Simple Summary
Acute myeloid leukemia (AML) with mutated RUNX1 (RUNX1mut) has an adverse prognosis based on the 2022 European LeukemiaNet risk stratification. However, the WHO classifications of 2022 removed RUNX1 mutations from the unique entity because of various prognoses and treatment outcomes. Intriguingly, the overall survival (OS) and relapse-free survival (RFS) outcomes were similar in patients who had de novo AML with intermediate-risk cytogenetics with and without RUNX1mut. Our study endorsed an unfavorable prognosis of this entity.
Abstract
Acute myeloid leukemia (AML) with mutated RUNX1 (RUNX1mut) is considered to have an unfavorable prognosis. However, recent studies have reported comparable survival outcomes with wild-type RUNX1 (RUNX1wt). To assess the clinical outcomes of AML with and without RUNX1mut, we performed a prospective cohort study and systematic review and meta-analysis. The study enrolled 135 patients (27 with RUNX1mut; 108 with RUNX1wt). There were no significant differences in the median OS and RFS of the RUNX1mut and RUNX1wt groups (9.1 vs. 12.2 months; p = 0.268 and 7.8 vs. 14.6 months; p = 0.481, respectively). A subgroup analysis of de novo AML patients with intermediate-risk cytogenetics showed similar outcomes. Our meta-analysis pooled data from 23 studies and our study. The complete remission rate was significantly lower in the RUNX1mut group (pooled odds ratio: 0.42). The OS, RFS, and event-free survival rates also favored the RUNX1wt group (pooled risk ratios: 1.36, 1.37, and 1.37, respectively). A subgroup analysis of de novo AML patients with intermediate-risk cytogenetics demonstrated nearly identical OS and RFS outcomes. This study confirms that patients with AML and RUNX1mut had poor prognoses. Nonetheless, in de novo AML with intermediate-risk cytogenetics, the survival outcomes of both groups were comparable.
1. Introduction
Acute myeloid leukemia (AML) is a hematologic malignancy that results from impaired proliferation and differentiation of hematopoietic stem cells, leading to an accumulation of abnormal blast cells in bone marrow [1]. AML’s high heterogeneity is reflected in different disease prognoses, which are influenced by host- and disease-related factors [1,2,3]. For instance, old age, multiple comorbidities, and secondary AML are unfavorable predictive factors and the main obstacles for candidates of intensive therapy [1]. With regard to disease-related factors, the most essential one is molecular abnormalities [4]. The 2022 European LeukemiaNet (ELN) recommendations emphasize that cytogenetic and molecular studies are mandatory for AML diagnosis [5]. Molecular testing helps define risk stratification and identify targeted therapy [5].
The RUNX1 gene, located on chromosome 21 (21q22), regulates human hematopoiesis of all lineages [6]. The gene has a variety of biological functions, including cell differentiation, proliferation, cell cycle, DNA repair, apoptosis, ribosomal biogenesis, and metabolism [7]. RUNX1 is a recommended gene mutation that should be screened for in AML patients at diagnosis [5]. In 2016, the World Health Organization (WHO) classifications introduced “AML with mutated RUNX1 (RUNX1mut)” as a provisional entity to AML with a recurrent genetic abnormalities subtype because RUNX1mut patients had distinct clinical and genetic markers [8,9,10]. More recently, the 2022 International Consensus Classification stated that patients with RUNX1 mutations fit in the entity of AML with myelodysplasia-related gene mutations and remain in the adverse risk group [5]. However, the WHO classifications of 2022 eliminated RUNX1 mutations from the unique entity, instead defining a standalone AML type due to various clinical and molecular features [11]. A previous meta-analysis demonstrated that AML patients with RUNX1mut had increased risks of developing discouraging survival outcomes [12]. However, several recent publications have shown different survival outcomes [13,14,15]. For instance, Quesada et al. demonstrated that the overall survival (OS) and event-free survival (EFS) rates of AML patients with and without RUNX1mut were comparable [14]. Similarly, Venugopal et al. showed that both groups had similar OS rates, even among young and old patients [15].
We aimed to determine the clinical outcomes for a Southeast Asian population, for which there are presently limited data. Due to the conflicting results from previous studies, we performed a prospective study to illustrate a survival analysis of Thai AML patients with RUNX1mut and RUNX1 wild-type (RUNX1wt). Furthermore, we performed a systematic review and meta-analysis to better understand the characteristics and prognosis of overall AML patients with RUNX1mut in real-world situations.
2. Materials and Methods
This was a prospective, single-center, observational study of newly diagnosed AML patients. We also conducted a systematic review and meta-analysis that integrated our findings with all published studies’ findings. This approach allowed us to comprehensively compare the clinical outcomes of patients with RUNX1mut and those with RUNX1wt.
2.1. Prospective Cohort Study
The study was conducted on newly diagnosed AML patients with and without RUNX1mut between January 2019 and April 2022. The patients attended Siriraj Hospital, Mahidol University, Thailand, an academic university hospital and the country’s largest acute leukemia referral center. Patients were enrolled if they were over 18 years old and had a diagnosis of AML requiring treatment and follow-up. We excluded patients diagnosed with acute promyelocytic leukemia or those who had not undergone molecular testing before treatment.
Our center’s next-generation sequencing technique examined entire coding regions of 25 genes recurrently mutated in myeloid neoplasms. These genes were as follows: ASXL1, CALR, CBL, CEBPA, CSF3R, DNMT3A, EZH2, FLT3, IDH1, IDH2, JAK2, KIT, KRAS, MPL, NPM1, NRAS, RUNX1, SETBP1, SF3B1, SH2B3, SRSF2, TET2, TP53, U2AF1, and ZRSR2. The initial process was DNA extraction using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The concentration of extracted genomic DNA (gDNA) was measured using a Nanodrop System (Thermo Fisher Scientific, Waltham, MA, USA) and a Qubit dsDNA HS assay (Qubit 3.0 Fluorimeter; Life Technologies, Carlsbad, CA, USA). The GeneRead QIAact Custom Panel (Qiagen, Hilden, Germany) was used in the QIAGEN GeneReader Next-Generation Sequencing System. The heterozygous variant allele frequency (VAF) detection cutoff was at least 3%. Each variant was manually analyzed and curated by the UCSC Genome Browser, the COSMIC database, and dbSNP [16,17,18]. DNA-based assays were also investigated in NPM1 and FLT3-ITD, as reported by Stone et al. [19].
The Siriraj Institutional Review Board approved this research, which followed the Declaration of Helsinki guidelines and all subsequent amendments. All patients gave informed consent for biobanking and the analyses. The study was registered at the Thai Clinical Trial Registry (TCTR20220921005).
The primary outcome of this study was the OS rate. The secondary outcomes were the complete remission (CR) and relapse-free survival (RFS) rates, and significant factors associated with OS and RFS.
Statistical Analysis for the Cohort Study
All data analyses were designed a priori and performed using IBM SPSS Statistics for Windows, version 20.0 (Armonk, NY, USA: IBM Corp.). The sample size calculation was based on the OS rate from our previous pilot study. Details of the calculation are demonstrated in Supplementary Data S1. Demographic and baseline characteristics were summarized descriptively using 2 categories (RUNX1mut and RUNX1wt AML). Continuous variables are presented as medians and interquartile ranges or means ± standard deviations, depending on the data type. The Mann–Whitney U test was employed to compare continuous data. Categorical data are presented as numbers and percentages and compared using Fisher’s exact or Chi-squared tests. A log-rank test was used to compare the factors correlated with OS and RFS, and the results are presented as a Kaplan–Meier survival curve. Cox proportional hazards analysis was used to compare the predictors of survival outcomes in the univariate and multivariate analyses. The independent variables with significance in the univariate analysis were entered into the multivariate model. The results are expressed as hazard ratios (HRs) and 95% confidence intervals (CIs). Statistical significance was determined as a probability (p) value of <0.05.
2.2. Systematic Review and Meta-Analysis
2.2.1. Data Sources and Searches
Three investigators (T.R., W.O., and T.S.) independently searched for published articles indexed in the MEDLINE, Embase, and Cochrane Library databases from inception to April 2022. The search strategy used the terms “acute myeloid leukemia” and “molecular”. Supplementary Data S2 details the strategy for each database. The study was performed following the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analysis) statement (Supplementary Data S3) [20].
2.2.2. Selection Criteria and Data Extraction
To qualify for the meta-analysis, studies had to be either randomized controlled studies or cohort studies (prospective or retrospective) and have 2 groups of patients. The first group was RUNX1mut patients, and the other group was patients with RUNX1wt. Selected studies needed to report at least 1 of these outcomes: CR, OS, RFS, or EFS rates. Subgroup analyses included patients with de novo AML and those with intermediate-risk stratification, according to the 2017 ELN recommendations [21]. Three investigators (T.R., W.O., and T.S.) independently selected relevant articles and performed data extractions. Disagreements or questions regarding the eligibility of individual articles were discussed between the 3 investigators until a consensus was reached. Two investigators (T.R. and W.O.) subsequently examined the baseline characteristic data and the outcomes of all included studies, with the extracted data cross-checked to avoid inaccuracies.
2.2.3. Quality Assessment
Quality assessment and risk of bias were assessed using Risk of Bias in Nonrandomized Studies of Interventions (ROBINS-I) [22].
2.2.4. Statistical Analysis for the Meta-Analysis
Review Manager 5.4 software from the Cochrane Collaboration (London, UK) was used to analyze the data. The effect was estimated and combined with 95% CIs using the Mantel–Haenszel method [23]. Cochran’s Q statistic was calculated, and the statistical heterogeneity among the studies was estimated using the I2 statistic. The 4 levels of heterogeneity were based on the value of I2 as follows: (1) insignificant heterogeneity (I2 of 0–25%); (2) low heterogeneity (I2 of 26–50%); (3) moderate heterogeneity (I2 of 51–75%); and (4) high heterogeneity (I2 of 76–100%) [24]. A random-effects model was applied based on the assumption that there was heterogeneity in each study due to individual patient characteristics, treatment differences, and disease risk stratification [24]. p values less than 0.05 were considered statistically significant. Funnel plots and Egger regression were used to detect publication biases [25]. The study protocol was registered with the International Prospective Register of Systematic Review (PROSPERO) network (CRD42022327857).
2.2.5. Terminology
Our patients’ diagnoses and risk stratification were based on cytogenetic and molecular findings according to the 2017 ELN classifications [21]. Secondary AML in this cohort consisted of cases of AML that had an antecedent hematological disorder, prior chemotherapy, or radiation therapy [26]. CR was defined as bone marrow blasts below 5%, the absence of extramedullary blasts, blasts with extramedullary disease with an absolute neutrophil count exceeding 1000/μL, and a platelet count of more than 100,000/μL [21]. The duration between diagnosis and either death or the last follow-up was defined as OS. The duration from the CR date to relapse or death from any cause gave the RFS. The EFS duration was measured from the date of diagnosis to treatment failure, disease relapse, or death from any cause [27].
3. Results
3.1. Prospective Cohort Study
The study enrolled 135 patients divided into two groups based on the presence of RUNX1 mutation. Among them, 27 patients (20%) were RUNX1mut patients, while 108 (80%) were RUNX1wt patients. The cohort’s median age was 55 years (interquartile range (IQR): 40–64). However, the patients in the RUNX1mut group were significantly older than those in the RUNX1wt group, with median ages of 62 years (IQR: 55–75) and 53 years (IQR: 40–61), respectively (p = 0.008). The proportions of men and women in both groups were approximately equal. Most patients had an intermediate cytogenetic risk (73.3%) according to the 2017 ELN classifications. The most common subtype of AML was de novo AML (81.5%). However, the RUNX1mut group had a higher proportion of secondary AML patients than de novo AML (40.7% and 13%). The baseline mutational status of patients showed the presence of FLT3-ITD (22.2%), DNMT3A (22.2%), NPM1 (17.3%), TP53 (9.6%), IDH2 (8.1%), biallelic CEBPA (5.2%), EZH2 (5.2%), IDH1 (4.4%), and ASXL1 (4.4%). Among the cases, 92 were treated with intensive chemotherapy regimens, consisting of 3 + 7 or 2 + 5 regimens, while 37 were treated with low-intensity therapy (hypomethylating agents, low-dose cytarabine, hydroxyurea, or palliative treatment). Only 16 patients (11.9%) from the entire cohort underwent allogeneic stem cell transplantation due to limited access to available matched donors in Thailand. None of the patients in our cohort received targeted therapies (FLT3, IDH1, IDH2, and BCL2 inhibitors). Table 1 summarizes the patients’ baseline characteristics, laboratory investigations, and treatments. The molecular mutation profiles of the RUNX1mut and RUNX1wt groups are compared in Supplementary Data S4.

Table 1.
Baseline characteristics of patients.
3.1.1. Treatment Responses and Clinical Outcomes
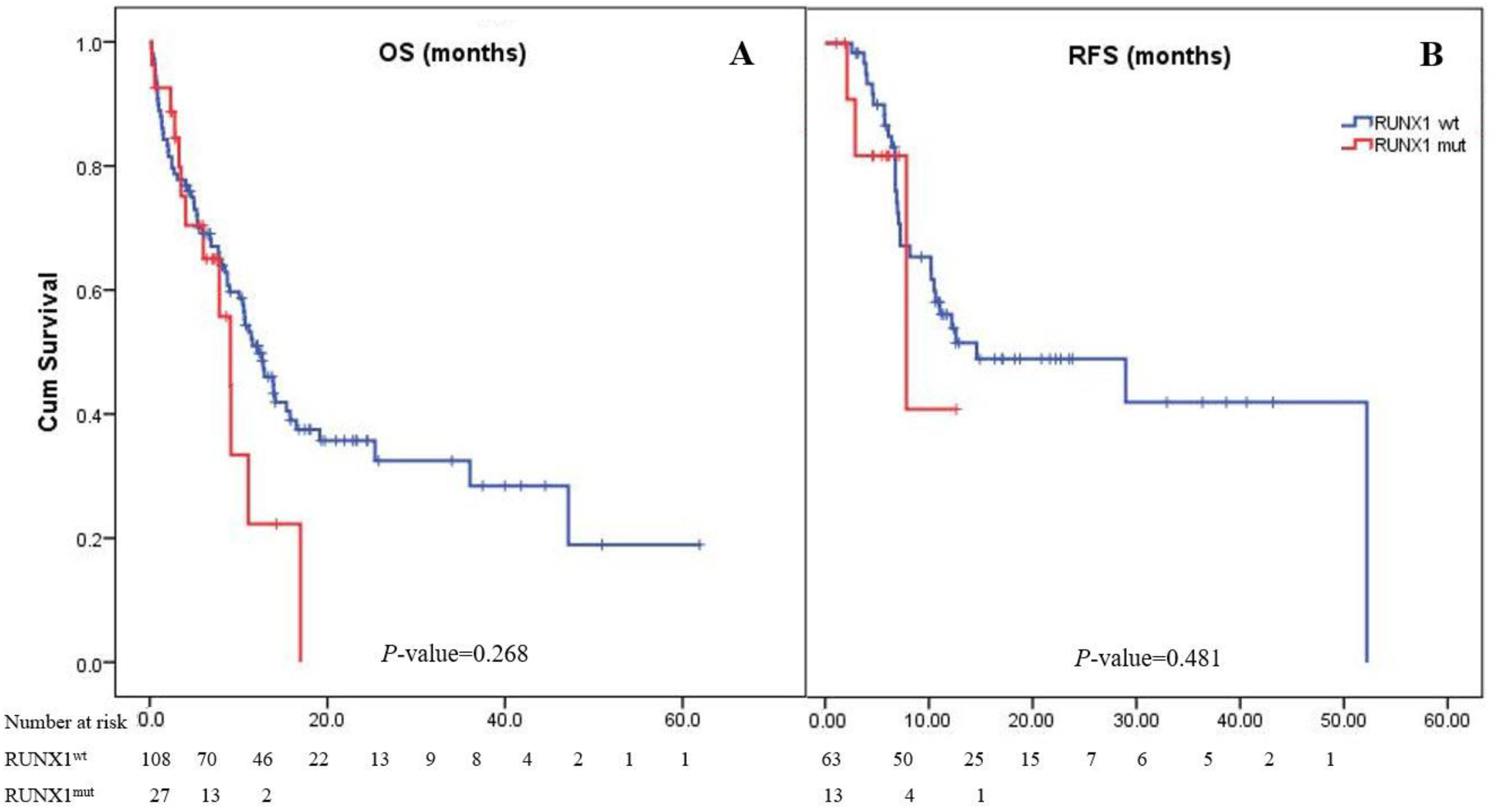
Most patients (82.6%) achieved CR after induction therapy. The CR rate in the RUNX1mut group was 68.4%, whereas the RUNX1wt group had a rate of 86.3% (pooled odds ratio (OR): 0.34; 95% CI: 0.11–1.11; p = 0.075). However, 52 (38.5%) patients experienced disease relapse (Table 1). The median follow-up period of the 135 patients in the study cohort was 2 years. The median OS of the patients in the cohort was 11.4 months (95% CI: 9.1–14.1), while the 1- and 2-year OS rates were 48% and 33%, respectively. The OS of patients with RUNX1mut (9.1 months [4.0–11.1]) was comparable to that of the RUNX1wt group (12.9 months [9.0–15.8]; p = 0.268; Figure 1). The overall RFS in this cohort was 14.6 months (95% CI: 0–31.7). The RUNX1mut AML patients had a slightly shorter RFS than the RUNX1wt patients (7.8 months [0.9–14.8] vs. 14.6 months [0–31.8]; p = 0.481; Figure 1).
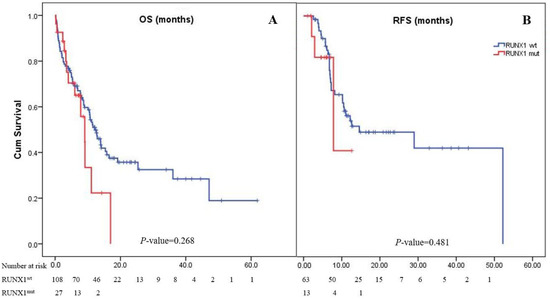
Figure 1.
Kaplan–Meier curves of overall survival (A) and relapse-free survival (B). Panel A shows Kaplan–Meier curves for overall survival corresponding to the presence or absence of the RUNX1 mutation. The median overall survival of AML patients with RUNX1mut was 9.1 months, while it was 12.9 months in patients with RUNX1wt (p = 0.268). Panel B shows Kaplan–Meier curves for relapse-free survival as assessed by the investigator. The median relapse-free survival was 7.8 months in the RUNX1mut group and 14.6 months in the RUNX1wt group (p = 0.481).
3.1.2. Factors Associated with Survival Outcomes
Comparing the factors associated with the OS of the patients from the multivariate analysis, FLT3-ITD, DNMT3A, EZH2, IDH1, and TP53 mutations were associated with poor prognoses, with HRs of 2.74, 2.19, 3.93, 3.72, and 3.15, respectively. Patients older than 60 years had poorer prognoses than those under 60 years (HR: 2.80; 95% CI: 1.44–5.45; p = 0.002). RUNX1 mutation was not associated with OS in our patients (HR: 1.42; 95% CI: 0.77–2.62; p = 0.268; Table 2).

Table 2.
Factors associated with overall survival and relapse-free survival.
Patients aged more than 60 years and the presence of poor-risk cytogenetics, the FLT3-ITD mutation, or the SRSF2 mutation were significantly associated with poorer outcomes in the RFS analysis. However, RUNX1 mutation had no impact on RFS (HR: 1.37; 95% CI: 0.41–4.62; p = 0.604; Table 2). Some factors influencing RFS outcomes might not have reached statistical significance due to the limited number of patients.
3.2. Systematic Review and Meta-Analysis
3.2.1. Study Identification, Selection, and Characteristics
We searched for potentially relevant articles published up to April 2022 in the Embase (21 058 articles), Medline (23 470 articles), and Cochrane databases (730 articles) and screened them for retrieval. Of these articles, 14 831 were excluded due to duplication, leaving 30 427 articles. Three investigators (T.R., W.O., and T.S.) performed title and abstract reviews of these papers. Articles were excluded if they met at least one of these three criteria: (1) their population differed from that evaluated in our study; (2) they did not fulfill the inclusion criteria; and (3) they did not report our outcomes of interest. This process eliminated 30 202 articles. The 225 remaining articles underwent full-length article reviews. In total, 24 studies were eventually included in our meta-analysis: 12 prospective studies [9,13,14,28,29,30,31,32,33,34,35,36], 11 retrospective studies [37,38,39,40,41,42,43,44,45,46,47]), and our prospective cohort study. Supplementary Data S5 illustrates the literature review and article selection process.
Funnel plots of the CR, OS, RFS, and EFS outcomes of the AML patients with RUNX1mut and RUNX1wt were relatively symmetrical and showed no publication bias (Supplementary Data S6). We did not conduct any other funnel plot symmetrical tests since such tests are not recommended when the standard errors of the intervention effect estimates are approximate [48]. For other biases, ROBINS-I was used for the assessments (Supplementary Data S7).
3.2.2. Baseline Patient Characteristics
A total of 8022 patients were included: 1093 RUNX1mut AML patients and 6929 RUNX1wt AML patients. The age of the participants varied markedly in both groups (from 11 to 92 years). Most patients (92.2%) had de novo AML, while 7.8% had secondary AML. According to the 2017 ELN risk classifications, the participants comprised 646 patients with favorable risk, 3861 patients with intermediate risk, and 1404 patients with poor risk. However, there were 2111 patients whose prognostic details were unavailable. Three common molecular mutations were the NPM1 mutation, FLT3-ITD mutation, and DNMT3A mutation. Most patients received standard induction chemotherapy, including the 3 + 7 regimen and the high-dose cytarabine regimen. The characteristics of the studies and patients are summarized in Table 3.

Table 3.
Patients’ baseline characteristics for studies included in the meta-analysis.
3.2.3. Treatment Response and Clinical Outcomes
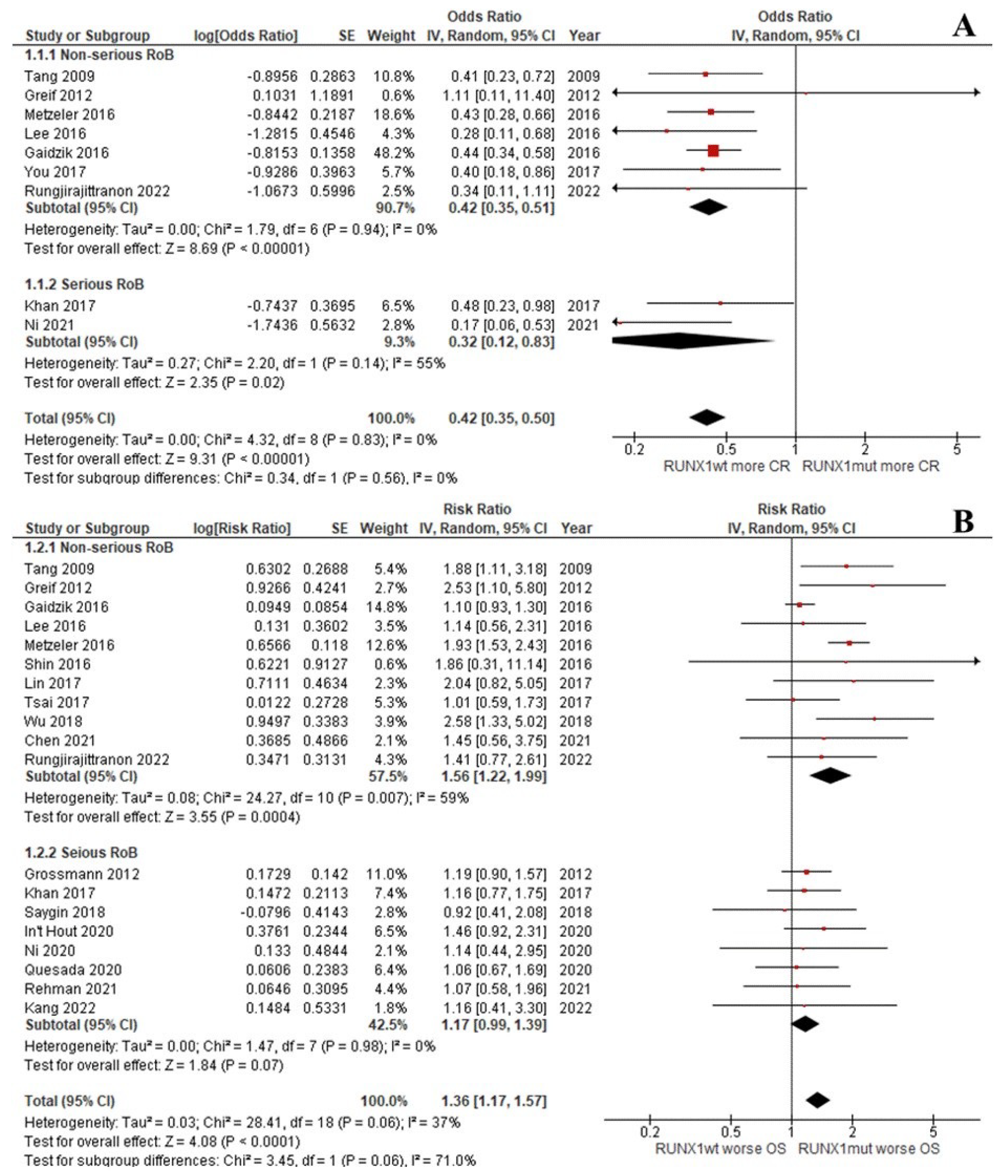
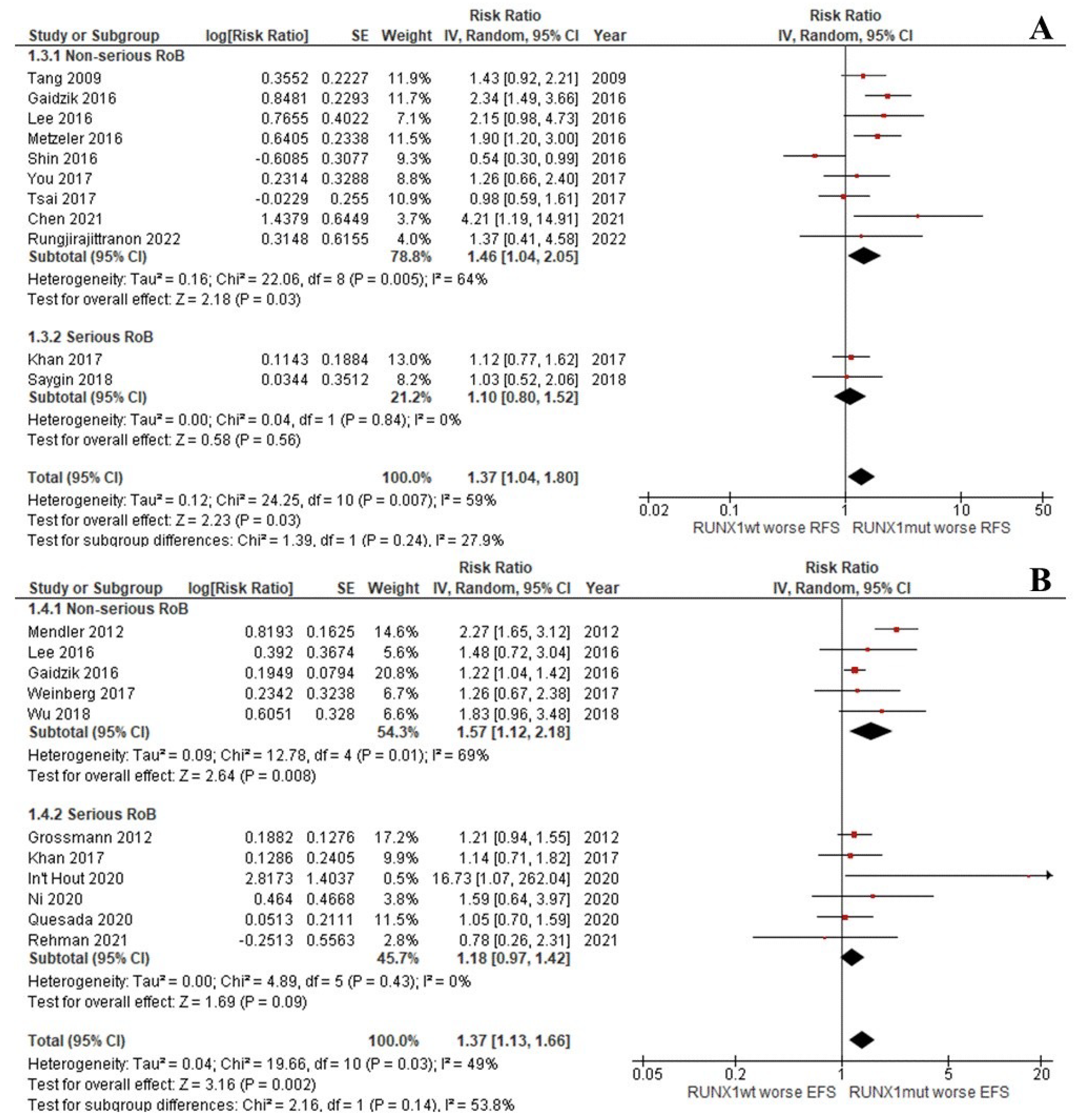
The CR rate of the patients with RUNX1mut was significantly lower than that of the RUNX1wt patients (OR: 0.42; 95% CI: 0.35–0.50; I2 = 0%; p < 0.00001; Figure 2A) [9,13,29,31,33,37,39,45]. Likewise, the OS, RFS, and EFS rates of the RUNX1mut group were significantly inferior to the corresponding values of RUNX1wt patients. For OS, the pooled risk ratio (RR) was 1.36 (95% CI: 1.17–1.57; I2 = 37%; p < 0.0001; Figure 2B) [9,13,14,29,31,32,34,35,36,37,38,39,40,42,43,44]. For RFS, the RR was 1.37 (95% CI: 1.04–1.80; I2 = 59%; p = 0.03; Figure 3A) [9,13,29,33,34,37,38,39,42,44]. For EFS, the RR was 1.37 (95% CI: 1.13–1.66; I2 = 49%; p = 0.002; Figure 3B) [9,13,30,32,35,36,37,41,43,46].
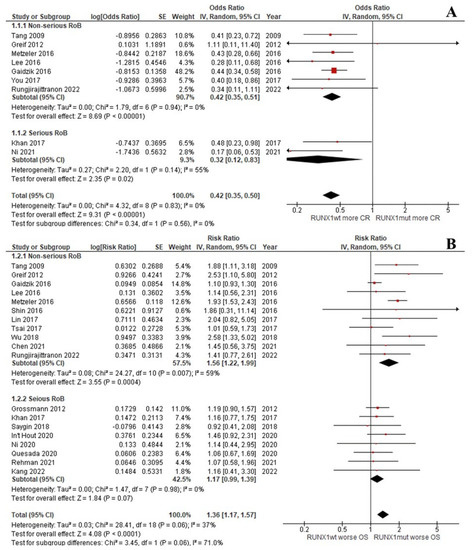
Figure 2.
Forest plot of the clinical outcomes of the RUNX1mut and RUNX1wt AML patients: (A) CR rate; (B) OS.
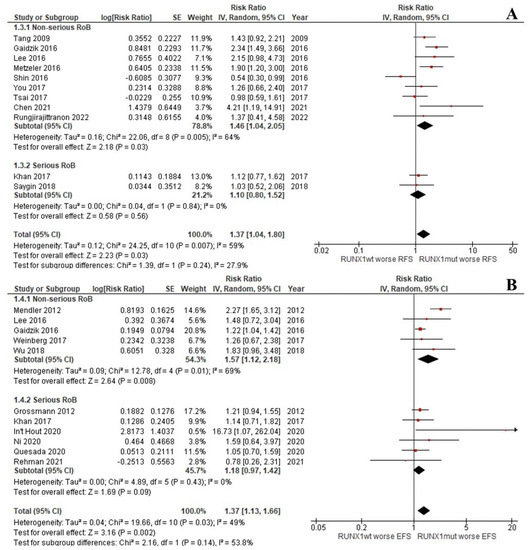
Figure 3.
Forest plot of the clinical outcomes of the RUNX1mut and RUNX1wt AML patients: (A) RFS; (B) EFS.
3.2.4. Subgroup Analyses According to AML Type
Adverse risk cytogenetics and secondary AML are unfavorable factors affecting patients’ prognoses. To better understand the clinical outcomes of AML with RUNX1mut, we focused on AML patients who did not have these dismal prognostic factors. Three subgroup analyses were performed: intermediate-risk cytogenetics, de novo AML, and de novo AML with intermediate-risk cytogenetics [14,28,29,30,31,33,34,35,36,37,38,41,42,44,46].
For AML patients with intermediate cytogenetic risks, our pooled data showed that patients in the RUNX1wt group achieved higher CR rate, with a pooled OR of 0.30 (95% CI: 0.20–0.45; I2 = 0%; p < 0.00001; Supplementary Data S8) [30,31,33,37]. Even though the OS and EFS rates were similar to those in the primary analysis [30,31,35,37,38,42,45], the RFS of the groups did not significantly differ (pooled RR: 1.25; 95% CI: 0.73–2.12; I2 = 70%; p = 0.42; Supplementary Data S8) [30,33,37,38,42].
Moving on to the outcomes of the de novo AML populations, the CR and OS rates were consistent with the full analysis [14,28,29,33,34,36,37,38,44]. The RFS and EFS rates of the RUNX1mut patients did not significantly differ between the groups, with pooled RRs of 1.23 (95% CI: 0.81–1.85; I2 = 57%; p = 0.33) and 1.25 (95% CI: 0.99–1.58; I2 = 0%; p = 0.06), respectively (Supplementary Data S9) [14,28,29,33,34,36,37,38,41,44].
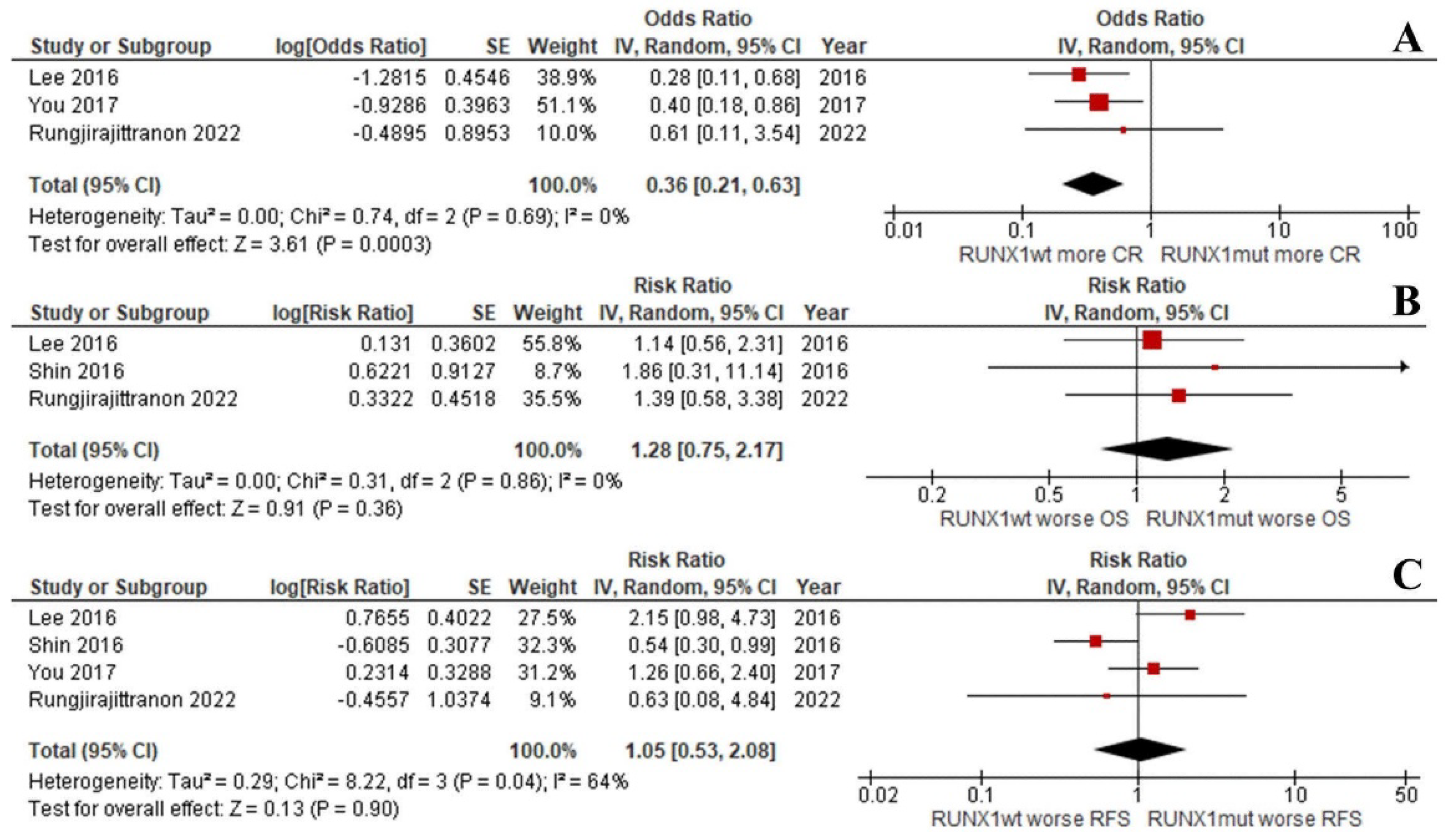
Interestingly, the OS and RFS of the groups were equal when focusing on de novo AML patients with intermediate-risk cytogenetics (RR: 1.28; 95% CI: 0.75–2.17; I2 = 0%; p = 0.36; Figure 4B and RR: 1.05; 95% CI: 0.53–2.08; I2 = 64%; p = 0.90; Figure 4C) [33,37,38]. This was despite the CR rate being significantly lower for the RUNX1mut patients (RR: 0.36; 95% CI: 0.21–0.63; I2 = 0%; p = 0.0003; Figure 4A) [37,38]. Unfortunately, the number of included studies was insufficient to compare the EFS outcomes of the two groups.
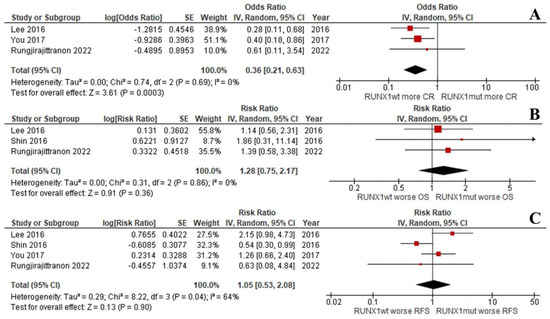
Figure 4.
Forest plot of the clinical outcomes of the RUNX1mut and RUNX1wt de novo AML patients with intermediate-risk cytogenetics: (A) CR rate; (B) OS; (C) RFS.
3.2.5. Sensitivity Analysis
4. Discussion
Our prospective study found higher proportions of AML patients with advanced age, secondary AML, and intermediate-risk cytogenetics among those with RUNX1mut than among those with RUNX1wt. These findings are consistent with the results of previous investigations [9,13,15,29,33]. Regarding molecular mutations, the ASLX1 mutation was found to be a significant co-mutation in the RUNX1mut arm. Our data show that FLT3-ITD and TP53 mutations correspond to worse prognostic factors for OS and RFS, in parallel with the risk stratification from the 2017 ELN recommendations [21]. Currently, the FLT3-ITD mutation is deemed an intermediate risk by the 2022 ELN recommendations [5]. However, our investigation found that this mutation was associated with poor outcomes. Unfortunately, the patients in our study with the FLT3-ITD mutation could not access an FLT3 inhibitor for their treatment because they could not afford the medication.
Furthermore, among the few patients with allogeneic stem cell transplantations in the present investigation’s cohort, the OS and RFS of the patients with RUNX1mut were comparable to those with RUNX1wt. We hypothesize that our results might represent the sole impact of RUNX1mut because less than 20% of the AML patients in our cohort had secondary AML, which was an independent risk factor for lower response rates, OS, and EFS [49]. Unlike AML with mutated TP53, which is a novel subtype with a homogeneously dismal prognosis, AML patients with RUNX1mut seemed to have a heterogeneous prognosis, depending on the host and disease factors [50,51,52,53,54,55].
A systematic review and meta-analysis conducted in 2018 revealed that OS and disease-free survival were almost twice as poor in patients with RUNX1mut; however, only a few studies were analyzed [12]. In addition, the meta-analysis did not conduct a subgroup analysis of AML patients without poor risk factors, such as adverse cytogenetic risk and secondary AML. Since that meta-analysis, several studies have been published. Our systematic review and meta-analysis gathered all existing studies; 4 to 245 RUNX1mut AML patients were in each study cohort, with a median of 26 cases per cohort. The number of RUNX1mut AML patients in the present investigation was as high as the median (26) of the included studies. Most of the included studies were conducted on Western populations. To better understand the prognosis of AML with RUNX1mut in non-high-risk groups, we conducted the present meta-analysis using the included studies combined with our cohort study’s results to better determine real-world outcomes.
As expected, RUNX1mut is a contributing factor to unpleasant prognoses. The de novo RUNX1mut AML patients with intermediate-risk cytogenetics had comparable OS and RFS to the RUNX1wt group, despite the RUNX1mut AML patients having a lower CR rate. As stated in the 2017 and 2022 ELN AML risk stratifications, patients with RUNX1mut should undergo allergenic stem cell transplantation [5,8]. Integrated, measurable residual disease (MRD) monitoring was recommended by the European LeukemiaNet MRD Working Party of 2021 as a surrogate marker for guiding the treatment of AML patients [56]. Patients with intermediate-risk cytogenetics will be considered for consolidation chemotherapy or autologous stem cell transplant if their MRD status is negative after two cycles of chemotherapy [56]. Venditti et al. demonstrated outcomes that supported the use of autologous stem cell transplantation in patients with favorable risk and intermediate risk with MRD negativity [57]. Concerning our study outcomes, autologous stem cell transplantation might be the treatment paradigm for RUNX1mut de novo AML patients with intermediate-risk cytogenetics who achieve negative MRD. This approach would be beneficial in countries where the availability of allogeneic stem cell transplantations is limited.
Our study had some limitations. First, the impact of co-genetic mutations could not be explored either in the prospective study or the meta-analysis. A previous study showed the significant impact of co-mutations on prognosis [4]. Unfortunately, several studies in our meta-analysis did not detail mutations other than RUNX1mut nor the impact of co-mutations. Second, some baseline characteristics, such as induction treatment regimens, MRD monitoring, and hematopoietic stem cell transplantation, were missing from observational studies. Third, limited studies were available for our subgroup analyses. Fourth, the subgroup analysis of the outcomes of patients who received either targeted therapy or allogeneic stem cell transplantation could not be evaluated in our meta-analysis due to a lack of data. Additionally, our prospective study included a small number of patients with relatively short follow-up periods. Lastly, we can see from Figure 1 that the hazards in patients with RUNX1wt and RUNX1mut were not proportionate. This finding opposes our assumption and may have led to an overestimated HR size.
5. Conclusions
Owing to the high proportion of secondary AML and elderly patients among AML patients with RUNX1mut, our study affirmed the poor prognosis of this mutation. However, the survival outcomes of de novo AML patients with intermediate-risk cytogenetics in the RUNX1wt and RUNX1mut groups might be similar.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cancers14215239/s1. Supplementary Data S1: Sample size calculation; Supplementary Data S2: Search Strategy; Supplementary Data S3: PRISMA diagram for the systematic review; Supplementary Data S4: Genetic profiling in this cohort study; Supplementary Data S5: The literature review and article selection process; Supplementary Data S6: Funnel plot for the meta-analysis of the risk ratio in OS; Supplementary Data S7: Study quality and risk of bias tool; Supplementary Data S8: Forest plot of the clinical outcomes of the RUNX1mut and RUNX1wt AML patients with intermediate-risk cytogenetics: (A) CR rate; (B) OS; (C) RFS; (D) EFS; Supplementary Data S9: Forest plot of the clinical outcomes of the RUNX1mut and RUNX1wt de novo AML patients: (A) CR rate; (B) OS; (C) RFS; (D) EFS.
Author Contributions
Conceptualization, T.R., T.S., S.K., N.L., W.R., P.V., B.M., K.K. and W.O.; data curation, T.R., T.S., W.R., P.V., B.M., K.K. and W.O.; formal analysis, N.L. and W.O.; funding acquisition, W.O.; investigation, T.R., T.S. and W.O.; methodology, T.R. and W.O.; project administration, W.O.; supervision, W.O.; validation, T.R.; writing—original draft, T.R. and T.S.; writing—review and editing, T.R., S.K., N.L., W.R. and W.O. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by the Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand.
Institutional Review Board Statement
This study was approved by the Ethics Committee for Research in Human Subjects of the Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand (Si 604/2018, approval date: 21 September 2018; and Si 461/2022, approval date: 17 June 2022).
Informed Consent Statement
Informed consent was obtained from all patients before being included in the prospective study of our cohort.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Acknowledgments
We thank Pattaraporn Tunsing for her assistance with the data collection and statistical analyses.
Conflicts of Interest
All authors declare no personal or professional conflict of interest, and further declare that no financial support was received from any companies that produce and/or distribute the drugs, devices, and materials described in this report.
Abbreviations
| AML | acute myeloid leukemia |
| CR | complete remission |
| CI | confidence interval |
| ECOG | Eastern Cooperative Oncology Group |
| EFS | event-free survival |
| ELN | European LeukemiaNet |
| HMAs | hypomethylating agents |
| HR | hazard ratio |
| IQR | interquartile range |
| OR | odds ratio |
| OS | overall survival |
| RFS | relapse-free survival |
| RR | risk ratio |
| RUNX1mut | RUNX1 mutation |
| RUNX1wt | RUNX1 wild type |
| WHO | World Health Organization |
References
- DiNardo, C.D.; Cortes, J.E. Mutations in AML: Prognostic and therapeutic implications. Hematol. Am. Soc. Hematol. Educ. Program. 2016, 2016, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.C. Genetic abnormalities and challenges in the treatment of acute myeloid leukemia. Genes Cancer 2011, 2, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, M.; Morita, K.; Yuanqing, Y.; Wang, W.; Burks, J.K.; Gumbs, C.; Little, L.; Tippen, S.; Thornton, R.; Coyle, M.; et al. Clinical heterogeneity of acute myeloid leukemia is associated with mutational heterogeneity. Blood 2018, 132 (Suppl. S1), 5240. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and Management of AML in Adults: 2022 ELN Recommendations from an International Expert Panel. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Okuda, T.; van Deursen, J.; Hiebert, S.W.; Grosveld, G.; Downing, J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 1996, 84, 321–330. [Google Scholar] [CrossRef]
- Yokota, A.; Huo, L.; Lan, F.; Wu, J.; Huang, G. The Clinical, Molecular, and Mechanistic Basis of RUNX1 Mutations Identified in Hematological Malignancies. Mol. Cells 2020, 43, 145–152. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Gaidzik, V.I.; Teleanu, V.; Papaemmanuil, E.; Weber, D.; Paschka, P.; Hahn, J.; Wallrabenstein, T.; Kolbinger, B.; Köhne, C.H.; Horst, H.A.; et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia 2016, 30, 2160–2168. [Google Scholar] [CrossRef]
- Haferlach, T.; Stengel, A.; Eckstein, S.; Perglerová, K.; Alpermann, T.; Kern, W.; Haferlach, C.; Meggendorfer, M. The new provisional WHO entity ’RUNX1 mutated AML’ shows specific genetics but no prognostic influence of dysplasia. Leukemia 2016, 30, 2109–2112. [Google Scholar] [CrossRef][Green Version]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef] [PubMed]
- Jalili, M.; Yaghmaie, M.; Ahmadvand, M.; Alimoghaddam, K.; Mousavi, S.A.; Vaezi, M.; Ghavamzadeh, A. Prognostic Value of RUNX1 Mutations in AML: A Meta-Analysis. Asian Pac. J. Cancer Prev. 2018, 19, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Cortes, J.; Kadia, T.; Naqvi, K.; Brandt, M.; Pierce, S.; Patel, K.P.; Borthakur, G.; Ravandi, F.; Konopleva, M.; et al. Clinical Outcomes and Co-Occurring Mutations in Patients with RUNX1-Mutated Acute Myeloid Leukemia. Int. J. Mol. Sci. 2017, 18, 1618. [Google Scholar] [CrossRef]
- Quesada, A.E.; Montalban-Bravo, G.; Luthra, R.; Patel, K.P.; Sasaki, K.; Bueso-Ramos, C.E.; Khoury, J.D.; Routbort, M.J.; Bassett, R.; Hidalgo-Lopez, J.E.; et al. Clinico-pathologic characteristics and outcomes of the World Health Organization (WHO) provisional entity de novo acute myeloid leukemia with mutated RUNX1. Mod. Pathol. 2020, 33, 1678–1689. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, S.; Loghavi, S.; Dinardo, C.; Konopleva, M.; Kadia, T.; Bhalla, K.; Issa, G.C.; Ravandi, F.; Daver, N.; Borthakur, G.; et al. RUNX1 Mutations in newly diagnosed acute myeloid leukemia do not adversely impact clinical outcomes in the modern era. HemaSphere 2021, 5 (Suppl. S2), 18–19. [Google Scholar]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Döhner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- Sterne, J.A.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef] [PubMed]
- Mathes, T.; Kuss, O. A comparison of methods for meta-analysis of a small number of studies with binary outcomes. Res. Synth. Methods 2018, 9, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Suurmond, R.; van Rhee, H.; Hak, T. Introduction, comparison, and validation of Meta-Essentials: A free and simple tool for meta-analysis. Res. Synth. Methods 2017, 8, 537–553. [Google Scholar] [CrossRef]
- Winer, E.S. Secondary Acute Myeloid Leukemia: A Primary Challenge of Diagnosis and Treatment. Hematol. Oncol. Clin. North Am. 2020, 34, 449–463. [Google Scholar] [CrossRef]
- Cheson, B.D.; Bennett, J.M.; Kopecky, K.J.; Büchner, T.; Willman, C.L.; Estey, E.H.; Schiffer, C.A.; Doehner, H.; Tallman, M.S.; Lister, T.A.; et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J. Clin. Oncol. 2003, 21, 4642–4649. [Google Scholar] [CrossRef]
- Schnittger, S.; Dicker, F.; Kern, W.; Wendland, N.; Sundermann, J.; Alpermann, T.; Haferlach, C.; Haferlach, T. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood 2011, 117, 2348–2357. [Google Scholar] [CrossRef]
- Tang, J.-L.; Hou, H.-A.; Chen, C.-Y.; Liu, C.-Y.; Chou, W.-C.; Tseng, M.-H.; Huang, C.-F.; Lee, F.-Y.; Liu, M.-C.; Yao, M.; et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: Prognostic implication and interaction with other gene alterations. Blood 2009, 114, 5352–5361. [Google Scholar] [CrossRef]
- Mendler, J.H.; Maharry, K.; Radmacher, M.D.; Mrózek, K.; Becker, H.; Metzeler, K.H.; Schwind, S.; Whitman, S.P.; Khalife, J.; Kohlschmidt, J.; et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J. Clin. Oncol. 2012, 30, 3109–3118. [Google Scholar] [CrossRef]
- Greif, P.A.; Konstandin, N.P.; Metzeler, K.H.; Herold, T.; Pasalic, Z.; Ksienzyk, B.; Dufour, A.; Schneider, F.; Schneider, S.; Kakadia, P.M.; et al. RUNX1 mutations in cytogenetically normal acute myeloid leukemia are associated with a poor prognosis and up-regulation of lymphoid genes. Haematologica 2012, 97, 1909–1915. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, V.; Schnittger, S.; Kohlmann, A.; Eder, C.; Roller, A.; Dicker, F.; Schmid, C.; Wendtner, C.-M.; Staib, P.; Serve, H.; et al. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood 2012, 120, 2963–2972. [Google Scholar] [CrossRef] [PubMed]
- You, E.; Cho, Y.-U.; Jang, S.; Seo, E.-J.; Lee, J.-H.; Lee, J.-H.; Lee, K.-H.; Koh, K.-N.; Im, H.-J.; Seo, J.-J.; et al. Frequency and Clinicopathologic Features of RUNX1 Mutations in Patients With Acute Myeloid Leukemia Not Otherwise Specified. Am. J. Clin. Pathol. 2017, 148, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-H.; Hou, H.-A.; Tang, J.-L.; Kuo, Y.-Y.; Chiu, Y.-C.; Lin, C.-C.; Liu, C.-Y.; Tseng, M.-H.; Lin, T.-Y.; Liu, M.-C.; et al. Prognostic impacts and dynamic changes of cohesin complex gene mutations in de novo acute myeloid leukemia. Blood Cancer J. 2017, 7, 663. [Google Scholar] [CrossRef]
- Wu, S.; Dai, Y.; Zhang, Y.; Wang, X.; Wang, L.; Ma, D.; Zhang, L.; Pang, Y.; Jiao, Y.; Niu, M.; et al. Mutational spectrum and prognostic stratification of intermediate-risk acute myeloid leukemia. Cancer Gene Ther. 2018, 25, 207–213. [Google Scholar] [CrossRef]
- Ni, J.; Hong, J.; Long, Z.; Li, Q.; Xia, R.; Zeng, Q. Mutation profile and prognostic relevance in elderly patients with de novo acute myeloid leukemia treated with decitabine-based chemotherapy. Int. J. Lab. Hematol. 2020, 42, 849–857. [Google Scholar] [CrossRef]
- Lee, S.-S.; Ahn, J.-S.; Kim, T.; Kim, H.-J.; Kim, Y.-K.; Ahn, S.-Y.; Jung, S.-H.; Yang, D.-H.; Lee, J.-J.; Park, H.J.; et al. RUNX1 Mutation in Cytogenetically Normal Acute Myeloid Leukaemia: Clinical Implications, Co-Mutation Analysis. Blood 2016, 128, 5253. [Google Scholar] [CrossRef]
- Shin, S.-Y.; Lee, S.-T.; Kim, H.-J.; Cho, E.H.; Kim, J.-W.; Park, S.; Jung, C.W.; Kim, S.-H. Mutation profiling of 19 candidate genes in acute myeloid leukemia suggests significance of DNMT3A mutations. Oncotarget 2016, 7, 54825–54837. [Google Scholar] [CrossRef]
- Metzeler, K.H.; Herold, T.; Rothenberg-Thurley, M.; Amler, S.; Sauerland, M.C.; Görlich, D.; Schneider, S.; Konstandin, N.P.; Dufour, A.; Bräundl, K.; et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 2016, 128, 686–698. [Google Scholar] [CrossRef]
- Lin, P.-H.; Li, H.-Y.; Fan, S.-C.; Yuan, T.-H.; Chen, M.; Hsu, Y.-H.; Yang, Y.-H.; Li, L.-Y.; Yeh, S.-P.; Bai, L.-Y.; et al. A targeted next-generation sequencing in the molecular risk stratification of adult acute myeloid leukemia: Implications for clinical practice. Cancer Med. 2017, 6, 349–360. [Google Scholar] [CrossRef]
- Weinberg, O.K.; Gibson, C.J.; Blonquist, T.M.; Neuberg, D.; Pozdnyakova, O.; Kuo, F.; Ebert, B.L.; Hasserjian, R.P. NPM1 mutation but not RUNX1 mutation or multilineage dysplasia defines a prognostic subgroup within de novo acute myeloid leukemia lacking recurrent cytogenetic abnormalities in the revised 2016 WHO classification. Am. J. Hematol. 2017, 92, E123–E124. [Google Scholar] [CrossRef] [PubMed]
- Saygin, C.; Hirsch, C.; Przychodzen, B.; Sekeres, M.A.; Hamilton, B.K.; Kalaycio, M.; Carraway, H.E.; Gerds, A.T.; Mukherjee, S.; Nazha, A.; et al. Mutations in DNMT3A, U2AF1, and EZH2 identify intermediate-risk acute myeloid leukemia patients with poor outcome after CR1. Blood Cancer J. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Hout, F.E.M.I.; Gerritsen, M.; Bullinger, L.; Van der Reijden, B.A.; Huls, G.; Vellenga, E.; Jansen, J.H. Transcription factor 4 (TCF4) expression predicts clinical outcome in RUNX1 mutated and translocated acute myeloid leukemia. Haematologica 2020, 105, e454–e457. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhu, H.; Qiao, C.; Zhao, S.; Liu, L.; Wang, Y.; Jin, H.; Qian, S.; Wu, Y. Next-generation sequencing reveals gene mutations landscape and clonal evolution in patients with acute myeloid leukemia. Hematology 2021, 26, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.F.; Ma, L.J.; Shi, L.L.; Shen, P.L.; Zhao, J.Q. [Clinical Characteristics of Acute Myeloid Leukemia Patients with RUNX1 Gene Mutation]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2021, 29, 1411–1416. [Google Scholar] [CrossRef]
- Rehman, A.; Akram, A.M.; Chaudhary, A.; Sheikh, N.; Hussain, Z.; Alsanie, W.F.; Rehman, R.A.; Hameed, N.; Saleem, T.; Zafar, A.; et al. RUNX1 mutation and elevated FLT3 gene expression cooperates to induce inferior prognosis in cytogenetically normal acute myeloid leukemia patients. Saudi J. Biol. Sci. 2021, 28, 4845–4851. [Google Scholar] [CrossRef]
- Kang, D.; Jung, J.; Park, S.; Cho, B.-S.; Kim, H.-J.; Kim, Y.; Lee, J.-M.; Kim, H.S.; Ahn, A.; Kim, M.; et al. Genetic Characteristics According to Subgroup of Acute Myeloid Leukemia with Myelodysplasia-Related Changes. J. Clin. Med. 2022, 11, 2378. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Sutton, A.J.; Ioannidis, J.P.A.; Terrin, N.; Jones, D.R.; Lau, J.; Carpenter, J.; Rücker, G.; Harbord, R.M.; Schmid, C.H.; et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 2011, 343, d4002. [Google Scholar] [CrossRef]
- Cheung, E.; Perissinotti, A.J.; Bixby, D.L.; Burke, P.W.; Pettit, K.M.; Benitez, L.L.; Brown, J.; Scappaticci, G.B.; Marini, B.L. The leukemia strikes back: A review of pathogenesis and treatment of secondary AML. Ann. Hematol. 2019, 98, 541–559. [Google Scholar] [CrossRef]
- Rücker, F.G.; Schlenk, R.F.; Bullinger, L.; Kayser, S.; Teleanu, V.; Kett, H.; Habdank, M.; Kugler, C.M.; Holzmann, K.; Gaidzik, V.I.; et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood 2012, 119, 2114–2121. [Google Scholar] [CrossRef]
- Middeke, J.M.; Herold, S.; Rücker-Braun, E.; Berdel, W.E.; Stelljes, M.; Kaufmann, M.; Schäfer-Eckart, K.; Baldus, C.D.; Stuhlmann, R.; Ho, A.D.; et al. TP53 mutation in patients with high-risk acute myeloid leukaemia treated with allogeneic haematopoietic stem cell transplantation. Br. J. Haematol. 2016, 172, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Montalban-Bravo, G.; Benton, C.B.; Wang, S.A.; Ravandi, F.; Kadia, T.; Cortes, J.; Daver, N.; Takahashi, K.; DiNardo, C.; Jabbour, E.; et al. More than 1 TP53 abnormality is a dominant characteristic of pure erythroid leukemia. Blood 2017, 129, 2584–2587. [Google Scholar] [CrossRef] [PubMed]
- Short, N.J.; Montalban-Bravo, G.; Hwang, H.; Ning, J.; Franquiz, M.J.; Kanagal-Shamanna, R.; Patel, K.P.; DiNardo, C.D.; Ravandi, F.; Garcia-Manero, G.; et al. Prognostic and therapeutic impacts of mutant TP53 variant allelic frequency in newly diagnosed acute myeloid leukemia. Blood Adv. 2020, 4, 5681–5689. [Google Scholar] [CrossRef] [PubMed]
- Grob, T.; Al Hinai, A.S.A.; Sanders, M.A.; Kavelaars, F.G.; Rijken, M.; Gradowska, P.L.; Biemond, B.J.; Breems, D.A.; Maertens, J.; van Marwijk Kooy, M.; et al. Molecular characterization of mutant TP53 acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood 2022, 139, 2347–2354. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, O.K.; Siddon, A.; Madanat, Y.F.; Gagan, J.; Arber, D.A.; Cin, P.D.; Narayanan, D.; Ouseph, M.M.; Kurzer, J.H.; Hasserjian, R.P. TP53 mutation defines a unique subgroup within complex karyotype de novo and therapy-related MDS/AML. Blood Adv. 2022, 6, 2847–2853. [Google Scholar] [CrossRef]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.-C.; et al. 2021 Update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [CrossRef]
- Venditti, A.; Piciocchi, A.; Candoni, A.; Melillo, L.; Calafiore, V.; Cairoli, R.; de Fabritiis, P.; Storti, G.; Salutari, P.; Lanza, F.; et al. GIMEMA AML1310 trial of risk-adapted, MRD-directed therapy for young adults with newly diagnosed acute myeloid leukemia. Blood 2019, 134, 935–945. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).