Analysis of Primary Cilium Expression and Hedgehog Pathway Activation in Mesothelioma Throws Back Its Complex Biology
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Immunofluorescence (IF)
2.3. Real-Time Reverse Transcriptase-Quantitative PCR (RT-qPCR)
2.4. Immunohistochemical Analysis
2.5. TCGA Database Analysis
3. Results
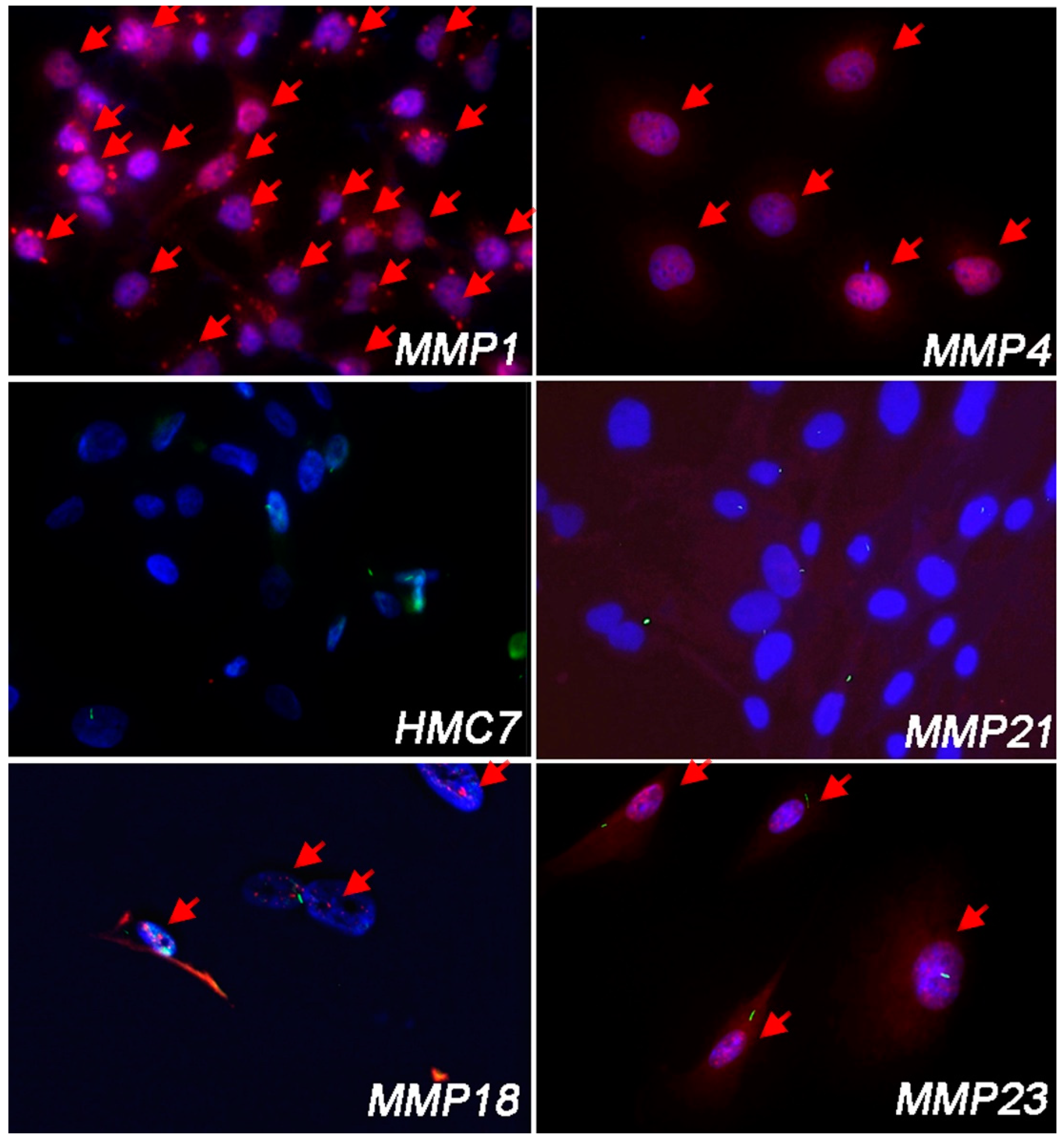
3.1. Malignant Pleural Mesothelioma (MPM) Shows an Heterogeneous Expression of Primary Cilium (PC)
3.2. Cell Lines Lacking PC Had Hh/GLI1 Pathway Activated
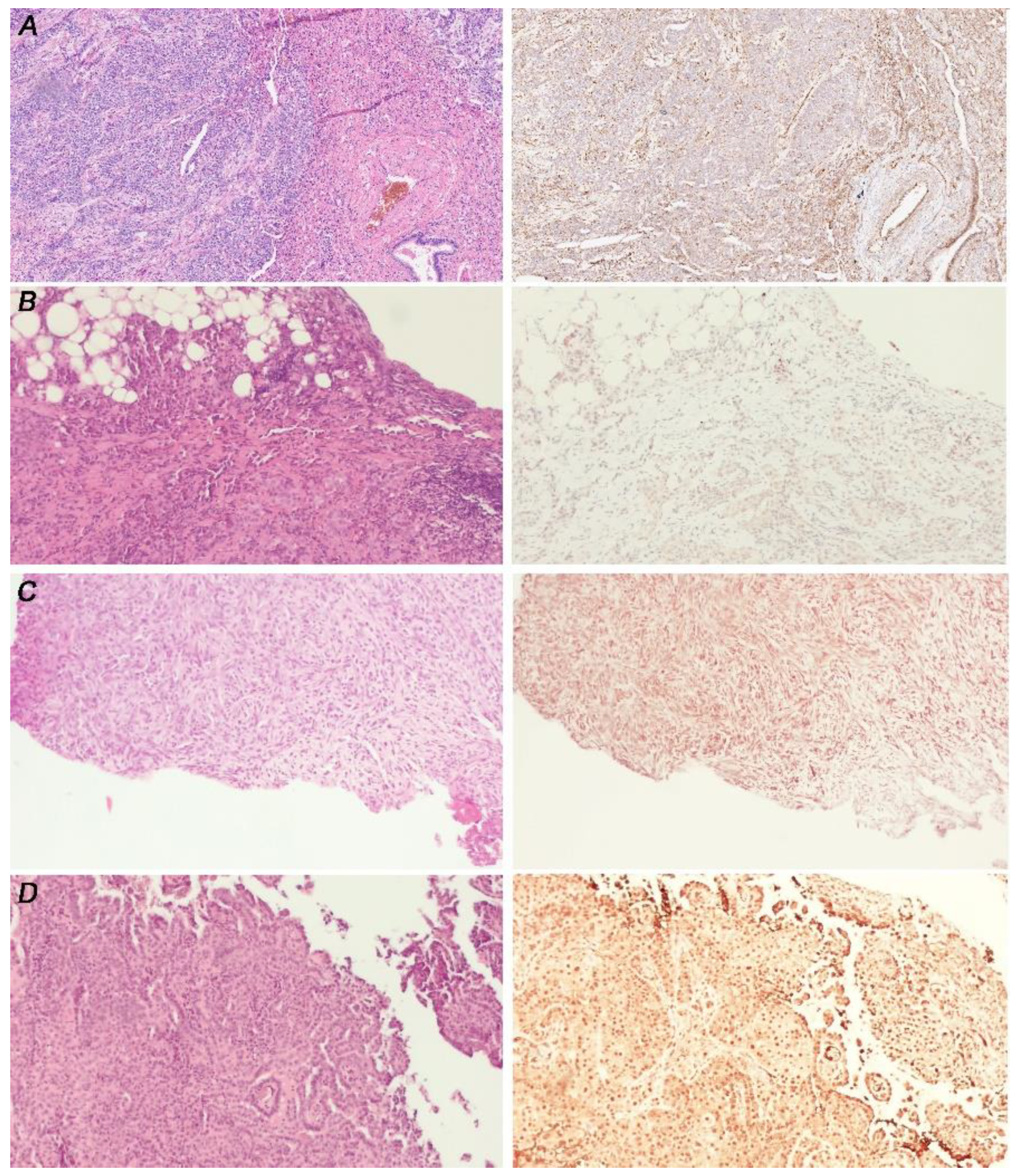
3.3. GLI1 in MPM Tissues Is Activated Independently from PC Expression
3.4. GLI1 Status Does Not Correlated with MPM Histotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Garcia, G., III; Raleigh, D.R.; Reiter, J.F. How the Ciliary Membrane Is Organized Inside-Out to Communicate Outside-In. Curr. Biol. 2018, 28, R421–R434. [Google Scholar] [CrossRef] [PubMed]
- Yanardag, S.; Pugacheva, E.N. Primary Cilium Is Involved in Stem Cell Differentiation and Renewal through the Regulation of Multiple Signaling Pathways. Cells 2021, 10, 1428. [Google Scholar] [CrossRef] [PubMed]
- Goetz, S.C.; Anderson, K.V. The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 2010, 11, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Gleeson, J.G. The primary cilium as a cellular signaling center: Lessons from disease. Curr. Opin. Genet. Dev. 2009, 19, 220–229. [Google Scholar] [CrossRef]
- Chai, J.Y.; Sugumar, V.; Alshawsh, M.A.; Wong, W.F.; Arya, A.; Chong, P.P.; Looi, C.Y. The Role of Smoothened-Dependent and -Independent Hedgehog Signaling Pathway in Tumorigenesis. Biomedicines 2021, 9, 1188. [Google Scholar] [CrossRef]
- Seeley, E.S.; Carrière, C.; Goetze, T.; Longnecker, D.S.; Korc, M. Pancreatic Cancer and Precursor Pancreatic Intraepithelial Neoplasia Lesions Are Devoid of Primary Cilia. Cancer Res. 2009, 69, 422–430. [Google Scholar] [CrossRef]
- Gradilone, S.A.; Radtke, B.N.; Bogert, P.S.; Huang, B.Q.; Gajdos, G.B.; LaRusso, N.F. HDAC6 Inhibition Restores Ciliary Expression and Decreases Tumor Growth. Cancer Res. 2013, 73, 2259–2270. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Dabiri, S.; Seeley, E.S. Primary cilium depletion typifies cutaneous melanoma in situ and malignant melanoma. PLoS ONE 2011, 6, e27410. [Google Scholar] [CrossRef]
- Hassounah, N.B.; Nagle, R.; Saboda, K.; Roe, D.J.; Dalkin, B.L.; McDermott, K.M. Primary cilia are lost in preinvasive and invasive prostate cancer. PLoS ONE 2013, 8, e68521. [Google Scholar] [CrossRef]
- Schraml, P.; Frew, I.J.; Thoma, C.R.; Boysen, G.; Struckmann, K.; Krek, W.; Moch, H. Sporadic clear cell renal cell carcinoma but not the papillary type is characterized by severely reduced frequency of primary cilia. Mod. Pathol. 2009, 22, 31–36. [Google Scholar] [CrossRef]
- Egeberg, D.L.; Lethan, M.; Manguso, R.; Schneider, L.; Awan, A.; Jørgensen, T.S.; Byskov, A.G.; Pedersen, L.B.; Christensen, S.T. Primary cilia and aberrant cell signaling in epithelial ovarian cancer. Cilia 2012, 1, 15. [Google Scholar] [CrossRef]
- Menzl, I.; Lebeau, L.; Pandey, R.; Hassounah, N.B.; Li, F.W.; Nagle, R.; Weihs, K.; McDermott, K.M. Loss of primary cilia occurs early in breast cancer development. Cilia 2014, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Coy, S.; Du, Z.; Sheu, S.-H.; Woo, T.; Rodriguez, F.J.; Kieran, M.W.; Santagata, S. Distinct patterns of primary and motile cilia in Rathke’s cleft cysts and craniopharyngioma subtypes. Mod. Pathol. 2016, 29, 1446–1459. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Y.; Seol, A.D.; So, P.-L.; Ermilov, A.N.; Bichakjian, C.K.; Epstein, E.H.J.; Dlugosz, A.A.; Reiter, J.F. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat. Med. 2009, 15, 1055–1061. [Google Scholar] [CrossRef]
- Koeniger, A.; Brichkina, A.; Nee, I.; Dempwolff, L.; Hupfer, A.; Galperin, I.; Finkernagel, F.; Nist, A.; Stiewe, T.; Adhikary, T.; et al. Activation of Cilia-Independent Hedgehog/GLI1 Signaling as a Novel Concept for Neuroblastoma Therapy. Cancers 2021, 13, 1908. [Google Scholar] [CrossRef] [PubMed]
- Sénicourt, B.; Boudjadi, S.; Carrier, J.C.; Beaulieu, J.-F. Neoexpression of a functional primary cilium in colorectal cancer cells. Heliyon 2016, 2, e00109. [Google Scholar] [CrossRef]
- Iruzubieta, P.; Monzón, M.; Castiella, T.; Ramírez, T.; Junquera, C. Hedgehog signalling pathway activation in gastrointestinal stromal tumours is mediated by primary cilia. Gastric Cancer 2020, 23, 64–72. [Google Scholar] [CrossRef]
- Shinmura, K.; Kusafuka, K.; Kawasaki, H.; Kato, H.; Hariyama, T.; Tsuchiya, K.; Kawanishi, Y.; Funai, K.; Misawa, K.; Mineta, H.; et al. Identification and characterization of primary cilia-positive salivary gland tumours exhibiting basaloid/myoepithelial differentiation. J. Pathol. 2021, 254, 519–530. [Google Scholar] [CrossRef]
- Scales, S.J.; de Sauvage, F.J. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 2009, 30, 303–312. [Google Scholar] [CrossRef]
- Sigafoos, A.N.; Paradise, B.D.; Fernandez-Zapico, M.E. Hedgehog/GLI Signaling Pathway: Transduction, Regulation, and Implications for Disease. Cancers 2021, 13, 3410. [Google Scholar] [CrossRef]
- Pak, E.; Segal, R.A. Hedgehog Signal Transduction: Key Players, Oncogenic Drivers, and Cancer Therapy. Dev. Cell 2016, 38, 333–344. [Google Scholar] [CrossRef]
- Johnson, R.L.; Rothman, A.L.; Xie, J.; Goodrich, L.V.; Bare, J.W.; Bonifas, J.M.; Quinn, A.G.; Myers, R.M.; Cox, D.R.; Epstein, E.H.J.; et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 1996, 272, 1668–1671. [Google Scholar] [CrossRef] [PubMed]
- Chuang, P.T.; McMahon, A.P. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 1999, 397, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Jeng, K.-S.; Chang, C.-F.; Lin, S.-S. Sonic Hedgehog Signaling in Organogenesis, Tumors, and Tumor Microenvironments. Int. J. Mol. Sci. 2020, 21, 758. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, H.J.; Pau, G.; Dijkgraaf, G.J.; Basset-Seguin, N.; Modrusan, Z.; Januario, T.; Tsui, V.; Durham, A.B.; Dlugosz, A.A.; Haverty, P.M.; et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell 2015, 27, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Pietrobono, S.; Gagliardi, S.; Stecca, B. Non-canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors Beyond Smoothened. Front. Genet. 2019, 10, 556. [Google Scholar] [CrossRef]
- IARC. Thoracic Tumours, WHO Calssification of Tumors, 5th ed.; IARC: Sydney, NSW, Australia, 2021.
- Abbott, D.M.; Bortolotto, C.; Benvenuti, S.; Lancia, A.; Filippi, A.R.; Stella, G.M. Malignant Pleural Mesothelioma: Genetic and Microenviromental Heterogeneity as an Unexpected Reading Frame and Therapeutic Challenge. Cancers 2020, 12, 1186. [Google Scholar] [CrossRef]
- Cersosimo, F.; Barbarino, M.; Lonardi, S.; Vermi, W.; Giordano, A.; Bellan, C.; Giurisato, E. Mesothelioma malignancy and the microenvironment: Molecular mechanisms. Cancers 2021, 13, 5664. [Google Scholar] [CrossRef]
- Zalcman, G.; Mazieres, J.; Margery, J.; Greillier, L.; Audigier-Valette, C.; Moro-Sibilot, D.; Molinier, O.; Corre, R.; Monnet, I.; Gounant, V.; et al. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): A randomised, controlled, open-label, phase 3 trial. Lancet 2016, 387, 1405–1414. [Google Scholar] [CrossRef]
- De Gooijer, C.J.; Borm, F.J.; Scherpereel, A.; Baas, P. Immunotherapy in Malignant Pleural Mesothelioma. Front. Oncol. 2020, 10, 187. [Google Scholar] [CrossRef]
- Li, H.; Lui, N.; Cheng, T.; Tseng, H.-H.K.; Yue, D.; Giroux-Leprieur, E.; Do, H.T.; Sheng, Q.; Jin, J.Q.; Luh, T.W.; et al. Gli as a novel therapeutic target in malignant pleural mesothelioma. PLoS ONE 2013, 8, e57346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, J.; Zhang, F.; Li, H.; Yue, D.; Wang, C.; Jablons, D.M.; He, B.; Lui, N. SMO expression level correlates with overall survival in patients with malignant pleural mesothelioma. J. Exp. Clin. Cancer Res. 2013, 32, 7. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, D.; Proto, C.; Botta, L.; Trama, A.; Tiseo, M.; Pasello, G.; Lo Russo, G.; Fabbri, A.; Imbimbo, M.; Busico, A.; et al. SMO mutations confer poor prognosis in malignant pleural mesothelioma. Transl. Lung Cancer Res. 2020, 9, 1940–1951. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Moura, U.; Opitz, I.; Soltermann, A.; Rehrauer, H.; Thies, S.; Weder, W.; Stahel, R.A.; Felley-Bosco, E. Role of hedgehog signaling in malignant pleural mesothelioma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 4646–4656. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Varona-Santos, J.; Singh, S.; Robbins, D.J.; Savaraj, N.; Nguyen, D.M. Targeting of the Hedgehog signal transduction pathway suppresses survival of malignant pleural mesothelioma cells in vitro. J. Thorac. Cardiovasc. Surg. 2014, 147, 508–516. [Google Scholar] [CrossRef]
- Meerang, M.; Bérard, K.; Felley-Bosco, E.; Lauk, O.; Vrugt, B.; Boss, A.; Kenkel, D.; Broggini-Tenzer, A.; Stahel, R.A.; Arni, S.; et al. Antagonizing the Hedgehog Pathway with Vismodegib Impairs Malignant Pleural Mesothelioma Growth In Vivo by Affecting Stroma. Mol. Cancer Ther. 2016, 15, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Dusek, C.O.; Hadden, M.K. Targeting the GLI family of transcription factors for the development of anti-cancer drugs. Expert Opin. Drug Discov. 2021, 16, 289–302. [Google Scholar] [CrossRef]
- Kotsiou, O.; Jagirdar, R.; Rouka, E.; Pitaraki, E.; Gourgoulianis, K.; Zarogiannis, S. Primary cilium regulates cell migration and mechanical contraction in mesothelioma cell models. Eur. Respir. J. 2021, 58, PA3103. [Google Scholar] [CrossRef]
- Felley-Bosco, E.; Opitz, I.; Meerang, M. Hedgehog Signaling in Malignant Pleural Mesothelioma. Genes 2015, 6, 500–511. [Google Scholar] [CrossRef]
- Barbarino, M.; Cesari, D.; Bottaro, M.; Luzzi, L.; Namagerdi, A.; Bertolino, F.M.; Bellan, C.; Proietti, F.; Somma, P.; Micheli, M.; et al. PRMT5 silencing selectively affects MTAP-deleted mesothelioma: In vitro evidence of a novel promising approach. J. Cell. Mol. Med. 2020, 24, 5565–5577. [Google Scholar] [CrossRef]
- Mukhopadhya, I.; Murray, G.I.; Berry, S.; Thomson, J.; Frank, B.; Gwozdz, G.; Ekeruche-Makinde, J.; Shattock, R.; Kelly, C.; Iannelli, F.; et al. Drug transporter gene expression in human colorectal tissue and cell lines: Modulation with antiretrovirals for microbicide optimization. J. Antimicrob. Chemother. 2016, 71, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Erhard, F. Estimating pseudocounts and fold changes for digital expression measurements. Bioinformatics 2018, 34, 4054–4063. [Google Scholar] [CrossRef] [PubMed]
- Broad Institute of Mit and Harvard; Broad Institute Tcga Genome Data Analysis Center. Paradigm Pathway Analysis of Mrnaseq Expression and Copy Number Data; Broad Institute of Mit and Harvard: Cambridge, MA, USA, 2014. [Google Scholar]
- Yoon, J.W.; Gallant, M.; Lamm, M.L.G.; Iannaccone, S.; Vieux, K.-F.; Proytcheva, M.; Hyjek, E.; Iannaccone, P.; Walterhouse, D. Noncanonical regulation of the Hedgehog mediator GLI1 by c-MYC in Burkitt lymphoma. Mol. Cancer Res. 2013, 11, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Dell’Anno, I.; Martin, S.A.; Barbarino, M.; Melani, A.; Silvestri, R.; Bottaro, M.; Paolicchi, E.; Corrado, A.; Cipollini, M.; Melaiu, O.; et al. Drug-repositioning screening identified fludarabine and risedronic acid as potential therapeutic compounds for malignant pleural mesothelioma. Investig. New Drugs 2021, 39, 644–657. [Google Scholar] [CrossRef] [PubMed]
- Dell’Anno, I.; Melani, A.; Martin, S.A.; Barbarino, M.; Silvestri, R.; Cipollini, M.; Giordano, A.; Mutti, L.; Nicolini, A.; Luzzi, L.; et al. A Drug Screening Revealed Novel Potential Agents against Malignant Pleural Mesothelioma. Cancers 2022, 14, 2527. [Google Scholar] [CrossRef] [PubMed]
- LoRusso, P.M.; Rudin, C.M.; Reddy, J.C.; Tibes, R.; Weiss, G.J.; Borad, M.J.; Hann, C.L.; Brahmer, J.R.; Chang, I.; Darbonne, W.C.; et al. Phase I Trial of Hedgehog Pathway Inhibitor Vismodegib (GDC-0449) in Patients with Refractory, Locally Advanced or Metastatic Solid Tumors. Clin. Cancer Res. 2011, 17, 2502–2511. [Google Scholar] [CrossRef] [PubMed]
- Popat, S.; Sharma, B.; MacMahon, S.; Nicholson, A.G.; Sharma, R.K.; Schuster, K.; Lang Lazdunski, L.; Fennell, D. Durable Response to Vismodegib in PTCH1 F1147fs Mutant Relapsed Malignant Pleural Mesothelioma: Implications for Mesothelioma Drug Treatment. JCO Precis. Oncol. 2021, 5, 39–43. [Google Scholar] [CrossRef]
- Rodon, J.; Tawbi, H.A.; Thomas, A.L.; Stoller, R.G.; Turtschi, C.P.; Baselga, J.; Sarantopoulos, J.; Mahalingam, D.; Shou, Y.; Moles, M.A.; et al. A Phase I, Multicenter, Open-Label, First-in-Human, Dose-Escalation Study of the Oral Smoothened Inhibitor Sonidegib (LDE225) in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2014, 20, 1900–1909. [Google Scholar] [CrossRef]
- Lim, C.B.; Prêle, C.M.; Cheah, H.M.; Cheng, Y.Y.; Klebe, S.; Reid, G.; Watkins, D.N.; Baltic, S.; Thompson, P.J.; Mutsaers, S.E. Mutational analysis of hedgehog signaling pathway genes in human malignant mesothelioma. PLoS ONE 2013, 8, e66685. [Google Scholar] [CrossRef]
- Rouka, E.; Hatzoglou, C.; Gourgoulianis, K.; Zarogiannis, S. Effect of primary cilium-associated genes expression on the survival of mesothelioma patients: In silico investigation of TCGA data. Eur. Respir. J. 2020, 56, 1135. [Google Scholar] [CrossRef]
- Curran, T. Reproducibility of academic preclinical translational research: Lessons from the development of Hedgehog pathway inhibitors to treat cancer. Open Biol. 2022, 8, 180098. [Google Scholar] [CrossRef] [PubMed]
- Lospinoso Severini, L.; Quaglio, D.; Basili, I.; Ghirga, F.; Bufalieri, F.; Caimano, M.; Balducci, S.; Moretti, M.; Romeo, I.; Loricchio, E.; et al. A Smo/Gli Multitarget Hedgehog Pathway Inhibitor Impairs Tumor Growth. Cancers 2019, 11, 1518. [Google Scholar] [CrossRef] [PubMed]
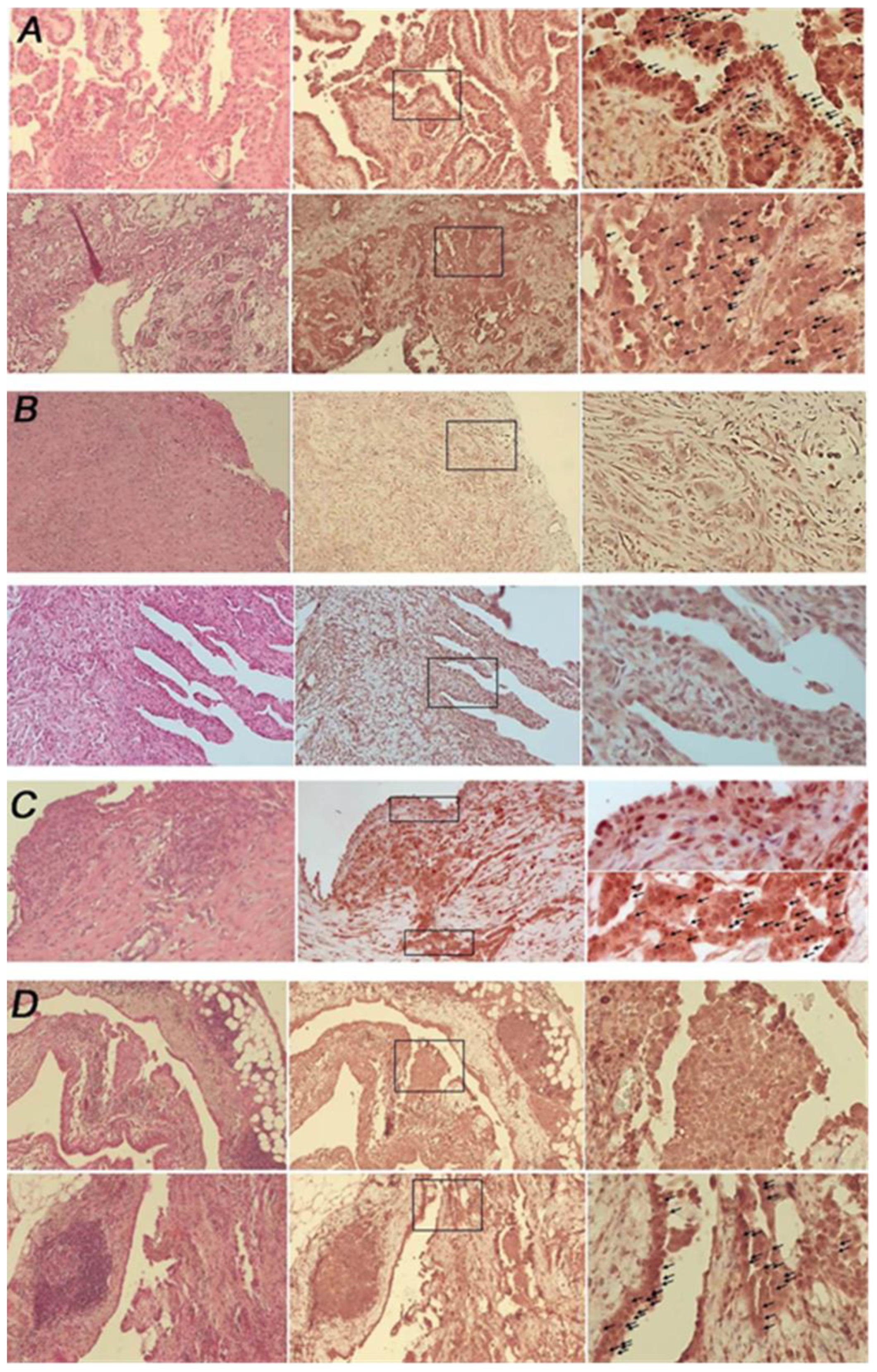



| Specimens’ Histotype | PC Status (%) |
|---|---|
| Epithelioid (n = 19) | Negative: 42.1% (n = 8) Positive: 36.8% (n = 7) Mix= 21.0% (n = 4) |
| Biphasic (n = 4) and desmoplastic (n = 1) | Negative = 100% |
| Pleuritis (n = 2) and reactive mesothelial hyperplasia (n = 2) | Positive = 100% |
| Pc-Positive (%) Non-Malignant Mesothelial Cells | |
| HMC13 | 34.7 |
| HMC12 | 46.7 |
| HMC7 | 40.3 |
| LP-9 | 68.6 |
| PC-Positive (%) MPM Cells | |
| MMP1 | 0% |
| MMP4 | 0% |
| MMP14 | 0% |
| MMP18A | 46.8% |
| MMP21 | 71.3% |
| MMP23 | 80.2% |
| MMP32 | 23.8% |
| MMP43 | 0% |
| GLI1 Score | PC Positive (%) (n = 5) | PC Negative (%) (n = 14) |
|---|---|---|
| 0 | 40 | 35.7 |
| 1 | 0 | 7.1 |
| 2 | 20 | 35.7 |
| 3 | 40 | 21.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbarino, M.; Bottaro, M.; Spagnoletti, L.; de Santi, M.M.; Guazzo, R.; Defraia, C.; Custoza, C.; Serio, G.; Iannelli, F.; Pesetti, M.; et al. Analysis of Primary Cilium Expression and Hedgehog Pathway Activation in Mesothelioma Throws Back Its Complex Biology. Cancers 2022, 14, 5216. https://doi.org/10.3390/cancers14215216
Barbarino M, Bottaro M, Spagnoletti L, de Santi MM, Guazzo R, Defraia C, Custoza C, Serio G, Iannelli F, Pesetti M, et al. Analysis of Primary Cilium Expression and Hedgehog Pathway Activation in Mesothelioma Throws Back Its Complex Biology. Cancers. 2022; 14(21):5216. https://doi.org/10.3390/cancers14215216
Chicago/Turabian StyleBarbarino, Marcella, Maria Bottaro, Laura Spagnoletti, Maria Margherita de Santi, Raffaella Guazzo, Chiara Defraia, Cosimo Custoza, Gabriella Serio, Francesco Iannelli, Matilde Pesetti, and et al. 2022. "Analysis of Primary Cilium Expression and Hedgehog Pathway Activation in Mesothelioma Throws Back Its Complex Biology" Cancers 14, no. 21: 5216. https://doi.org/10.3390/cancers14215216
APA StyleBarbarino, M., Bottaro, M., Spagnoletti, L., de Santi, M. M., Guazzo, R., Defraia, C., Custoza, C., Serio, G., Iannelli, F., Pesetti, M., Aiello, R., Rosati, D., Zanfrini, E., Luzzi, L., Bellan, C., & Giordano, A. (2022). Analysis of Primary Cilium Expression and Hedgehog Pathway Activation in Mesothelioma Throws Back Its Complex Biology. Cancers, 14(21), 5216. https://doi.org/10.3390/cancers14215216









