The Urothelial Transcriptomic Response to Interferon Gamma: Implications for Bladder Cancer Prognosis and Immunotherapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. mRNA Analysis
2.3. Publicly Available Bladder Cancer Cohort Data
2.4. IFNγ-Signature Generation
2.5. IFNγ-Signature Analysis
3. Results
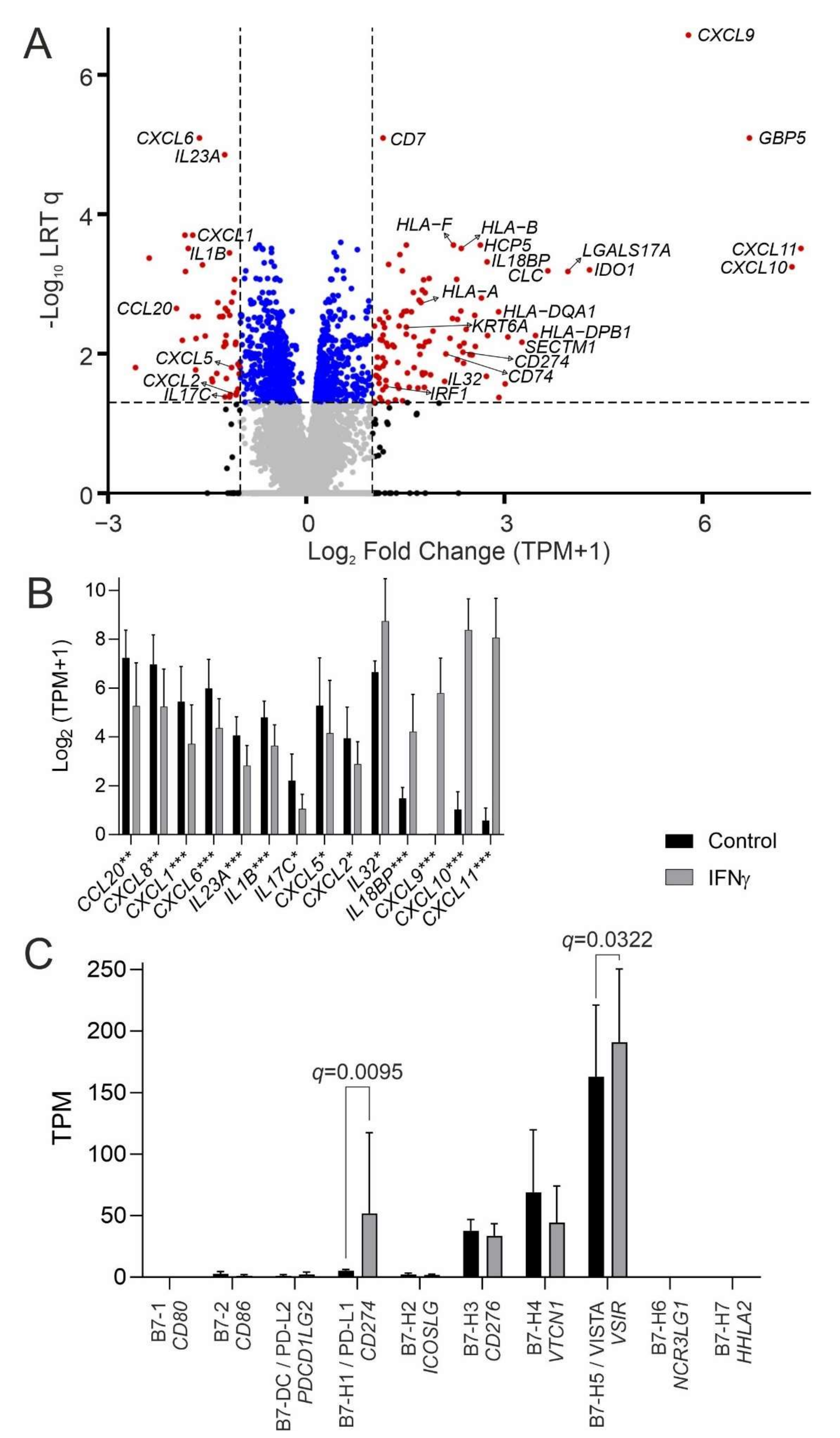
3.1. The Urothelial IFNγ Response
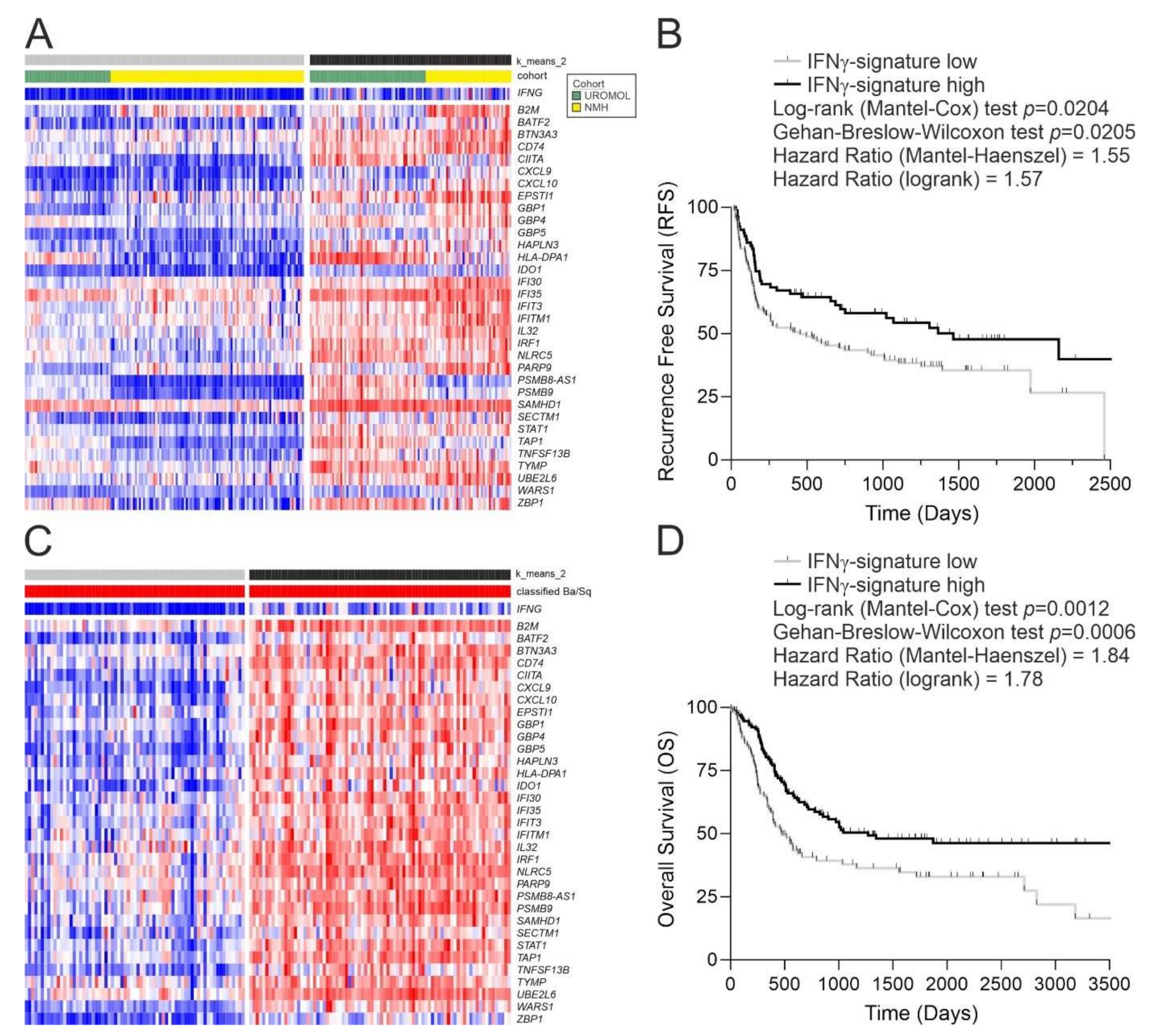
3.2. Application of the IFNγ Response Signature to Tumours
3.3. Bladder Cancer Neoantigens
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antonelli, A.C.; Binyamin, A.; Hohl, T.M.; Glickman, M.S.; Redelman-Sidi, G. Bacterial immunotherapy for cancer induces CD4-dependent tumor-specific immunity through tumor-intrinsic interferon-gamma signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 18627–18637. [Google Scholar] [CrossRef] [PubMed]
- Prescott, S.; James, K.; Hargreave, T.B.; Chisholm, G.D.; Smyth, J.F. Radio-immunoassay detection of interferon-gamma in urine after intravesical Evans BCG therapy. J. Urol. 1990, 144, 1248–1251. [Google Scholar] [CrossRef]
- De Reijke, T.M.; de Boer, E.C.; Kurth, K.H.; Schamhart, D.H. Urinary cytokines during intravesical bacillus Calmette-Guerin therapy for superficial bladder cancer: Processing, stability and prognostic value. J. Urol. 1996, 155, 477–482. [Google Scholar] [CrossRef]
- Taniguchi, K.; Koga, S.; Nishikido, M.; Yamashita, S.; Sakuragi, T.; Kanetake, H.; Saito, Y. Systemic immune response after intravesical instillation of bacille Calmette-Guerin (BCG) for superficial bladder cancer. Clin. Exp. Immunol. 1999, 115, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Riemensberger, J.; Bohle, A.; Brandau, S. IFN-gamma and IL-12 but not IL-10 are required for local tumour surveillance in a syngeneic model of orthotopic bladder cancer. Clin. Exp. Immunol. 2002, 127, 20–26. [Google Scholar] [CrossRef]
- Sato, Y.; Bolzenius, J.K.; Eteleeb, A.M.; Su, X.; Maher, C.A.; Sehn, J.K.; Arora, V.K. CD4+ T cells induce rejection of urothelial tumors after immune checkpoint blockade. JCI Insight 2018, 3, 23. [Google Scholar] [CrossRef]
- Baker, S.C.; Shabir, S.; Southgate, J. Biomimetic urothelial tissue models for the in vitro evaluation of barrier physiology and bladder drug efficacy. Mol. Pharm. 2014, 11, 1964–1970. [Google Scholar] [CrossRef]
- Southgate, J.; Hutton, K.A.; Thomas, D.F.; Trejdosiewicz, L.K. Normal human urothelial cells in vitro: Proliferation and induction of stratification. Lab. Investig. 1994, 71, 583–594. [Google Scholar]
- Cross, W.R.; Eardley, I.; Leese, H.J.; Southgate, J. A biomimetic tissue from cultured normal human urothelial cells: Analysis of physiological function. Am. J. Physiol. Renal. Physiol. 2005, 289, F459–F468. [Google Scholar] [CrossRef] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 2017, 14, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Groeneveld, C.S.; Jordan, B.; Lin, X.; McLaughlin, K.A.; Das, A.; Fall, L.A.; Fantini, D.; Taxter, T.J.; Mogil, L.S.; et al. Identification of Differential Tumor Subtypes of T1 Bladder Cancer. Eur. Urol. 2020, 78, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540–556. [Google Scholar] [CrossRef] [Green Version]
- Lindskrog, S.V.; Prip, F.; Lamy, P.; Taber, A.; Groeneveld, C.S.; Birkenkamp-Demtroder, K.; Jensen, J.B.; Strandgaard, T.; Nordentoft, I.; Christensen, E.; et al. An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat. Commun. 2021, 12, 2301. [Google Scholar] [CrossRef] [PubMed]
- Sjodahl, G.; Eriksson, P.; Liedberg, F.; Hoglund, M. Molecular classification of urothelial carcinoma: Global mRNA classification versus tumour-cell phenotype classification. J. Pathol. 2017, 242, 113–125. [Google Scholar] [CrossRef] [Green Version]
- Kamoun, A.; Reynies, A.; Allory, Y.; Sjodahl, G.; Gordon Robertson, A.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2019, 24, E210006. [Google Scholar] [CrossRef] [Green Version]
- Smith, N.J.; Varley, C.L.; Eardley, I.; Feather, S.; Trejdosiewicz, L.K.; Southgate, J. Toll-like receptor responses of normal human urothelial cells to bacterial flagellin and lipopolysaccharide. J. Urol. 2011, 186, 1084–1092. [Google Scholar] [CrossRef]
- Kuo, P.T.; Zeng, Z.; Salim, N.; Mattarollo, S.; Wells, J.W.; Leggatt, G.R. The Role of CXCR3 and Its Chemokine Ligands in Skin Disease and Cancer. Front. Med. 2018, 5, 271. [Google Scholar] [CrossRef] [Green Version]
- Baker, S.C.; Mason, A.S.; Slip, R.G.; Skinner, K.T.; Macdonald, A.; Masood, O.; Harris, R.S.; Fenton, T.R.; Periyasamy, M.; Ali, S.; et al. Induction of APOBEC3-mediated genomic damage in urothelium implicates BK polyomavirus (BKPyV) as a hit-and-run driver for bladder cancer. Oncogene 2022, 41, 2139–2151. [Google Scholar] [CrossRef]
- Stenehjem, D.D.; Tran, D.; Nkrumah, M.A.; Gupta, S. PD1/PDL1 inhibitors for the treatment of advanced urothelial bladder cancer. Onco. Targets Ther. 2018, 11, 5973–5989. [Google Scholar] [CrossRef] [Green Version]
- Seo, W.I.; Lee, C.H.; Jung, S.J.; Lee, D.S.; Park, H.Y.; Jeong, D.H.; Kim, W.; Chung, J.I.; Choi, I. Expression of VISTA on tumor-infiltrating immune cells correlated with short intravesical recurrence in non-muscle-invasive bladder cancer. Cancer Immunol. Immunother. 2021, 70, 3113–3122. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, M.J.; Meng, X.Y.; Fontugne, J.; Chen, C.L.; Radvanyi, F.; Bernard-Pierrot, I. Identification of new driver and passenger mutations within APOBEC-induced hotspot mutations in bladder cancer. Genome. Med. 2020, 12, 85. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baker, S.C.; Mason, A.S.; Slip, R.G.; Eriksson, P.; Sjödahl, G.; Trejdosiewicz, L.K.; Southgate, J. The Urothelial Transcriptomic Response to Interferon Gamma: Implications for Bladder Cancer Prognosis and Immunotherapy. Cancers 2022, 14, 5295. https://doi.org/10.3390/cancers14215295
Baker SC, Mason AS, Slip RG, Eriksson P, Sjödahl G, Trejdosiewicz LK, Southgate J. The Urothelial Transcriptomic Response to Interferon Gamma: Implications for Bladder Cancer Prognosis and Immunotherapy. Cancers. 2022; 14(21):5295. https://doi.org/10.3390/cancers14215295
Chicago/Turabian StyleBaker, Simon C., Andrew S. Mason, Raphael G. Slip, Pontus Eriksson, Gottfrid Sjödahl, Ludwik K. Trejdosiewicz, and Jennifer Southgate. 2022. "The Urothelial Transcriptomic Response to Interferon Gamma: Implications for Bladder Cancer Prognosis and Immunotherapy" Cancers 14, no. 21: 5295. https://doi.org/10.3390/cancers14215295
APA StyleBaker, S. C., Mason, A. S., Slip, R. G., Eriksson, P., Sjödahl, G., Trejdosiewicz, L. K., & Southgate, J. (2022). The Urothelial Transcriptomic Response to Interferon Gamma: Implications for Bladder Cancer Prognosis and Immunotherapy. Cancers, 14(21), 5295. https://doi.org/10.3390/cancers14215295







