MicroRNAs’ Crucial Role in Salivary Gland Cancers’ Onset and Prognosis
Abstract
Simple Summary
Abstract
1. Introduction
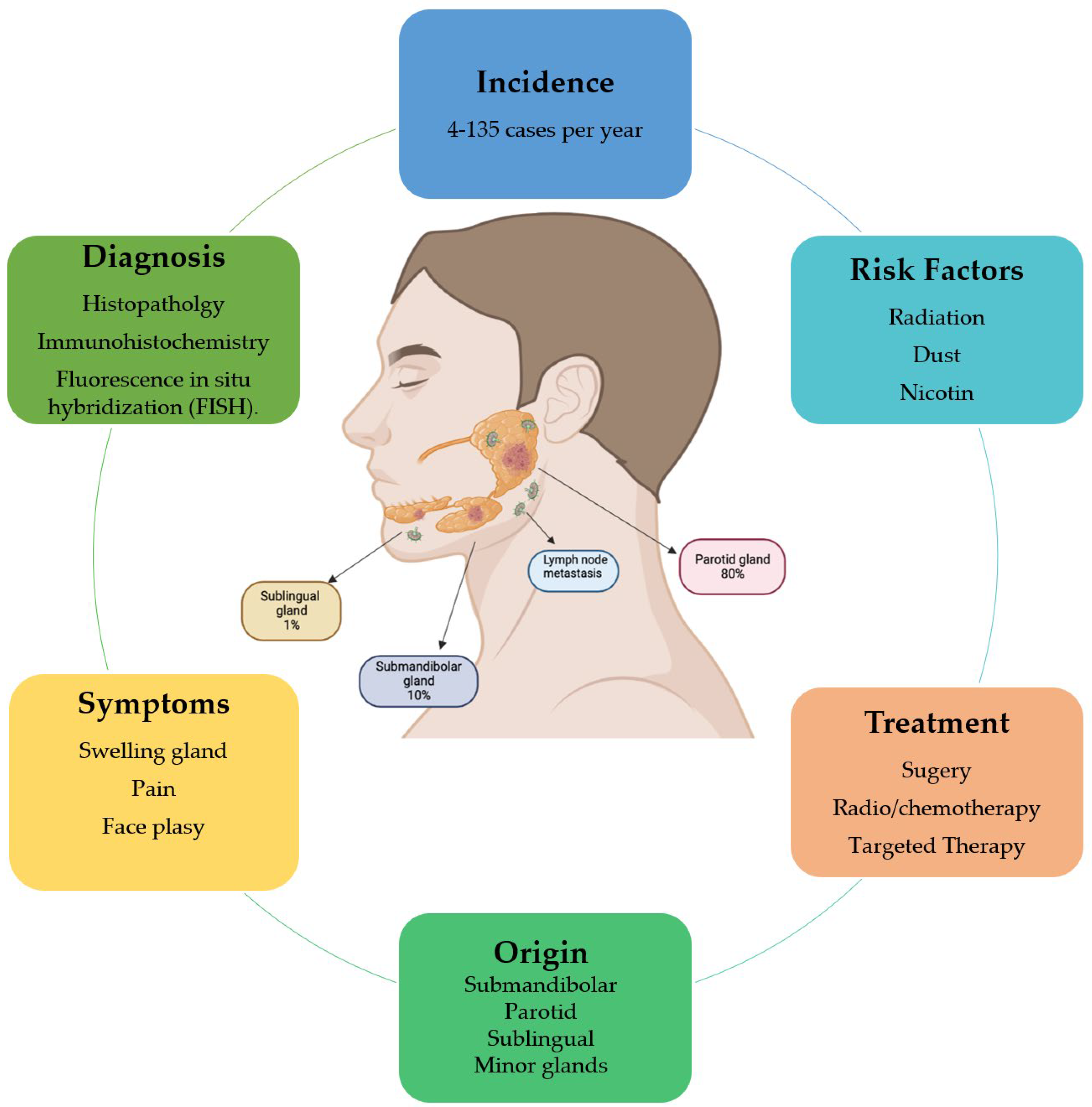
1.1. Characteristics of Salivary Gland Tumors
1.2. Features of MicroRNAs
2. Liquid Biopsy: Kill Two Birds with One Stone
3. MicroRNAs’ Role in Salivary Gland Cancers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pedersen, A.M.L.; Sørensen, C.E.; Proctor, G.B.; Carpenter, G.; Ekström, J. Salivary secretion in health and disease. J. Oral Rehabilit. 2018, 45, 730–746. [Google Scholar] [CrossRef] [PubMed]
- Porcheri, C.; Mitsiadis, T.A. Physiology, Pathology and Regeneration of Salivary Glands. Cells 2019, 8, 976. [Google Scholar] [CrossRef] [PubMed]
- Ogle, O.E. Salivary Gland Diseases. Dent. Clin. N. Am. 2020, 64, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Gatta, G.; Guzzo, M.; Locati, L.D.; McGurk, M.; Prott, F.J. Major and minor salivary gland tumours. Crit. Rev. Oncol. 2020, 152, 102959. [Google Scholar] [CrossRef]
- Jałocha-Kaczka, A.; Kolary-Siekierska, K.; Miłoński, J.; Olszewski, J. Clinic and own experience in the treatment of large salivary glands’ tumors. Otolaryngol. Pol. 2020, 74, 1–5. [Google Scholar] [CrossRef]
- Goyal, G.; Mehdi, S.A.; Ganti, A.K. Salivary Gland Cancers: Biology and Systemic Therapy. Oncology 2015, 29, 773–780. [Google Scholar]
- Lan, L.-F.; Gao, C.-K.; Ma, C.-W. Prediction of Minor Salivary Gland Carcinoma: A Novel Nomogram and Risk Classification System for Overall Survival and Cancer-Specific Survival. Otolaryngol. Neck Surg. 2020, 164, 359–368. [Google Scholar] [CrossRef]
- El-Naggar, A.K. Editor’s perspective on the 4th edition of the WHO head and neck tumor classification. J. Egypt. Natl. Cancer Inst. 2017, 29, 65–66. [Google Scholar] [CrossRef]
- Son, E.; Panwar, A.; Mosher, C.H.; Lydiatt, D. Cancers of the Major Salivary Gland. J. Oncol. Pract. 2018, 14, 99–108. [Google Scholar] [CrossRef]
- Kessler, A.T.; Bhatt, A. Review of the Major and Minor Salivary Glands, Part 2: Neoplasms and Tumor-like Lesions. J. Clin. Imaging Sci. 2018, 8, 48. [Google Scholar] [CrossRef]
- Al-Zaher, N.; Obeid, A.; Al-Salam, S.; Al-Kayyali, B.S. Acinic cell carcinoma of the salivary glands: A literature review. Hematol. Stem Cell Ther. 2009, 2, 259–264. [Google Scholar] [CrossRef]
- Lee, D.Y.; Brayer, K.J.; Mitani, Y.; Burns, E.A.; Rao, P.H.; Bell, D.; Williams, M.D.; Ferrarotto, R.; Pytynia, K.B.; El-Naggar, A.K.; et al. Oncogenic Orphan Nuclear Receptor NR4A3 Interacts and Cooperates with MYB in Acinic Cell Carcinoma. Cancers 2020, 12, 2433. [Google Scholar] [CrossRef]
- Keller, G.; Steinmann, D.; Quaas, A.; Grünwald, V.; Janssen, S.; Hussein, K. New concepts of personalized therapy in salivary gland carcinomas. Oral Oncol. 2017, 68, 103–113. [Google Scholar] [CrossRef]
- Lassche, G.; van Boxtel, W.; Ligtenberg, M.J.; Grunsven, A.C.V.E.-V.; van Herpen, C.M. Advances and challenges in precision medicine in salivary gland cancer. Cancer Treat. Rev. 2019, 80, 101906. [Google Scholar] [CrossRef]
- Kurzrock, R.; Bowles, D.; Kang, H.; Meric-Bernstam, F.; Hainsworth, J.; Spigel, D.; Bose, R.; Burris, H.; Sweeney, C.; Beattie, M.; et al. Targeted therapy for advanced salivary gland carcinoma based on molecular profiling: Results from MyPathway, a phase IIa multiple basket study. Ann. Oncol. 2020, 31, 412–421. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A High-Resolution Structure of the Pre-microRNA Nuclear Export Machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.-J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Gu, W.; Xu, Y.; Xie, X.; Wang, T.; Ko, J.-H.; Zhou, T. The role of RNA structure at 5′ untranslated region in microRNA-mediated gene regulation. RNA 2014, 20, 1369–1375. [Google Scholar] [CrossRef]
- Forman, J.J.; Coller, H.A. The code within the code: microRNAs target coding regions. Cell Cycle 2010, 9, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Lucas, A.S.; Wang, Z.; Liu, Y. Identifying microRNA targets in different gene regions. BMC Bioinform. 2014, 15 (Suppl. 7), S4. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Munker, R.; Calin, G.A. MicroRNA profiling in cancer. Clin. Sci. 2011, 121, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Summerer, I.; Unger, K.; Braselmann, H.; Schuettrumpf, L.; Maihöfer, C.; Baumeister, P.; Kirchner, T.R.; Niyazi, M.; Sage, E.H.; Specht, H.M.; et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br. J. Cancer 2015, 113, 76–82. [Google Scholar] [CrossRef]
- Kawasaki, H.; Takeuchi, T.; Ricciardiello, F.; Lombardi, A.; Biganzoli, E.; Fornili, M.; De Bortoli, D.; Mesolella, M.; Cossu, A.M.; Scrima, M.; et al. Definition of miRNA Signatures of Nodal Metastasis in LCa: miR-449a Targets Notch Genes and Suppresses Cell Migration and Invasion. Mol. Ther. Nucleic Acids 2020, 20, 711–724. [Google Scholar] [CrossRef]
- Falco, M.; Palma, G.; Rea, D.; De Biase, D.; Scala, S.; D’Aiuto, M.; Facchini, G.; Perdonà, S.; Barbieri, A.; Arra, C. Tumour biomarkers: Homeostasis as a novel prognostic indicator. Open Biol. 2016, 6. [Google Scholar] [CrossRef]
- Malathi, N.; Mythili, S.; Vasanthi, H.R. Salivary Diagnostics: A Brief Review. ISRN Dent. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Liang, Y.; Ridzon, D.; Wong, L.; Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007, 8, 166. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Zhu, C.; Ren, C.; Han, J.; Ding, Y.; Du, J.; Dai, N.; Dai, J.; Ma, H.; Hu, Z.; Shen, H.; et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br. J. Cancer 2014, 110, 2291–2299. [Google Scholar] [CrossRef]
- Humphrey, S.P.; Williamson, R.T. A review of saliva: Normal composition, flow, and function. J. Prosthet. Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; Majem, B.; Muinelo-Romay, L.; Álvarez-Castro, A.; Santamaría, A.; Gil-Moreno, A.; López, R.L.; Suárez-Cunqueiro, M.M. Human salivary microRNAs in Cancer. J. Cancer 2018, 9, 638–649. [CrossRef]
- Wang, S.; Claret, F.-X.; Wu, W. MicroRNAs as Therapeutic Targets in Nasopharyngeal Carcinoma. Front. Oncol. 2019, 9, 756. [Google Scholar] [CrossRef]
- Donati, S.; Ciuffi, S.; Brandi, M.L. Human Circulating miRNAs Real-time qRT-PCR-based Analysis: An Overview of Endogenous Reference Genes Used for Data Normalization. Int. J. Mol. Sci. 2019, 20, 4353. [Google Scholar] [CrossRef]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, Characterization, and Clinical Utility for Oral Cancer Detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef]
- Severino, P.; Oliveira, L.; Andreghetto, F.M.; Torres, N.; Curioni, O.A.; Cury, P.M.; Toporcov, T.N.; Paschoal, A.R.; Durham, A.M. Small RNAs in metastatic and non-metastatic oral squamous cell carcinoma. BMC Med. Genom. 2015, 8, 31. [Google Scholar] [CrossRef]
- Liu, C.-J.; Lin, S.-C.; Yang, C.-C.; Cheng, H.-W.; Chang, K.-W. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck 2011, 34, 219–224. [Google Scholar] [CrossRef]
- Wang, L.; Chen, Y.; Yan, Y.; Guo, X.; Fang, Y.; Su, Y.; Pathak, J.L.; Ge, L. miR-146a Overexpression in Oral Squamous Cell Carcinoma Potentiates Cancer Cell Migration and Invasion Possibly via Targeting HTT. Front. Oncol. 2020, 10, 585976. [Google Scholar] [CrossRef]
- Hung, P.-S.; Liu, C.-J.; Chou, C.-S.; Kao, S.-Y.; Yang, C.-C.; Chang, K.-W.; Chiu, T.-H.; Lin, S.-C. miR-146a Enhances the Oncogenicity of Oral Carcinoma by Concomitant Targeting of the IRAK1, TRAF6 and NUMB Genes. PLoS ONE 2013, 8, e79926. [Google Scholar] [CrossRef] [PubMed]
- Gau, V.; Wong, D. Oral Fluid Nanosensor Test (OFNASET) with Advanced Electrochemical-Based Molecular Analysis Platform. Ann. N. Y. Acad. Sci. 2007, 1098, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Mitani, Y.; Roberts, D.B.; Fatani, H.; Weber, R.S.; Kies, M.S.; Lippman, S.M.; El-Naggar, A.K. MicroRNA Profiling of Salivary Adenoid Cystic Carcinoma: Association of miR-17-92 Upregulation with Poor Outcome. PLoS ONE 2013, 8, e66778. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhao, X.; Dong, Z.; Cao, G.; Zhang, S. Identification of microRNA profiles in salivary adenoid cystic carcinoma cells during metastatic progression. Oncol. Lett. 2014, 7, 2029–2034. [Google Scholar] [CrossRef]
- Matse, J.H.; Yoshizawa, J.; Wang, X.; Elashoff, D.; Bolscher, J.G.; Veerman, E.C.; Bloemena, E.; Wong, D.T. Discovery and Prevalidation of Salivary Extracellular microRNA Biomarkers Panel for the Noninvasive Detection of Benign and Malignant Parotid Gland Tumors. Clin. Cancer Res. 2013, 19, 3032–3038. [Google Scholar] [CrossRef]
- Cinpolat, O.; Unal, Z.N.; Ismi, O.; Gorur, A.; Unal, M. Comparison of microRNA profiles between benign and malignant salivary gland tumors in tissue, blood and saliva samples: A prospective, case-control study. Braz. J. Otorhinolaryngol. 2017, 83, 276–284. [Google Scholar] [CrossRef]
- Santos, P.R.B.; Coutinho-Camillo, C.M.; Soares, F.A.; Freitas, V.; Vilas-Bôas, D.S.; Xavier, F.; Rocha, C.A.G.; de Araújo, I.B.; dos Santos, J.N. MicroRNAs expression pattern related to mast cell activation and angiogenesis in paraffin-embedded salivary gland tumors. Pathol. Res. Pract. 2017, 213, 1470–1476. [Google Scholar] [CrossRef]
- Zhang, X.; Cairns, M.; Rose, B.; O’Brien, C.; Shannon, K.; Clark, J.; Gamble, J.; Tran, N. Alterations in miRNA processing and expression in pleomorphic adenomas of the salivary gland. Int. J. Cancer 2009, 124, 2855–2863. [Google Scholar] [CrossRef]
- Sun, J.-Y.; Huang, Y.; Li, J.-P.; Zhang, X.; Wang, L.; Meng, Y.-L.; Yan, B.; Bian, Y.-Q.; Zhao, J.; Wang, W.-Z.; et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem. Biophys. Res. Commun. 2012, 420, 787–792. [Google Scholar] [CrossRef]
- Li, B.H.; Zhou, J.S.; Ye, F.; Cheng, X.D.; Zhou, C.Y.; Lu, W.G.; Xie, X. Reduced miR-100 expression in cervical cancer and precursors and its carcinogenic effect through targeting PLK1 protein. Eur. J. Cancer 2011, 47, 2166–2174. [Google Scholar] [CrossRef]
- Denaro, M.; Navari, E.; Ugolini, C.; Seccia, V.; Donati, V.; Casani, A.P.; Basolo, F. A microRNA signature for the differential diagnosis of salivary gland tumors. PLoS ONE 2019, 14, e0210968. [Google Scholar] [CrossRef]
- Wang, W.-W.; Chen, B.; Lei, C.-B.; Liu, G.-X.; Wang, Y.-G.; Yi, C.; Wang, Y.-Y.; Zhang, S.-Y. miR-582-5p inhibits invasion and migration of salivary adenoid cystic carcinoma cells by targeting FOXC1. Jpn. J. Clin. Oncol. 2017, 47, 690–698. [Google Scholar] [CrossRef]
- Naakka, E.; Barros-Filho, M.C.; Adnan-Awad, S.; Al-Samadi, A.; Marchi, F.A.; Kuasne, H.; Korelin, K.; Suleymanova, I.; Brown, A.L.; Scapulatempo-Neto, C.; et al. miR-22 and miR-205 Drive Tumor Aggressiveness of Mucoepidermoid Carcinomas of Salivary Glands. Front. Oncol. 2022, 11, 786150. [Google Scholar] [CrossRef]
- Boštjančič, E.; Hauptman, N.; Grošelj, A.; Glavač, D.; Volavšek, M. Expression, Mutation, and Amplification Status of EGFR and Its Correlation with Five miRNAs in Salivary Gland Tumours. BioMed Res. Int. 2017, 2017, 9150402. [Google Scholar] [CrossRef]
- Matse, J.H.; Yoshizawa, J.; Wang, X.; Elashoff, D.; Bolscher, J.G.M.; Veerman, E.C.I.; Leemans, C.R.; Pegtel, M.D.; Wong, D.T.W.; Bloemena, E. Human Salivary Micro-RNA in Patients with Parotid Salivary Gland Neoplasms. PLoS ONE 2015, 10, e0142264. [Google Scholar] [CrossRef]
- Andreasen, S.; Tan, Q.; Agander, T.K.; Hansen, T.V.O.; Steiner, P.; Bjørndal, K.; Høgdall, E.; Larsen, S.R.; Erentaite, D.; Olsen, C.H.; et al. MicroRNA dysregulation in adenoid cystic carcinoma of the salivary gland in relation to prognosis and gene fusion status: A cohort study. Virchows Arch. 2018, 473, 329–340. [Google Scholar] [CrossRef]


| Malignant Types | Benign Types |
|---|---|
| Mucoepideroid carcinoma | Pleomorphic adenoma |
| Adenoid cystic carcinoma | Myoepithelioma |
| Acinic cell carcinoma | Basal cell adenoma |
| Polymorphous adenocarcinoma | Whartin tumor |
| Clear cell carcinoma | Oncocytoma |
| Basal cell adenocarcinoma | Lymphadenoma |
| Intraductal carcinoma | Cystadenoma |
| Adenocarcinoma | Sialadenoma papilliferum |
| Salivary duct carcinoma | Ductal papillomas |
| Myoepithelial carcinoma | Sebaceous adenoma |
| Epithelial–myoepithelial carcinoma | Canalicular adenoma and other ductal adenomas |
| Carcinoma ex pleomorphic adenoma | |
| Secretory carcinoma | |
| Sebaceous adenocarcinoma | |
| Carcinosarcoma | |
| Poorly differentiated carcinoma Undifferentiated carcinoma Large-cell neuroendocrine carcinoma Small-cell neuroendocrine carcinoma | |
| Lymphoepithelial carcinoma | |
| Squamous cell carcinoma | |
| Oncocytic carcinoma | |
| Sialoblastoma |
| MicroRNA | SGC Subtype | Function | Levels 1 | References |
|---|---|---|---|---|
| hsa-let-7a | SGTs | Prognostic | + | [44,55] |
| hsa-let7g | SGTs | Diagnostic | − | [46] |
| hsa-let7g-5p | SGTs | Prognostic | − | [52,56] |
| hsa-miR-100 | SGTs | Prognostic | − | [45,52] |
| hsa-miR-103a-3p | PGT | Prognostic | + | [56] |
| hsa-miR-106 | ACC | Prognostic | + | [44] |
| hsa-miR-106a | SGTs | Prognostic | + | [52] |
| hsa-miR-106b | SGTs | Prognostic | + | [52] |
| hsa-miR-1180 | ACC | Prognostic | + | [57] |
| hsa-miR-1233 | PGT | Diagnostic | + | [56] |
| hsa-miR-125a-5p | ACC | Prognostic | + | [30] |
| hsa-miR-125b | SGTs | Prognostic | − | [45,52] |
| hsa-miR-1267 | PGT | Diagnostic | + | [56] |
| hsa-mir-132 | AP ACC MEC | Prognostic | + | [48] |
| hsa-miR-132 | PGT | Diagnostic | − | [46,48] |
| hsa-miR-133b | SGTs | Prognostic | + | [54] |
| hsa-miR-135a-5p | SGTs | Diagnostic | − | [52] |
| hsa-miR-140 | SGTs | Prognostic | − | [46,52,55] |
| hsa-miR-140-5p | PGT | Diagnostic | − | [46] |
| hsa-miR-140-5p | SGTs | Prognostic | − | [46,52] |
| hsa-miR-146b | SGTs | Diagnostic | + | [47] |
| hsa-miR-150 | ACC | Prognostic | + | [44] |
| hsa-miR-152 | ACC | Prognostic | + | [57] |
| hsa-miR-15b | PGT | Diagnostic | − | [46] |
| hsa-mir-16 | AP ACC MEC | Prognostic | + | [48] |
| hsa-miR-17 | AP ACC MEC | Prognostic | + | [44,48] |
| hsa-miR-17 | SGTs | Diagnostic | + | [48] |
| hsa-mir-181a-2 | ACC | Prognostic | + | [57] |
| hsa-miR-1825 | PGT | Diagnostic | + | [56] |
| hsa-mir-195 | AP ACC MEC | Prognostic | − | [48] |
| hsa-miR-195 | SGTs | Diagnostic | − | [52] |
| hsa-miR-195-5p | SGTs | Diagnostic | − | [52] |
| hsa-miR-199a | SGTs | Diagnostic | + | [47] |
| hsa-miR-199a-5p | SGTs | Diagnostic | + | [52] |
| hsa-miR-205 | MEC | Prognostic | + | [54] |
| hsa-miR-20a | ACC | Prognostic | + | [44,52] |
| hsa-mir-21 | ACC | Prognostic | + | [57] |
| hsa-miR-21 | SGTs | Diagnostic | + | [47,52] |
| hsa-miR-211 | PGT | Diagnostic | + | [56] |
| hsa-miR-211-3p | ACC | Prognostic | − | [45] |
| hsa-miR-22 | MEC | Prognostic | + | [54] |
| hsa-mir-221 | AP ACC MEC | Prognostic | − | [48] |
| hsa-miR-221 | AP ACC MEC | Diagnostic | − | [48] |
| hsa-miR-222 | PGT | Diagnostic | − | [46] |
| hsa-miR-222-3p | SGTs | Diagnostic | + | [52] |
| hsa-miR-222-3p | SGTs | Prognostic | + | [46,52] |
| hsa-miR-30e | SGTs | Diagnostic | + | [47] |
| hsa-miR-31 | SGTs | Diagnostic | + | [47] |
| hsa-miR-3125 | MEC | Prognostic | − | [54] |
| hsa-miR-3131 | ACC | Prognostic | − | [45] |
| hsa-miR-320a | ACC | Prognostic | + | [50] |
| hsa-miR-320e | SGTs | Diagnostic | + | [52] |
| hsa-miR-345 | SGTs | Diagnostic | + | [47] |
| hsa-miR-374 | SGTs | Diagnostic | + | [56] |
| hsa-miR-374 | SGTs | Prognostic | + | [46,52] |
| hsa-miR-374c | ACC | Prognostic | + | [57] |
| hsa-miR-375 | ACC | Prognostic | − | [44,45,52] |
| hsa-miR-4430 | ACC | Prognostic | + | [45] |
| hsa-miR-4487 | ACC | Prognostic | + | [45] |
| hsa-miR-455-3p | ACC | Prognostic | + | [44] |
| hsa-miR-4676 | ACC | Prognostic | + | [57] |
| hsa-miR-4717-5p | ACC | Prognostic | + | [57] |
| hsa-miR-486-3p | ACC | Prognostic | + | [45] |
| hsa-miR-5191 | ACC | Prognostic | − | [45] |
| hsa-miR-577 | PGT | Diagnostic | + | [46] |
| hsa-miR-582-5p | MEC | Prognostic | − | [54] |
| hsa-mir-6865 | ACC | Prognostic | + | [57] |
| hsa-miR-9 | AP ACC MEC | Prognostic | − | [48] |
| hsa-miR-9 | AP ACC MEC | Diagnostic | − | [48,52] |
| hsa-miR-93 | SGTs | Prognostic | + | [52] |
| hsa-miR-99a | SGTs | Prognostic | − | [45,52] |
| hsa-miR-99b | SGTs | Prognostic | + | [55] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bocchetti, M.; Grisolia, P.; Melisi, F.; Ferraro, M.G.; De Luca, P.; Camaioni, A.; Falco, M.; Abate, M.; Misso, G.; Alfano, R.; et al. MicroRNAs’ Crucial Role in Salivary Gland Cancers’ Onset and Prognosis. Cancers 2022, 14, 5304. https://doi.org/10.3390/cancers14215304
Bocchetti M, Grisolia P, Melisi F, Ferraro MG, De Luca P, Camaioni A, Falco M, Abate M, Misso G, Alfano R, et al. MicroRNAs’ Crucial Role in Salivary Gland Cancers’ Onset and Prognosis. Cancers. 2022; 14(21):5304. https://doi.org/10.3390/cancers14215304
Chicago/Turabian StyleBocchetti, Marco, Piera Grisolia, Federica Melisi, Maria Grazia Ferraro, Pietro De Luca, Angelo Camaioni, Michela Falco, Marianna Abate, Gabriella Misso, Roberto Alfano, and et al. 2022. "MicroRNAs’ Crucial Role in Salivary Gland Cancers’ Onset and Prognosis" Cancers 14, no. 21: 5304. https://doi.org/10.3390/cancers14215304
APA StyleBocchetti, M., Grisolia, P., Melisi, F., Ferraro, M. G., De Luca, P., Camaioni, A., Falco, M., Abate, M., Misso, G., Alfano, R., Accardo, N., Oliva, F., Cossu, A. M., Caraglia, M., Scrima, M., & Ricciardiello, F. (2022). MicroRNAs’ Crucial Role in Salivary Gland Cancers’ Onset and Prognosis. Cancers, 14(21), 5304. https://doi.org/10.3390/cancers14215304








