigenetics 2019, 11, 93. [Google Scholar] [CrossRef]
- Rodrigues Filho, E.M.; Simon, D.; Ikuta, N.; Klovan, C.; Dannebrock, F.A.; Oliveira de Oliveira, C.; Regner, A. Elevated cell-free plasma DNA level as an independent predictor of mortality in patients with severe traumatic brain injury. J. Neurotrauma 2014, 31, 1639–1646. [Google Scholar] [CrossRef]
- O’Connell, G.C.; Petrone, A.B.; Tennant, C.S.; Lucke-Wold, N.; Kabbani, Y.; Tarabishy, A.R.; Chantler, P.D.; Barr, T.L. Circulating extracellular DNA levels are acutely elevated in ischaemic stroke and associated with innate immune system activation. Brain Inj. 2017, 31, 1369–1375. [Google Scholar] [CrossRef]
- Tug, S.; Helmig, S.; Deichmann, E.R.; Schmeier-Jurchott, A.; Wagner, E.; Zimmermann, T.; Radsak, M.; Giacca, M.; Simon, P. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc. Immunol. Rev. 2015, 21, 164–173. [Google Scholar] [PubMed]
- De Vlaminck, I.; Martin, L.; Kertesz, M.; Patel, K.; Kowarsky, M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl. Acad. Sci. USA 2015, 112, 13336–13341. [Google Scholar] [CrossRef] [PubMed]
- Phuong, N.T.N.; Manh, D.H.; Dumre, S.P.; Mizukami, S.; Weiss, L.N.; Van Thuong, N.; Ha, T.T.N.; Phuc, L.H.; Van An, T.; Tieu, T.M.; et al. Plasma cell-free DNA: A potential biomarker for early prediction of severe dengue. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 10. [Google Scholar] [CrossRef]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaat4921. [Google Scholar] [CrossRef] [PubMed]
- Underhill, H.R.; Kitzman, J.O.; Hellwig, S.; Welker, N.C.; Daza, R.; Baker, D.N.; Gligorich, K.M.; Rostomily, R.C.; Bronner, M.P.; Shendure, J. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016, 12, e1006162. [Google Scholar] [CrossRef] [PubMed]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Kloten, V.; Ruchel, N.; Bruchle, N.O.; Gasthaus, J.; Freudenmacher, N.; Steib, F.; Mijnes, J.; Eschenbruch, J.; Binnebosel, M.; Knuchel, R.; et al. Liquid biopsy in colon cancer: Comparison of different circulating DNA extraction systems following absolute quantification of KRAS mutations using Intplex allele-specific PCR. Oncotarget 2017, 8, 86253–86263. [Google Scholar] [CrossRef]
- Perez-Barrios, C.; Nieto-Alcolado, I.; Torrente, M.; Jimenez-Sanchez, C.; Calvo, V.; Gutierrez-Sanz, L.; Palka, M.; Donoso-Navarro, E.; Provencio, M.; Romero, A. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: Impact on biomarker testing. Transl. Lung Cancer Res. 2016, 5, 665–672. [Google Scholar] [CrossRef]
- Sorber, L.; Zwaenepoel, K.; Deschoolmeester, V.; Roeyen, G.; Lardon, F.; Rolfo, C.; Pauwels, P. A Comparison of Cell-Free DNA Isolation Kits: Isolation and Quantification of Cell-Free DNA in Plasma. J. Mol. Diagn. 2017, 19, 162–168. [Google Scholar] [CrossRef]
- Warton, K.; Graham, L.J.; Yuwono, N.; Samimi, G. Comparison of 4 commercial kits for the extraction of circulating DNA from plasma. Cancer Genet. 2018, 228–229, 143–150. [Google Scholar] [CrossRef]
- Diefenbach, R.J.; Lee, J.H.; Kefford, R.F.; Rizos, H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 2018, 228–229, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Saelee, S.L.; Lovejoy, A.F.; Hinzmann, B.; Mayol, K.; Huynh, S.; Harrell, A.; Lefkowitz, J.; Deodhar, N.; Garcia-Montoya, G.; Yaung, S.J.; et al. Quantitative PCR-Based Method to Assess Cell-Free DNA Quality, Adjust Input Mass, and Improve Next-Generation Sequencing Assay Performance. J. Mol. Diagn. 2022, 24, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Deveson, I.W.; Mercer, T.; Johann, D.J., Jr.; Jones, W.; Tong, W.; Xu, J. Ultra-deep sequencing data from a liquid biopsy proficiency study demonstrating analytic validity. Sci. Data 2022, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Mellert, H.; Reese, J.; Jackson, L.; Maxwell, V.; Tschida, C.; Pestano, G.A. Targeted Next-Generation Sequencing of Liquid Biopsy Samples from Patients with NSCLC. Diagnostics 2021, 11, 155. [Google Scholar] [CrossRef]
- Garcia-Pardo, M.; Makarem, M.; Li, J.J.N.; Kelly, D.; Leighl, N.B. Integrating circulating-free DNA (cfDNA) analysis into clinical practice: Opportunities and challenges. Br. J. Cancer 2022, 127, 592–602. [Google Scholar] [CrossRef]
- Raman, L.; Van der Linden, M.; Van der Eecken, K.; Vermaelen, K.; Demedts, I.; Surmont, V.; Himpe, U.; Dedeurwaerdere, F.; Ferdinande, L.; Lievens, Y.; et al. Shallow whole-genome sequencing of plasma cell-free DNA accurately differentiates small from non-small cell lung carcinoma. Genome Med. 2020, 12, 35. [Google Scholar] [CrossRef]
- Wang, Y.; Lee, C.G. MicroRNA and cancer--focus on apoptosis. J. Cell. Mol. Med. 2009, 13, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. miR2 Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, M.X.; Yan, G.Y. RWRMDA: Predicting novel human microRNA-disease associations. Mol. Biosyst. 2012, 8, 2792–2798. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014, 42, D1070–D1074. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, J.; Cui, Q. Could circulating miRNAs contribute to cancer therapy? Trends Mol. Med. 2013, 19, 71–73. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Bauer, A.; Elsharawy, A.; Haas, J.; Backes, C.; Wendschlag, A.; Giese, N.; Tjaden, C.; Ott, K.; et al. Toward the blood-borne miRNome of human diseases. Nat. Methods 2011, 8, 841–843. [Google Scholar] [CrossRef]
- Mo, M.H.; Chen, L.; Fu, Y.; Wang, W.; Fu, S.W. Cell-free Circulating miRNA Biomarkers in Cancer. J. Cancer 2012, 3, 432–448. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Bidard, F.C.; Peeters, D.J.; Fehm, T.; Nole, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Ni, X.; Zhuo, M.; Su, Z.; Duan, J.; Gao, Y.; Wang, Z.; Zong, C.; Bai, H.; Chapman, A.R.; Zhao, J.; et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc. Natl. Acad. Sci. USA 2013, 110, 21083–21088. [Google Scholar] [CrossRef]
- Lohr, J.G.; Adalsteinsson, V.A.; Cibulskis, K.; Choudhury, A.D.; Rosenberg, M.; Cruz-Gordillo, P.; Francis, J.M.; Zhang, C.Z.; Shalek, A.K.; Satija, R.; et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 2014, 32, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Kalinsky, K.; Mayer, J.A.; Xu, X.; Pham, T.; Wong, K.L.; Villarin, E.; Pircher, T.J.; Brown, M.; Maurer, M.A.; Bischoff, F.Z. Correlation of hormone receptor status between circulating tumor cells, primary tumor, and metastasis in breast cancer patients. Clin. Transl. Oncol. 2015, 17, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Ting, D.T.; Stott, S.L.; Wittner, B.S.; Ozsolak, F.; Paul, S.; Ciciliano, J.C.; Smas, M.E.; Winokur, D.; Gilman, A.J.; et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature 2012, 487, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Wallwiener, M.; Hartkopf, A.D.; Riethdorf, S.; Nees, J.; Sprick, M.R.; Schonfisch, B.; Taran, F.A.; Heil, J.; Sohn, C.; Pantel, K.; et al. The impact of HER2 phenotype of circulating tumor cells in metastatic breast cancer: A retrospective study in 107 patients. BMC Cancer 2015, 15, 403. [Google Scholar] [CrossRef] [PubMed]
- Kalinich, M.; Bhan, I.; Kwan, T.T.; Miyamoto, D.T.; Javaid, S.; LiCausi, J.A.; Milner, J.D.; Hong, X.; Goyal, L.; Sil, S.; et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, 1123–1128. [Google Scholar] [CrossRef]
- Keller, L.; Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 2019, 19, 553–567. [Google Scholar] [CrossRef]
- Bardelli, A.; Pantel, K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef]
- Lianidou, E.S. Gene expression profiling and DNA methylation analyses of CTCs. Mol. Oncol. 2016, 10, 431–442. [Google Scholar] [CrossRef]
- Boral, D.; Vishnoi, M.; Liu, H.N.; Yin, W.; Sprouse, M.L.; Scamardo, A.; Hong, D.S.; Tan, T.Z.; Thiery, J.P.; Chang, J.C.; et al. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 2017, 8, 196. [Google Scholar] [CrossRef]
- Jakabova, A.; Bielcikova, Z.; Pospisilova, E.; Matkowski, R.; Szynglarewicz, B.; Staszek-Szewczyk, U.; Zemanova, M.; Petruzelka, L.; Eliasova, P.; Kolostova, K.; et al. Molecular characterization and heterogeneity of circulating tumor cells in breast cancer. Breast Cancer Res. Treat. 2017, 166, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Markou, A.; Strati, A.; Malamos, N.; Georgoulias, V.; Lianidou, E.S. Molecular characterization of circulating tumor cells in breast cancer by a liquid bead array hybridization assay. Clin. Chem. 2011, 57, 421–430. [Google Scholar] [CrossRef]
- Strati, A.; Markou, A.; Parisi, C.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Lianidou, E. Gene expression profile of circulating tumor cells in breast cancer by RT-qPCR. BMC Cancer 2011, 11, 422. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Desitter, I.; Guerrouahen, B.S.; Benali-Furet, N.; Wechsler, J.; Janne, P.A.; Kuang, Y.; Yanagita, M.; Wang, L.; Berkowitz, J.A.; Distel, R.J.; et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011, 31, 427–441. [Google Scholar]
- Liu, Y.; Li, R.; Zhang, L.; Guo, S. Nanomaterial-Based Immunocapture Platforms for the Recognition, Isolation, and Detection of Circulating Tumor Cells. Front. Bioeng. Biotechnol. 2022, 10, 850241. [Google Scholar] [CrossRef]
- Riethdorf, S.; O’Flaherty, L.; Hille, C.; Pantel, K. Clinical applications of the CellSearch platform in cancer patients. Adv. Drug Deliv. Rev. 2018, 125, 102–121. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Pixberg, C.F.; Schulz, W.A.; Stoecklein, N.H.; Neves, R.P. Characterization of DNA Methylation in Circulating Tumor Cells. Genes 2015, 6, 1053–1075. [Google Scholar] [CrossRef]
- Chimonidou, M.; Strati, A.; Tzitzira, A.; Sotiropoulou, G.; Malamos, N.; Georgoulias, V.; Lianidou, E.S. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin. Chem. 2011, 57, 1169–1177. [Google Scholar] [CrossRef]
- Sigin, V.O.; Kalinkin, A.I.; Kuznetsova, E.B.; Simonova, O.A.; Chesnokova, G.G.; Litviakov, N.V.; Slonimskaya, E.M.; Tsyganov, M.M.; Ibragimova, M.K.; Volodin, I.V.; et al. DNA methylation markers panel can improve prediction of response to neoadjuvant chemotherapy in luminal B breast cancer. Sci. Rep. 2020, 10, 9239. [Google Scholar] [CrossRef]
- Kanwal, R.; Gupta, K.; Gupta, S. Cancer epigenetics: An introduction. Methods Mol. Biol. 2015, 1238, 3–25. [Google Scholar] [CrossRef]
- Liu, L.; Toung, J.M.; Jassowicz, A.F.; Vijayaraghavan, R.; Kang, H.; Zhang, R.; Kruglyak, K.M.; Huang, H.J.; Hinoue, T.; Shen, H.; et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann. Oncol. 2018, 29, 1445–1453. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; Consortium, C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Chimonidou, M.; Tzitzira, A.; Strati, A.; Sotiropoulou, G.; Sfikas, C.; Malamos, N.; Georgoulias, V.; Lianidou, E. CST6 promoter methylation in circulating cell-free DNA of breast cancer patients. Clin. Biochem. 2013, 46, 235–240. [Google Scholar] [CrossRef]
- Chimonidou, M.; Strati, A.; Malamos, N.; Georgoulias, V.; Lianidou, E.S. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin. Chem. 2013, 59, 270–279. [Google Scholar] [CrossRef]
- Duffy, M.J.; Napieralski, R.; Martens, J.W.; Span, P.N.; Spyratos, F.; Sweep, F.C.; Brunner, N.; Foekens, J.A.; Schmitt, M.; Group, E.P. Methylated genes as new cancer biomarkers. Eur. J. Cancer 2009, 45, 335–346. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Muralidharan-Chari, V.; Clancy, J.W.; Sedgwick, A.; D’Souza-Schorey, C. Microvesicles: Mediators of extracellular communication during cancer progression. J. Cell Sci. 2010, 123, 1603–1611. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Maiso, P.; Azab, A.K.; Tai, Y.T.; Reagan, M.; Azab, F.; Flores, L.M.; Campigotto, F.; Weller, E.; et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J. Clin. Investig. 2013, 123, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- Longjohn, M.N.; Hudson, J.B.J.; Smith, N.C.; Rise, M.L.; Moorehead, P.C.; Christian, S.L. Deciphering the messages carried by extracellular vesicles in hematological malignancies. Blood Rev. 2021, 46, 100734. [Google Scholar] [CrossRef] [PubMed]
- Jurj, A.; Zanoaga, O.; Braicu, C.; Lazar, V.; Tomuleasa, C.; Irimie, A.; Berindan-Neagoe, I. A Comprehensive Picture of Extracellular Vesicles and Their Contents. Molecular Transfer to Cancer Cells. Cancers 2020, 12, 298. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Ghanam, J.; Chetty, V.K.; Barthel, L.; Reinhardt, D.; Hoyer, P.F.; Thakur, B.K. DNA in extracellular vesicles: From evolution to its current application in health and disease. Cell Biosci. 2022, 12, 37. [Google Scholar] [CrossRef]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis. Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef]
- Yeh, Y.Y.; Ozer, H.G.; Lehman, A.M.; Maddocks, K.; Yu, L.; Johnson, A.J.; Byrd, J.C. Characterization of CLL exosomes reveals a distinct microRNA signature and enhanced secretion by activation of BCR signaling. Blood 2015, 125, 3297–3305. [Google Scholar] [CrossRef]
- Berardocco, M.; Radeghieri, A.; Busatto, S.; Gallorini, M.; Raggi, C.; Gissi, C.; D’Agnano, I.; Bergese, P.; Felsani, A.; Berardi, A.C. RNA-seq reveals distinctive RNA profiles of small extracellular vesicles from different human liver cancer cell lines. Oncotarget 2017, 8, 82920–82939. [Google Scholar] [CrossRef]
- Lunavat, T.R.; Cheng, L.; Kim, D.K.; Bhadury, J.; Jang, S.C.; Lasser, C.; Sharples, R.A.; Lopez, M.D.; Nilsson, J.; Gho, Y.S.; et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells--Evidence of unique microRNA cargos. RNA Biol. 2015, 12, 810–823. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017, 18, 1153. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- Paolino, G.; Huber, V.; Camerini, S.; Casella, M.; Macone, A.; Bertuccini, L.; Iosi, F.; Moliterni, E.; Cecchetti, S.; Ruspantini, I.; et al. The Fatty Acid and Protein Profiles of Circulating CD81-Positive Small Extracellular Vesicles Are Associated with Disease Stage in Melanoma Patients. Cancers 2021, 13, 4157. [Google Scholar] [CrossRef]
- Schey, K.L.; Luther, J.M.; Rose, K.L. Proteomics characterization of exosome cargo. Methods 2015, 87, 75–82. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5 th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Vasioukhin, V.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Stroun, M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br. J. Haematol. 1994, 86, 774–779. [Google Scholar] [CrossRef]
- Rogers, A.; Joe, Y.; Manshouri, T.; Dey, A.; Jilani, I.; Giles, F.; Estey, E.; Freireich, E.; Keating, M.; Kantarjian, H.; et al. Relative increase in leukemia-specific DNA in peripheral blood plasma from patients with acute myeloid leukemia and myelodysplasia. Blood 2004, 103, 2799–2801. [Google Scholar] [CrossRef]
- Zajec, M.; Langerhorst, P.; VanDuijn, M.M.; Gloerich, J.; Russcher, H.; van Gool, A.J.; Luider, T.M.; Joosten, I.; de Rijke, Y.B.; Jacobs, J.F.M. Mass Spectrometry for Identification, Monitoring, and Minimal Residual Disease Detection of M-Proteins. Clin. Chem. 2020, 66, 421–433. [Google Scholar] [CrossRef]
- Greystoke, A.; O’Connor, J.P.; Linton, K.; Taylor, M.B.; Cummings, J.; Ward, T.; Maders, F.; Hughes, A.; Ranson, M.; Illidge, T.M.; et al. Assessment of circulating biomarkers for potential pharmacodynamic utility in patients with lymphoma. Br. J. Cancer 2011, 104, 719–725. [Google Scholar] [CrossRef]
- Cheng, S.H.; Lau, K.M.; Li, C.K.; Chan, N.P.; Ip, R.K.; Cheng, C.K.; Lee, V.; Shing, M.M.; Leung, A.W.; Ha, S.Y.; et al. Minimal residual disease-based risk stratification in Chinese childhood acute lymphoblastic leukemia by flow cytometry and plasma DNA quantitative polymerase chain reaction. PLoS ONE 2013, 8, e69467. [Google Scholar] [CrossRef] [PubMed]
- Kanakry, J.A.; Li, H.; Gellert, L.L.; Lemas, M.V.; Hsieh, W.S.; Hong, F.; Tan, K.L.; Gascoyne, R.D.; Gordon, L.I.; Fisher, R.I.; et al. Plasma Epstein-Barr virus DNA predicts outcome in advanced Hodgkin lymphoma: Correlative analysis from a large North American cooperative group trial. Blood 2013, 121, 3547–3553. [Google Scholar] [CrossRef] [PubMed]
- Hohaus, S.; Giachelia, M.; Massini, G.; Mansueto, G.; Vannata, B.; Bozzoli, V.; Criscuolo, M.; D’Alo, F.; Martini, M.; Larocca, L.M.; et al. Cell-free circulating DNA in Hodgkin’s and non-Hodgkin’s lymphomas. Ann. Oncol. 2009, 20, 1408–1413. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, D.M.; Esfahani, M.S.; Scherer, F.; Soo, J.; Jin, M.C.; Liu, C.L.; Newman, A.M.; Duhrsen, U.; Huttmann, A.; Casasnovas, O.; et al. Dynamic Risk Profiling Using Serial Tumor Biomarkers for Personalized Outcome Prediction. Cell 2019, 178, 699–713.e19. [Google Scholar] [CrossRef]
- Kurtz, D.M.; Green, M.R.; Bratman, S.V.; Scherer, F.; Liu, C.L.; Kunder, C.A.; Takahashi, K.; Glover, C.; Keane, C.; Kihira, S.; et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 2015, 125, 3679–3687. [Google Scholar] [CrossRef]
- Roschewski, M.; Dunleavy, K.; Pittaluga, S.; Moorhead, M.; Pepin, F.; Kong, K.; Shovlin, M.; Jaffe, E.S.; Staudt, L.M.; Lai, C.; et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: A correlative biomarker study. Lancet Oncol. 2015, 16, 541–549. [Google Scholar] [CrossRef]
- Biancon, G.; Gimondi, S.; Vendramin, A.; Carniti, C.; Corradini, P. Noninvasive Molecular Monitoring in Multiple Myeloma Patients Using Cell-Free Tumor DNA: A Pilot Study. J. Mol. Diagn. 2018, 20, 859–870. [Google Scholar] [CrossRef]
- Vrabel, D.; Sedlarikova, L.; Besse, L.; Rihova, L.; Bezdekova, R.; Almasi, M.; Kubaczkova, V.; Brozova, L.; Jarkovsky, J.; Plonkova, H.; et al. Dynamics of tumor-specific cfDNA in response to therapy in multiple myeloma patients. Eur. J. Haematol. 2020, 104, 190–197. [Google Scholar] [CrossRef]
- Ferrajoli, A.; Shanafelt, T.D.; Ivan, C.; Shimizu, M.; Rabe, K.G.; Nouraee, N.; Ikuo, M.; Ghosh, A.K.; Lerner, S.; Rassenti, L.Z.; et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 2013, 122, 1891–1899. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Chen, C.; Runnels, J.; Leleu, X.; Azab, F.; Azab, A.K.; Jia, X.; Ngo, H.T.; Melhem, M.R.; et al. microRNA expression in the biology, prognosis, and therapy of Waldenstrom macroglobulinemia. Blood 2009, 113, 4391–4402. [Google Scholar] [CrossRef]
- Cui, B.; Chen, L.; Zhang, S.; Mraz, M.; Fecteau, J.F.; Yu, J.; Ghia, E.M.; Zhang, L.; Bao, L.; Rassenti, L.Z.; et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood 2014, 124, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Belov, L.; Matic, K.J.; Hallal, S.; Best, O.G.; Mulligan, S.P.; Christopherson, R.I. Extensive surface protein profiles of extracellular vesicles from cancer cells may provide diagnostic signatures from blood samples. J. Extracell. Vesicles 2016, 5, 25355. [Google Scholar] [CrossRef] [PubMed]
- De Luca, L.; D’Arena, G.; Simeon, V.; Trino, S.; Laurenzana, I.; Caivano, A.; La Rocca, F.; Villani, O.; Mansueto, G.; Deaglio, S.; et al. Characterization and prognostic relevance of circulating microvesicles in chronic lymphocytic leukemia. Leuk. Lymphoma 2017, 58, 1424–1432. [Google Scholar] [CrossRef] [PubMed]
- Szczepanski, M.J.; Szajnik, M.; Welsh, A.; Whiteside, T.L.; Boyiadzis, M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica 2011, 96, 1302–1309. [Google Scholar] [CrossRef]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidus, J.; Chang, B.H.; Kurre, P. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci. Rep. 2015, 5, 11295. [Google Scholar] [CrossRef]
- Manier, S.; Liu, C.J.; Avet-Loiseau, H.; Park, J.; Shi, J.; Campigotto, F.; Salem, K.Z.; Huynh, D.; Glavey, S.V.; Rivotto, B.; et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017, 129, 2429–2436. [Google Scholar] [CrossRef] [PubMed]
- van Eijndhoven, M.A.; Zijlstra, J.M.; Groenewegen, N.J.; Drees, E.E.; van Niele, S.; Baglio, S.R.; Koppers-Lalic, D.; van der Voorn, H.; Libregts, S.F.; Wauben, M.H.; et al. Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients. JCI Insight 2016, 1, e89631. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Thompson, B.; Leleu, X.; Azab, A.K.; Azab, F.; Runnels, J.; Jia, X.; Ngo, H.T.; Melhem, M.R.; et al. MicroRNAs 15 a and 16 regulate tumor proliferation in multiple myeloma. Blood 2009, 113, 6669–6680. [Google Scholar] [CrossRef]
- Trino, S.; Lamorte, D.; Caivano, A.; De Luca, L.; Sgambato, A.; Laurenzana, I. Clinical relevance of extracellular vesicles in hematological neoplasms: From liquid biopsy to cell biopsy. Leukemia 2021, 35, 661–678. [Google Scholar] [CrossRef]
- Caivano, A.; Laurenzana, I.; De Luca, L.; La Rocca, F.; Simeon, V.; Trino, S.; D’Auria, F.; Traficante, A.; Maietti, M.; Izzo, T.; et al. High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumour. Biol. 2015, 36, 9739–9752. [Google Scholar] [CrossRef] [PubMed]
- Di Noto, G.; Bugatti, A.; Zendrini, A.; Mazzoldi, E.L.; Montanelli, A.; Caimi, L.; Rusnati, M.; Ricotta, D.; Bergese, P. Merging colloidal nanoplasmonics and surface plasmon resonance spectroscopy for enhanced profiling of multiple myeloma-derived exosomes. Biosens. Bioelectron. 2016, 77, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.R.; Luk, F.; Brown, R.D.; Suen, H.; Kwan, Y.; Bebawy, M. Isolation of Human CD138(+) Microparticles from the Plasma of Patients with Multiple Myeloma. Neoplasia 2016, 18, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Morandi, F.; Marimpietri, D.; Horenstein, A.L.; Bolzoni, M.; Toscani, D.; Costa, F.; Castella, B.; Faini, A.C.; Massaia, M.; Pistoia, V.; et al. Microvesicles released from multiple myeloma cells are equipped with ectoenzymes belonging to canonical and non-canonical adenosinergic pathways and produce adenosine from ATP and NAD. Oncoimmunology 2018, 7, e1458809. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Tian, M.Y.; Deng, L.; Wang, Y.S.; Xing, R.; Liu, H.; Fu, R. The potential diagnostic power of CD138+ microparticles from the plasma analysis for multiple myeloma clinical monitoring. Hematol. Oncol. 2019, 37, 401–408. [Google Scholar] [CrossRef]
- Zhang, L.; Lei, Q.; Wang, H.; Xu, C.; Liu, T.; Kong, F.; Yang, C.; Yan, G.; Sun, L.; Zhao, A.; et al. Tumor-derived extracellular vesicles inhibit osteogenesis and exacerbate myeloma bone disease. Theranostics 2019, 9, 196–209. [Google Scholar] [CrossRef]
- Rajeev Krishnan, S.; De Rubis, G.; Suen, H.; Joshua, D.; Lam Kwan, Y.; Bebawy, M. A liquid biopsy to detect multidrug resistance and disease burden in multiple myeloma. Blood Cancer J. 2020, 10, 37. [Google Scholar] [CrossRef]
- Aung, T.; Chapuy, B.; Vogel, D.; Wenzel, D.; Oppermann, M.; Lahmann, M.; Weinhage, T.; Menck, K.; Hupfeld, T.; Koch, R.; et al. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc. Natl. Acad. Sci. USA 2011, 108, 15336–15341. [Google Scholar] [CrossRef]
- Nadali, G.; Tavecchia, L.; Zanolin, E.; Bonfante, V.; Viviani, S.; Camerini, E.; Musto, P.; Di Renzo, N.; Carotenuto, M.; Chilosi, M.; et al. Serum level of the soluble form of the CD30 molecule identifies patients with Hodgkin’s disease at high risk of unfavorable outcome. Blood 1998, 91, 3011–3016. [Google Scholar] [CrossRef]
- Boysen, J.; Nelson, M.; Magzoub, G.; Maiti, G.P.; Sinha, S.; Goswami, M.; Vesely, S.K.; Shanafelt, T.D.; Kay, N.E.; Ghosh, A.K. Dynamics of microvesicle generation in B-cell chronic lymphocytic leukemia: Implication in disease progression. Leukemia 2017, 31, 350–360. [Google Scholar] [CrossRef]
- Caivano, A.; La Rocca, F.; Simeon, V.; Girasole, M.; Dinarelli, S.; Laurenzana, I.; De Stradis, A.; De Luca, L.; Trino, S.; Traficante, A.; et al. MicroRNA-155 in serum-derived extracellular vesicles as a potential biomarker for hematologic malignancies—A short report. Cell. Oncol. 2017, 40, 97–103. [Google Scholar] [CrossRef]
- Bouyssou, J.M.; Liu, C.J.; Bustoros, M.; Sklavenitis-Pistofidis, R.; Aljawai, Y.; Manier, S.; Yosef, A.; Sacco, A.; Kokubun, K.; Tsukamoto, S.; et al. Profiling of circulating exosomal miRNAs in patients with Waldenstrom Macroglobulinemia. PLoS ONE 2018, 13, e0204589. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, S.; Urzi, O.; Conigliaro, A.; Bosco, G.L.; Parisi, S.; Carlisi, M.; Siragusa, S.; Raimondi, L.; Luca, A.; Giavaresi, G.; et al. Extracellular Vesicle microRNAs Contribute to the Osteogenic Inhibition of Mesenchymal Stem Cells in Multiple Myeloma. Cancers 2020, 12, 449. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Li, Y.C.; Geng, C.Y.; Zhou, H.X.; Gao, W.; Chen, W.M. Serum exosomal microRNAs as novel biomarkers for multiple myeloma. Hematol. Oncol. 2019, 37, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, D.M.; Scherer, F.; Jin, M.C.; Soo, J.; Craig, A.F.M.; Esfahani, M.S.; Chabon, J.J.; Stehr, H.; Liu, C.L.; Tibshirani, R.; et al. Circulating Tumor DNA Measurements As Early Outcome Predictors in Diffuse Large B-Cell Lymphoma. J. Clin. Oncol. 2018, 36, 2845–2853. [Google Scholar] [CrossRef] [PubMed]
- Rocci, A.; Hofmeister, C.C.; Geyer, S.; Stiff, A.; Gambella, M.; Cascione, L.; Guan, J.; Benson, D.M.; Efebera, Y.A.; Talabere, T.; et al. Circulating miRNA markers show promise as new prognosticators for multiple myeloma. Leukemia 2014, 28, 1922–1926. [Google Scholar] [CrossRef]
- Provencio, M.; Rodriguez, M.; Cantos, B.; Sabin, P.; Quero, C.; Garcia-Arroyo, F.R.; Rueda, A.; Maximiano, C.; Rodriguez-Abreu, D.; Sanchez, A.; et al. mRNA in exosomas as a liquid biopsy in non-Hodgkin Lymphoma: A multicentric study by the Spanish Lymphoma Oncology Group. Oncotarget 2017, 8, 50949–50957. [Google Scholar] [CrossRef]
- Braoudaki, M.; Lambrou, G.I.; Vougas, K.; Karamolegou, K.; Tsangaris, G.T.; Tzortzatou-Stathopoulou, F. Protein biomarkers distinguish between high- and low-risk pediatric acute lymphoblastic leukemia in a tissue specific manner. J. Hematol. Oncol. 2013, 6, 52. [Google Scholar] [CrossRef]
- Kardava, L.; Yang, Q.; St Leger, A.; Foon, K.A.; Lentzsch, S.; Vallejo, A.N.; Milcarek, C.; Borghesi, L. The B lineage transcription factor E2 A regulates apoptosis in chronic lymphocytic leukemia (CLL) cells. Int. Immunol. 2011, 23, 375–384. [Google Scholar] [CrossRef]
- Scherer, F.; Kurtz, D.M.; Diehn, M.; Alizadeh, A.A. High-throughput sequencing for noninvasive disease detection in hematologic malignancies. Blood 2017, 130, 440–452. [Google Scholar] [CrossRef]
- Gao, Y.J.; He, Y.J.; Yang, Z.L.; Shao, H.Y.; Zuo, Y.; Bai, Y.; Chen, H.; Chen, X.C.; Qin, F.X.; Tan, S.; et al. Increased integrity of circulating cell-free DNA in plasma of patients with acute leukemia. Clin. Chem. Lab. Med. 2010, 48, 1651–1656. [Google Scholar] [CrossRef]
- Nakamura, S.; Yokoyama, K.; Yusa, N.; Ogawa, M.; Takei, T.; Kobayashi, A.; Ito, M.; Shimizu, E.; Kasajima, R.; Wada, Y.; et al. Circulating tumor DNA dynamically predicts response and/or relapse in patients with hematological malignancies. Int. J. Hematol. 2018, 108, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Yokoyama, K.; Shimizu, E.; Yusa, N.; Kondoh, K.; Ogawa, M.; Takei, T.; Kobayashi, A.; Ito, M.; Isobe, M.; et al. Prognostic impact of circulating tumor DNA status post-allogeneic hematopoietic stem cell transplantation in AML and MDS. Blood 2019, 133, 2682–2695. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.H.; Shim, J.H.; Ryu, K.J.; Kim, Y.J.; Choi, M.E.; Yoon, S.E.; Cho, J.; Park, B.; Park, W.Y.; Kim, W.S.; et al. Clinical relevance of serum-derived exosomal messenger RNA sequencing in patients with non-Hodgkin lymphoma. J. Cancer 2022, 13, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- Suehara, Y.; Sakata-Yanagimoto, M.; Hattori, K.; Kusakabe, M.; Nanmoku, T.; Sato, T.; Noguchi, M.; Chiba, S. Mutations found in cell-free DNAs of patients with malignant lymphoma at remission can derive from clonal hematopoiesis. Cancer Sci. 2019, 110, 3375–3381. [Google Scholar] [CrossRef]
- Blombery, P.A.; Ryland, G.L.; Markham, J.; Guinto, J.; Wall, M.; McBean, M.; Jones, K.; Thompson, E.R.; Cameron, D.L.; Papenfuss, A.T.; et al. Detection of clinically relevant early genomic lesions in B-cell malignancies from circulating tumour DNA using a single hybridisation-based next generation sequencing assay. Br. J. Haematol. 2018, 183, 146–149. [Google Scholar] [CrossRef]
- Bessi, L.; Viailly, P.J.; Bohers, E.; Ruminy, P.; Maingonnat, C.; Bertrand, P.; Vasseur, N.; Beaussire, L.; Cornic, M.; Etancelin, P.; et al. Somatic mutations of cell-free circulating DNA detected by targeted next-generation sequencing and digital droplet PCR in classical Hodgkin lymphoma. Leuk. Lymphoma 2019, 60, 498–502. [Google Scholar] [CrossRef]
- Shi, Y.; Su, H.; Song, Y.; Jiang, W.; Sun, X.; Qian, W.; Zhang, W.; Gao, Y.; Jin, Z.; Zhou, J.; et al. Circulating tumor DNA predicts response in Chinese patients with relapsed or refractory classical hodgkin lymphoma treated with sintilimab. EBioMedicine 2020, 54, 102731. [Google Scholar] [CrossRef]
- Rushton, C.K.; Arthur, S.E.; Alcaide, M.; Cheung, M.; Jiang, A.; Coyle, K.M.; Cleary, K.L.S.; Thomas, N.; Hilton, L.K.; Michaud, N.; et al. Genetic and evolutionary patterns of treatment resistance in relapsed B-cell lymphoma. Blood Adv. 2020, 4, 2886–2898. [Google Scholar] [CrossRef]
- Liu, H.; Yang, C.; Zhao, X.; Le, J.; Wu, G.; Wei, J.; Liang, Y.; Qian, W. Genotyping on ctDNA Identifies Shifts in Mutation Spectrum Between Newly Diagnosed and Relapse/Refractory DLBCL. Onco. Targets Ther. 2020, 13, 10797–10806. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, W.; Li, J.; Xiong, J.; Liu, J.; Chen, T.; Wen, Q.; Zeng, Y.; Gao, L.; Gao, L.; et al. Plasma circulating tumor DNA assessment reveals KMT2 D as a potential poor prognostic factor in extranodal NK/T-cell lymphoma. Biomark. Res. 2020, 8, 27. [Google Scholar] [CrossRef]
- Shin, S.H.; Kim, Y.J.; Lee, D.; Cho, D.; Ko, Y.H.; Cho, J.; Park, W.Y.; Park, D.; Kim, S.J.; Kim, W.S. Analysis of circulating tumor DNA by targeted ultra-deep sequencing across various non-Hodgkin lymphoma subtypes. Leuk. Lymphoma 2019, 60, 2237–2246. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Pang, D.; Guo, H.; Jiang, X.; Liu, S.; Huang, L.; Wei, X.; Liang, Z.; Wang, X.; Li, W. Clinicopathological Characteristics and Mutational Profiling of Adult T-Cell Lymphoblastic Lymphoma in a Chinese Population. Cancer Manag. Res. 2020, 12, 3003–3012. [Google Scholar] [CrossRef] [PubMed]
- Paiva, B.; Puig, N.; Cedena, M.T.; Rosinol, L.; Cordon, L.; Vidriales, M.B.; Burgos, L.; Flores-Montero, J.; Sanoja-Flores, L.; Lopez-Anglada, L.; et al. Measurable Residual Disease by Next-Generation Flow Cytometry in Multiple Myeloma. J. Clin. Oncol. 2020, 38, 784–792. [Google Scholar] [CrossRef]
- Tosetti, F.; Vene, R.; Camodeca, C.; Nuti, E.; Rossello, A.; D’Arrigo, C.; Galante, D.; Ferrari, N.; Poggi, A.; Zocchi, M.R. Specific ADAM10 inhibitors localize in exosome-like vesicles released by Hodgkin lymphoma and stromal cells and prevent sheddase activity carried to bystander cells. Oncoimmunology 2018, 7, e1421889. [Google Scholar] [CrossRef] [PubMed]
- Rigolin, G.M.; Maffei, R.; Rizzotto, L.; Ciccone, M.; Sofritti, O.; Daghia, G.; Cibien, F.; Cavazzini, F.; Marasca, R.; Cuneo, A. Circulating endothelial cells in patients with chronic lymphocytic leukemia: Clinical-prognostic and biologic significance. Cancer 2010, 116, 1926–1937. [Google Scholar] [CrossRef]
- Sukmawati, D.; Tanaka, R. Introduction to next generation of endothelial progenitor cell therapy: A promise in vascular medicine. Am. J. Transl. Res. 2015, 7, 411–421. [Google Scholar] [PubMed]
- Yu, D.D.; Liu, H.L.; Bai, Y.L.; Wu, B.; Chen, W.H.; Ren, J.H.; Zhang, T.; Yang, K.Y.; Wu, G. Correlation between increased circulating endothelial progenitor cells and stage of non-Hodgkin lymphoma. J. Huazhong Univ. Sci. Technol. Med. Sci. 2013, 33, 284–287. [Google Scholar] [CrossRef]
- Bhaskar, A.; Gupta, R.; Kumar, L.; Sharma, A.; Sharma, M.C.; Kalaivani, M.; Thakur, S.C. Circulating endothelial progenitor cells as potential prognostic biomarker in multiple myeloma. Leuk. Lymphoma 2012, 53, 635–640. [Google Scholar] [CrossRef]
- Moschetta, M.; Mishima, Y.; Kawano, Y.; Manier, S.; Paiva, B.; Palomera, L.; Aljawai, Y.; Calcinotto, A.; Unitt, C.; Sahin, I.; et al. Targeting vasculogenesis to prevent progression in multiple myeloma. Leukemia 2016, 30, 1103–1115. [Google Scholar] [CrossRef]
- Moschetta, M.; Mishima, Y.; Aljawai, Y.; Sahin, I.; Calcinotto, A.; Manier, S.; Maiso, P.; Sacco, A.; Bellone, M.; Roccaro, A.M.; et al. Bone Marrow Mobilization Of Endothelial Progenitor Cells Represents An Early Pathogenic Event During Multiple Myeloma Progression. Blood 2013, 122, 680. [Google Scholar] [CrossRef]
- Spina, V.; Bruscaggin, A.; Cuccaro, A.; Martini, M.; Di Trani, M.; Forestieri, G.; Manzoni, M.; Condoluci, A.; Arribas, A.; Terzi-Di-Bergamo, L.; et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood 2018, 131, 2413–2425. [Google Scholar] [CrossRef] [PubMed]
- Lakhotia, R.; Melani, C.; Dunleavy, K.; Pittaluga, S.; Saba, N.; Lindenberg, L.; Mena, E.; Bergvall, E.; Lucas, A.N.; Jacob, A.; et al. Circulating tumor DNA predicts therapeutic outcome in mantle cell lymphoma. Blood Adv. 2022, 6, 2667–2680. [Google Scholar] [CrossRef] [PubMed]
- Scherer, F.; Kurtz, D.M.; Newman, A.M.; Stehr, H.; Craig, A.F.; Esfahani, M.S.; Lovejoy, A.F.; Chabon, J.J.; Klass, D.M.; Liu, C.L.; et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med. 2016, 8, 364ra155. [Google Scholar] [CrossRef] [PubMed]
- Camus, V.; Viennot, M.; Lequesne, J.; Viailly, P.J.; Bohers, E.; Bessi, L.; Marcq, B.; Etancelin, P.; Dubois, S.; Picquenot, J.M.; et al. Targeted genotyping of circulating tumor DNA for classical Hodgkin lymphoma monitoring: A prospective study. Haematologica 2021, 106, 154–162. [Google Scholar] [CrossRef]
- Mithraprabhu, S.; Khong, T.; Ramachandran, M.; Chow, A.W.S.; Klarica, D.; Mai, L.; Walsh, S.; Broemeling, D.; Marziali, A.; Wiggin, M.; et al. Mutational Characterisation and Tracking Disease Progression Using Circulating Cell-Free Tumor DNA in Multiple Myeloma Patients. Blood 2016, 128, 3280. [Google Scholar] [CrossRef]
- Mithraprabhu, S.; Khong, T.; Ramachandran, M.; Chow, A.; Klarica, D.; Mai, L.; Walsh, S.; Broemeling, D.; Marziali, A.; Wiggin, M.; et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia 2017, 31, 1695–1705. [Google Scholar] [CrossRef]
- Mithraprabhu, S.; Morley, R.; Khong, T.; Kalff, A.; Bergin, K.; Hocking, J.; Savvidou, I.; Bowen, K.M.; Ramachandran, M.; Choi, K.; et al. Monitoring tumour burden and therapeutic response through analysis of circulating tumour DNA and extracellular RNA in multiple myeloma patients. Leukemia 2019, 33, 2022–2033. [Google Scholar] [CrossRef]
- Oberle, A.; Brandt, A.; Voigtlaender, M.; Thiele, B.; Radloff, J.; Schulenkorf, A.; Alawi, M.; Akyuz, N.; Marz, M.; Ford, C.T.; et al. Monitoring multiple myeloma by next-generation sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica 2017, 102, 1105–1111. [Google Scholar] [CrossRef]
- Guo, G.; Raje, N.S.; Seifer, C.; Kloeber, J.; Isenhart, R.; Ha, G.; Yee, A.J.; O’Donnell, E.K.; Tai, Y.T.; Richardson, P.G.; et al. Genomic discovery and clonal tracking in multiple myeloma by cell-free DNA sequencing. Leukemia 2018, 32, 1838–1841. [Google Scholar] [CrossRef]
- Bianchi, G.; Kyle, R.A.; Larson, D.R.; Witzig, T.E.; Kumar, S.; Dispenzieri, A.; Morice, W.G.; Rajkumar, S.V. High levels of peripheral blood circulating plasma cells as a specific risk factor for progression of smoldering multiple myeloma. Leukemia 2013, 27, 680–685. [Google Scholar] [CrossRef]
- Gonsalves, W.I.; Rajkumar, S.V.; Gupta, V.; Morice, W.G.; Timm, M.M.; Singh, P.P.; Dispenzieri, A.; Buadi, F.K.; Lacy, M.Q.; Kapoor, P.; et al. Quantification of clonal circulating plasma cells in newly diagnosed multiple myeloma: Implications for redefining high-risk myeloma. Leukemia 2014, 28, 2060–2065. [Google Scholar] [CrossRef] [PubMed]
- Sanoja-Flores, L.; Flores-Montero, J.; Garces, J.J.; Paiva, B.; Puig, N.; Garcia-Mateo, A.; Garcia-Sanchez, O.; Corral-Mateos, A.; Burgos, L.; Blanco, E.; et al. Next generation flow for minimally-invasive blood characterization of MGUS and multiple myeloma at diagnosis based on circulating tumor plasma cells (CTPC). Blood Cancer J. 2018, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Rajkumar, S.V.; Kyle, R.A.; Lacy, M.Q.; Dispenzieri, A.; Fonseca, R.; Lust, J.A.; Gertz, M.A.; Greipp, P.R.; Witzig, T.E. Prognostic value of circulating plasma cells in monoclonal gammopathy of undetermined significance. J. Clin. Oncol. 2005, 23, 5668–5674. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.J.; Yu, J.; Li, J.Y.; Liu, Y.T.; Zhong, R.Q. Circulating microRNA expression is associated with genetic subtype and survival of multiple myeloma. Med. Oncol. 2012, 29, 2402–2408. [Google Scholar] [CrossRef] [PubMed]
- Deligezer, U.; Yaman, F.; Erten, N.; Dalay, N. Frequent copresence of methylated DNA and fragmented nucleosomal DNA in plasma of lymphoma patients. Clin. Chim. Acta 2003, 335, 89–94. [Google Scholar] [CrossRef]
- Shi, H.; Guo, J.; Duff, D.J.; Rahmatpanah, F.; Chitima-Matsiga, R.; Al-Kuhlani, M.; Taylor, K.H.; Sjahputera, O.; Andreski, M.; Wooldridge, J.E.; et al. Discovery of novel epigenetic markers in non-Hodgkin’s lymphoma. Carcinogenesis 2007, 28, 60–70. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, X.; Liu, J. The role of circulating miRNAs in multiple myeloma. Sci. China Life Sci. 2015, 58, 1262–1269. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, L.; Xiang, B.; Zhu, H.; Wu, Y.; Chen, M.; Guan, P.; Zou, X.; Valencia, C.A.; Dong, B.; et al. Potential role of exosome-associated microRNA panels and in vivo environment to predict drug resistance for patients with multiple myeloma. Oncotarget 2016, 7, 30876–30891. [Google Scholar] [CrossRef]
- Feng, Y.; Zhong, M.; Zeng, S.; Wang, L.; Liu, P.; Xiao, X.; Liu, Y. Exosome-derived miRNAs as predictive biomarkers for diffuse large B-cell lymphoma chemotherapy resistance. Epigenomics 2019, 11, 35–51. [Google Scholar] [CrossRef]
- Wu, R.; Guo, S.; Cai, J.; Huang, X.; Yang, J.; Wery, J.-P.; Li, H.Q.-x. Biomarker Predictive Ibrutinib Response Using Profiled ABC-DLBCL Patient Derived Xenografts. Blood 2015, 126, 2759. [Google Scholar] [CrossRef]
- Mondello, P.; Brea, E.J.; De Stanchina, E.; Toska, E.; Chang, A.Y.; Fennell, M.; Seshan, V.; Garippa, R.; Scheinberg, D.A.; Baselga, J.; et al. Panobinostat acts synergistically with ibrutinib in diffuse large B cell lymphoma cells with MyD88 L265 P mutations. JCI Insight 2017, 2, e90196. [Google Scholar] [CrossRef] [PubMed]
- Drandi, D.; Decruyenaere, P.; Ferrante, M.; Offner, F.; Vandesompele, J.; Ferrero, S. Nucleic Acid Biomarkers in Waldenstrom Macroglobulinemia and IgM-MGUS: Current Insights and Clinical Relevance. Diagnostics 2022, 12, 969. [Google Scholar] [CrossRef]
- Bagratuni, T.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; Mavrianou-Koutsoukou, N.; Liacos, C.; Patseas, D.; Kanellias, N.; Migkou, M.; Ziogas, D.C.; Eleutherakis-Papaiakovou, E.; et al. Detection of MYD88 and CXCR4 mutations in cell-free DNA of patients with IgM monoclonal gammopathies. Leukemia 2018, 32, 2617–2625. [Google Scholar] [CrossRef] [PubMed]
- Treon, S.P.; Cao, Y.; Xu, L.; Yang, G.; Liu, X.; Hunter, Z.R. Somatic mutations in MYD88 and CXCR4 are determinants of clinical presentation and overall survival in Waldenstrom macroglobulinemia. Blood 2014, 123, 2791–2796. [Google Scholar] [CrossRef] [PubMed]
- Treon, S.P.; Xu, L.; Hunter, Z. MYD88 Mutations and Response to Ibrutinib in Waldenstrom’s Macroglobulinemia. N. Engl. J. Med. 2015, 373, 584–586. [Google Scholar] [CrossRef] [PubMed]
- Treon, S.P.; Xu, L.; Yang, G.; Zhou, Y.; Liu, X.; Cao, Y.; Sheehy, P.; Manning, R.J.; Patterson, C.J.; Tripsas, C.; et al. MYD88 L265 P somatic mutation in Waldenstrom’s macroglobulinemia. N. Engl. J. Med. 2012, 367, 826–833. [Google Scholar] [CrossRef]
- Xu, L.; Hunter, Z.R.; Yang, G.; Cao, Y.; Liu, X.; Manning, R.; Tripsas, C.; Chen, J.; Patterson, C.J.; Kluk, M.; et al. Detection of MYD88 L265 P in peripheral blood of patients with Waldenstrom’s Macroglobulinemia and IgM monoclonal gammopathy of undetermined significance. Leukemia 2014, 28, 1698–1704. [Google Scholar] [CrossRef]
- Yang, G.; Zhou, Y.; Liu, X.; Xu, L.; Cao, Y.; Manning, R.J.; Patterson, C.J.; Buhrlage, S.J.; Gray, N.; Tai, Y.T.; et al. A mutation in MYD88 (L265 P) supports the survival of lymphoplasmacytic cells by activation of Bruton tyrosine kinase in Waldenstrom macroglobulinemia. Blood 2013, 122, 1222–1232. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Jimenez, C.; Maiso, P.; Moschetta, M.; Mishima, Y.; Aljawai, Y.; Sahin, I.; Kuhne, M.; Cardarelli, P.; et al. C1013 G/CXCR4 acts as a driver mutation of tumor progression and modulator of drug resistance in lymphoplasmacytic lymphoma. Blood 2014, 123, 4120–4131. [Google Scholar] [CrossRef]
- Luanpitpong, S.; Janan, M.; Thumanu, K.; Poohadsuan, J.; Rodboon, N.; Klaihmon, P.; Issaragrisil, S. Deciphering the Elevated Lipid via CD36 in Mantle Cell Lymphoma with Bortezomib Resistance Using Synchrotron-Based Fourier Transform Infrared Spectroscopy of Single Cells. Cancers 2019, 11, 576. [Google Scholar] [CrossRef]
| Pros | Cons |
|---|---|
| Constant tissue production | Costs |
| Fast process | Not reliable yet |
| No unfit patients | Several confounding factors |
| Fewer complications | Low availability of specialized machine that would improve the accuracy |
| Simple procedure | Not all components are detectable in early and MRD settings |
| Easy repeatability | - |
| Subclonal genetic makeup | - |
| Metastasis data | - |
| Possibility of observing evolution over time | - |
| More accurate with regard to prognosis and treatment response | - |
| Early diagnosis, even without knowing the primary tumor location | - |
| Target | Technology Used | ClinicalTrials.gov IDs | Study Title | Study Endpoints |
|---|---|---|---|---|
| ctDNA | Genetic tests NOS | NCT03023202 | UWCCC Precision Medicine Molecular Tumor Board Registry | Frequency of acceptance of molecular tumor board recommendations. Benefits of PMMTB-recommended treatment. Correlation of mutations with protein overexpression, circulating tumor DNA, and spheroid culture investigations. |
| ctDNA, RNA | NGS, RNA sequencing | NCT01775072 | Genomic Profiling in Cancer Patients | Nature and the frequency of “actionable” oncogenic mutations. |
| miRNA | PCR | NCT02791217 | Identification of Hematological Malignancies and Therapy Predication Using microRNAs as a Diagnostic Tool | Molecular characteristics (GEP, miRNA); EFS; OS. |
| ctDNA, RNA | Genetic tests NOS | NCT01792882 | Prospective Collection of Surplus Surgical Tumor Tissues and Pre-surgical Blood Samples | Tumor genetic sequence variation. Transcription profile. Epigenetic modification. |
| ctDNA | Genetic tests NOS | NCT01137643 | Tissue, Blood, and Body Fluid Sample Collection from Patients With Hematologic Cancer | Development of a centralized, quality-controlled, quality-assured facility for the procurement, processing, storage, and distribution of normal and malignant tissue specimens and corresponding blood specimens. |
| ctDNA, RNA | NGS, RNA sequencing | NCT02213822 | Molecular Testing of Cancer by Integrated Genomic, Transcriptomic, and Proteomic Analysis | Frequency of “actionable” oncogenic mutations; prevalence of genomic, transcriptomic, and proteomic abnormalities. |
| Omics | Genetic tests NOS | NCT04298892 | Integrated Multiomics and Multilevel Characterization of Haematological Disorders and Malignancies | Hematologic diseases characterization. Response/resistance to ex vivo drug treatments. Biomarkers of drug-related toxicity. Association between biological and molecular features with patient’s clinical features. MRD. Recurrence/MRD patterns after treatments. Prognostic and early diagnostic biomarkers. Identification of circulating and tissue molecular markers. Technological advancement. |
| Epigenomics | Genetic tests NOS | NCT04264767 | Characterization of Methylation Patterns in Cancer and Non-Cancer cfDNA | Characterization of methylation patterns that will discriminate cancer and normal samples and the origin of cancer. |
| ctDNA | Genetic tests NOS | NCT01772771 | Molecular Testing for the MD Anderson Cancer Center Personalized Cancer Therapy Program | Frequency and distribution of mutations and co-mutations between different tumor types and levels of clinical-pathological factors. |
| ctDNA, epigenomics | Genetic tests NOS | NCT03727009 | Blood Sample Collection to Evaluate Biomarkers in Subjects with Untreated Hematologic Malignancies | Blood-based biomarkers associated with genetic and epigenetic alterations. |
| CTCs | Magnetic nanoparticles coated with antibodies | NCT04290923 | Determination of Blood Tumor Cells | CTC counting |
| ctDNA | NGS | NCT02534649 | Bergonie Institut Profiling: Fighting Cancer by Matching Molecular Alterations and Drugs in Early Phase Trials | Efficacy of LBx in terms of frequency of genomic alteration, molecular profiling, failure rate of molecular screenings, and safety of the procedures. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savino, F.D.; Rigali, F.; Giustini, V.; D’Aliberti, D.; Spinelli, S.; Piazza, R.; Sacco, A.; Roccaro, A.M. Liquid Biopsy in Cancer: Focus on Lymphoproliferative Disorders. Cancers 2022, 14, 5378. https://doi.org/10.3390/cancers14215378
Savino FD, Rigali F, Giustini V, D’Aliberti D, Spinelli S, Piazza R, Sacco A, Roccaro AM. Liquid Biopsy in Cancer: Focus on Lymphoproliferative Disorders. Cancers. 2022; 14(21):5378. https://doi.org/10.3390/cancers14215378
Chicago/Turabian StyleSavino, Francesco D., Fabio Rigali, Viviana Giustini, Deborah D’Aliberti, Silvia Spinelli, Rocco Piazza, Antonio Sacco, and Aldo M. Roccaro. 2022. "Liquid Biopsy in Cancer: Focus on Lymphoproliferative Disorders" Cancers 14, no. 21: 5378. https://doi.org/10.3390/cancers14215378
APA StyleSavino, F. D., Rigali, F., Giustini, V., D’Aliberti, D., Spinelli, S., Piazza, R., Sacco, A., & Roccaro, A. M. (2022). Liquid Biopsy in Cancer: Focus on Lymphoproliferative Disorders. Cancers, 14(21), 5378. https://doi.org/10.3390/cancers14215378








