Tumor-Associated Macrophages Promote Metastasis of Oral Squamous Cell Carcinoma via CCL13 Regulated by Stress Granule
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Patients and Specimens
2.3. Multiplex Immunohistochemistry (mIHC)
2.4. Immunohistochemistry (IHC) and Immunofluorescence Staining (IF) of Tumor Tissues
2.5. Cell Immunofluorescence (IF)
2.6. Flow Cytometry
2.7. siRNA Transfection for Macrophages
2.8. Plasmid Construct
2.9. Transwell Migration Assay and Wound Healing Assay
2.10. ELISA
2.11. Real-Time Quantitative PCR (RT-qPCR) and Western Blot
2.12. RNA Sequencing
2.13. Coimmunoprecipitation and Mass Spectrometry
2.14. Fluorescence In Situ Hybridization (FISH)
2.15. RNA Immunoprecipitation (RIP)
2.16. RNA Stability Assay
2.17. H&E Staining
2.18. Orthotopic Metastasis Model in the Mouse Tongue
2.19. Statistical Analysis
3. Results
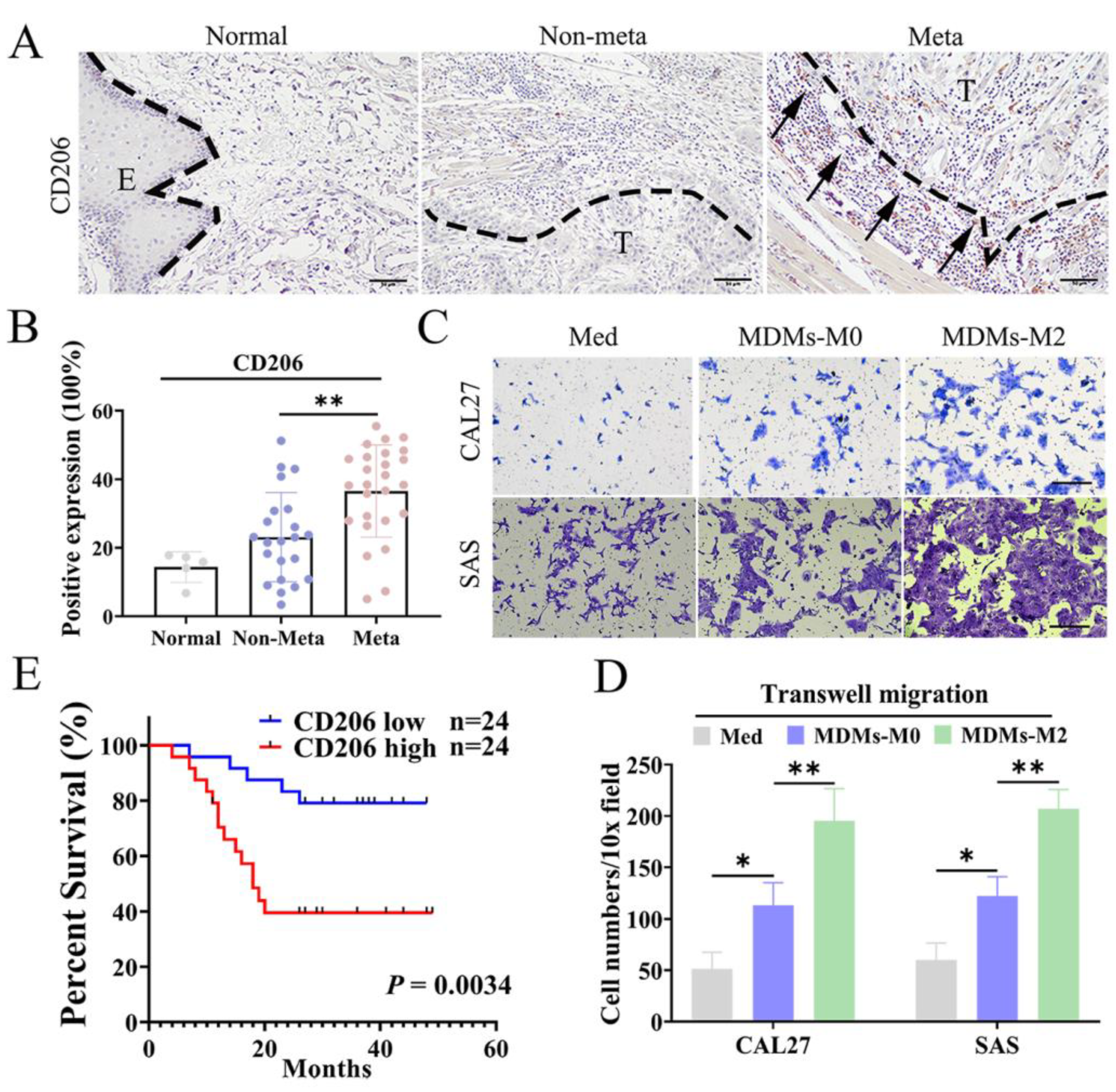
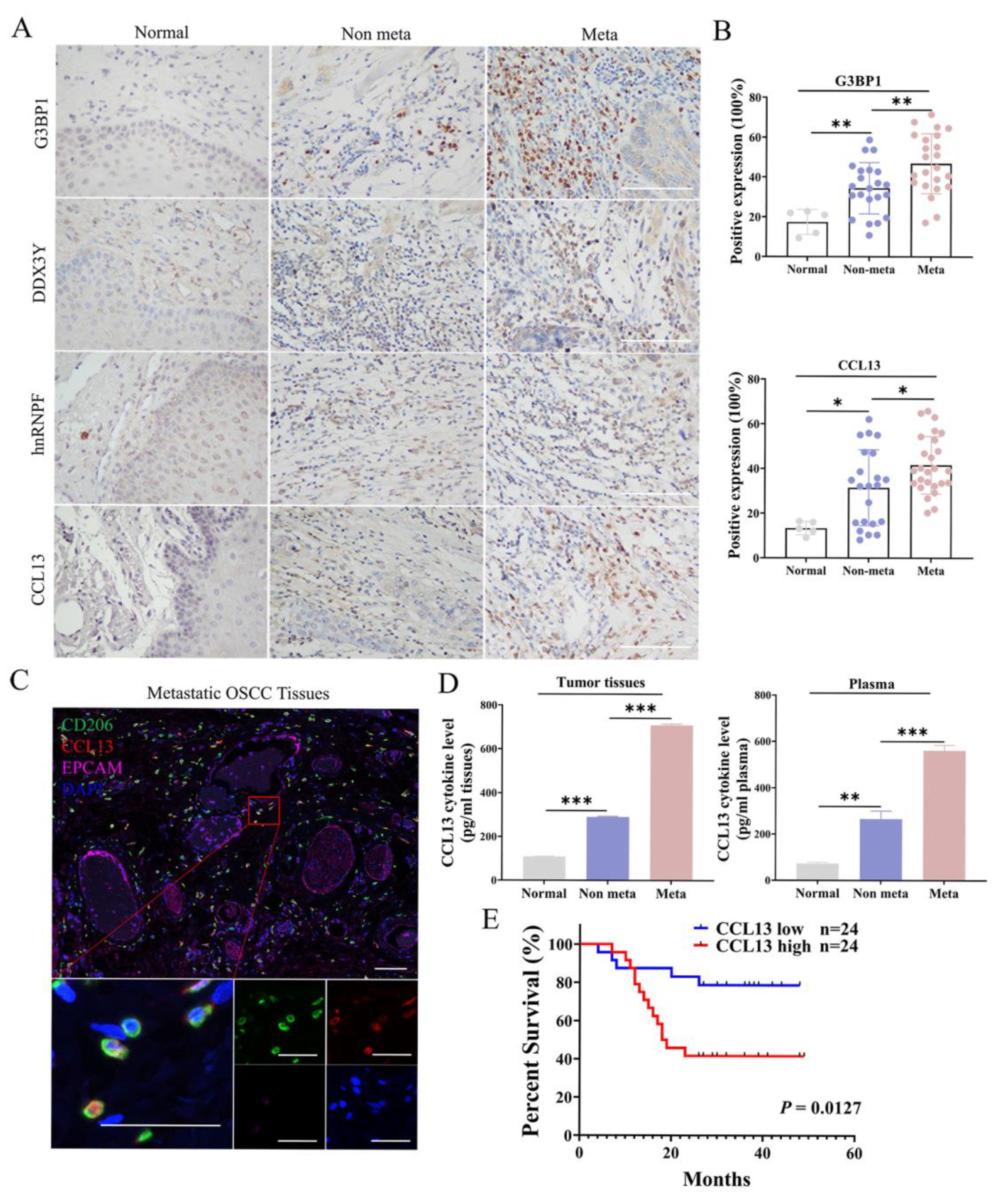
3.1. M2 TAMs Correlated with Metastasis and Poor Prognosis of OSCC Patients
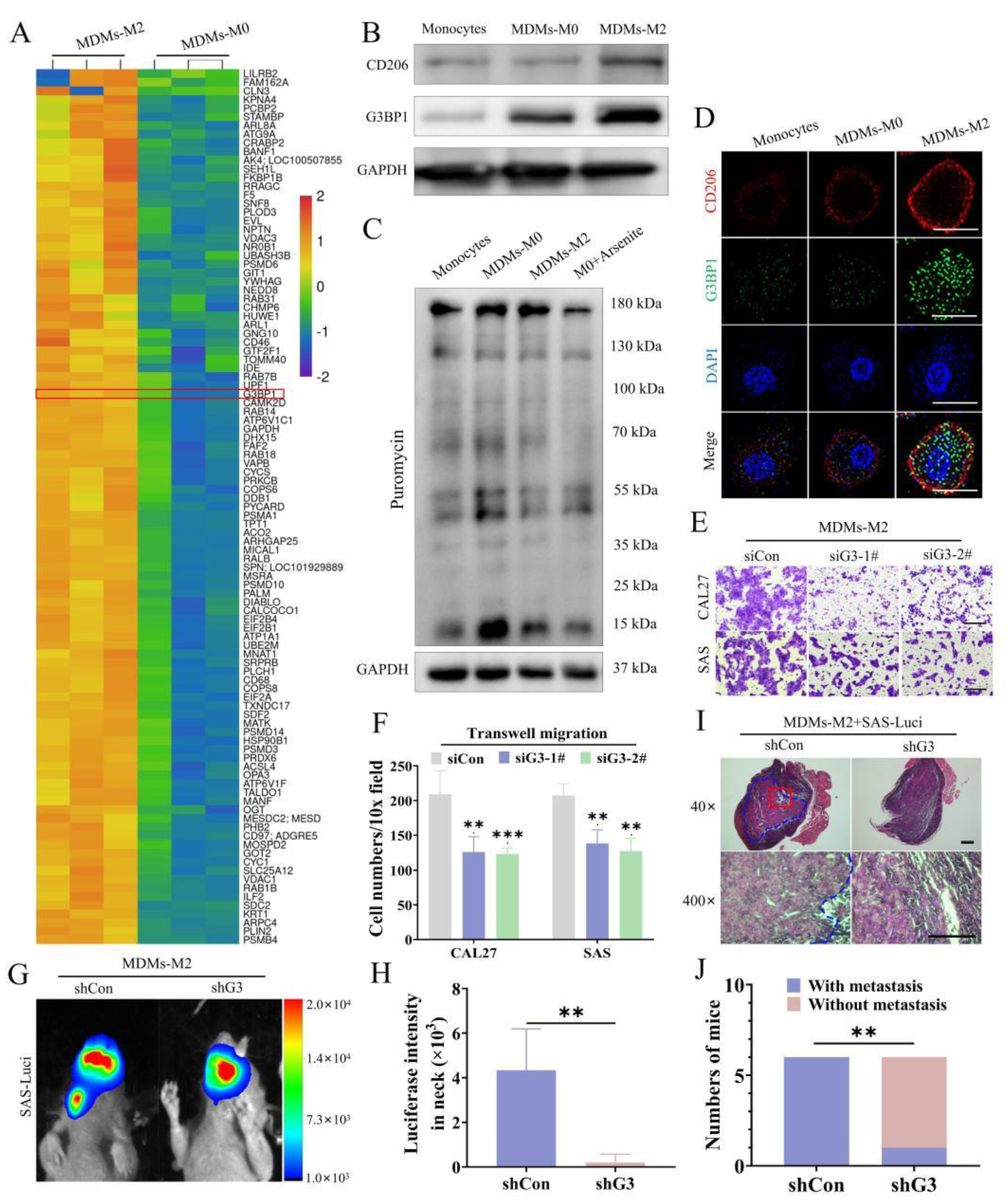
3.2. Inhibition of SG Formation Attenuated the Protumor Effects of M2 TAMs
3.3. Disruption of SG Decreased the Production of CCL13 in M2 TAMs
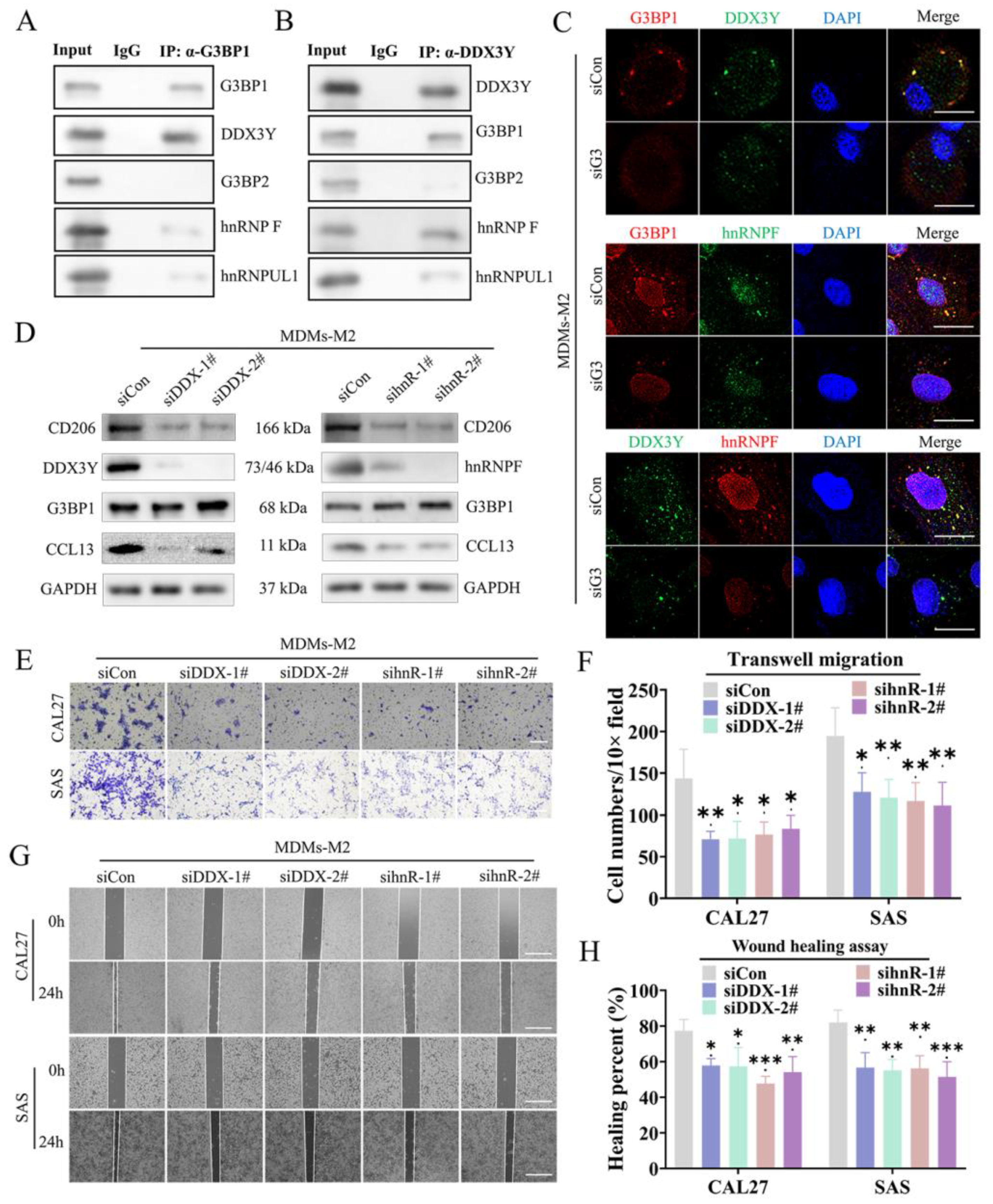
3.4. The SG Mediated the Direct Connection between G3BP1 and DDX3Y-hnRNPF to Regulate the Expression of CCL13
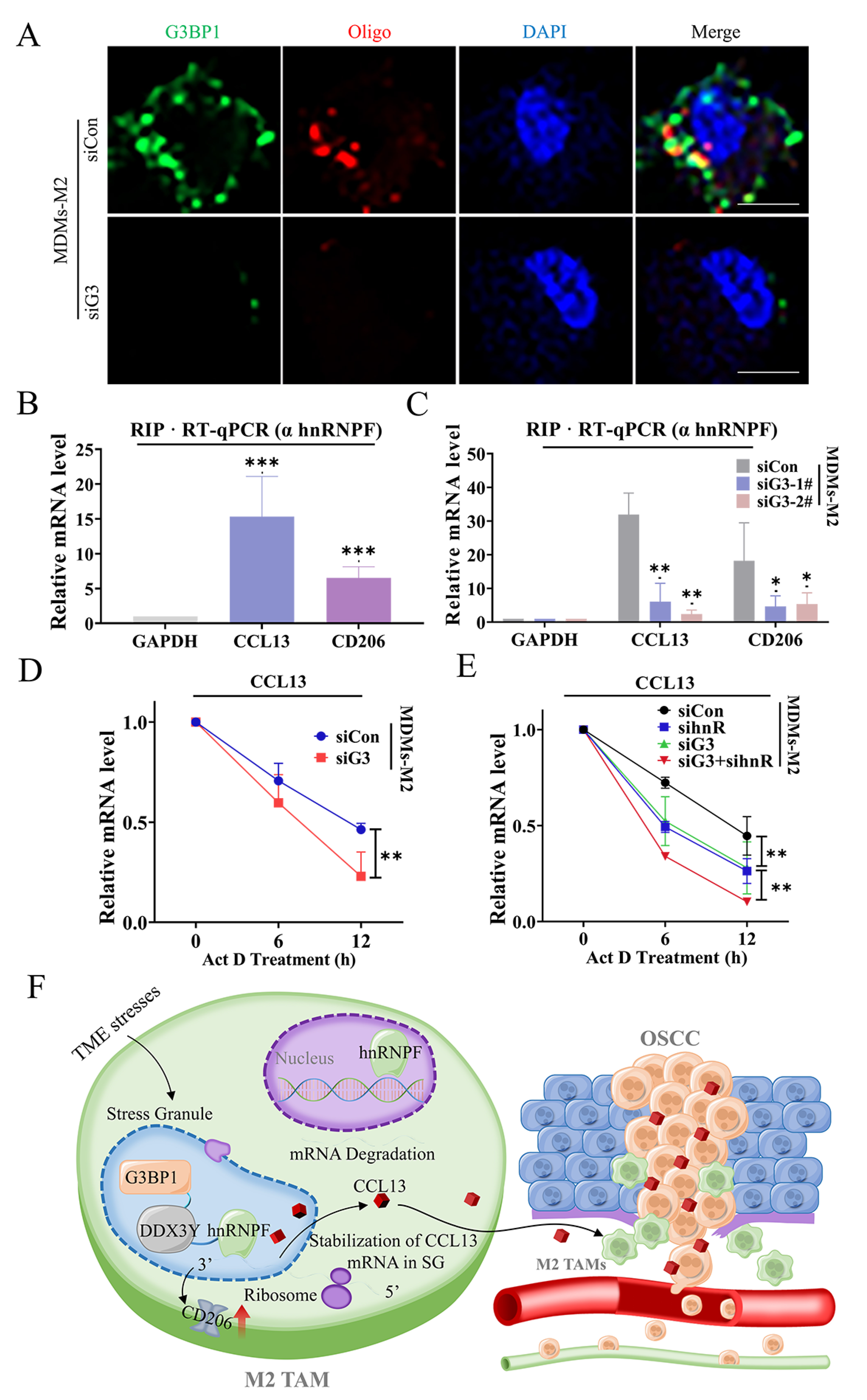
3.5. SG Enhanced DDX3Y/hnRNPF-Mediated CCL13 mRNA Stability via Binding with Its Transcripts
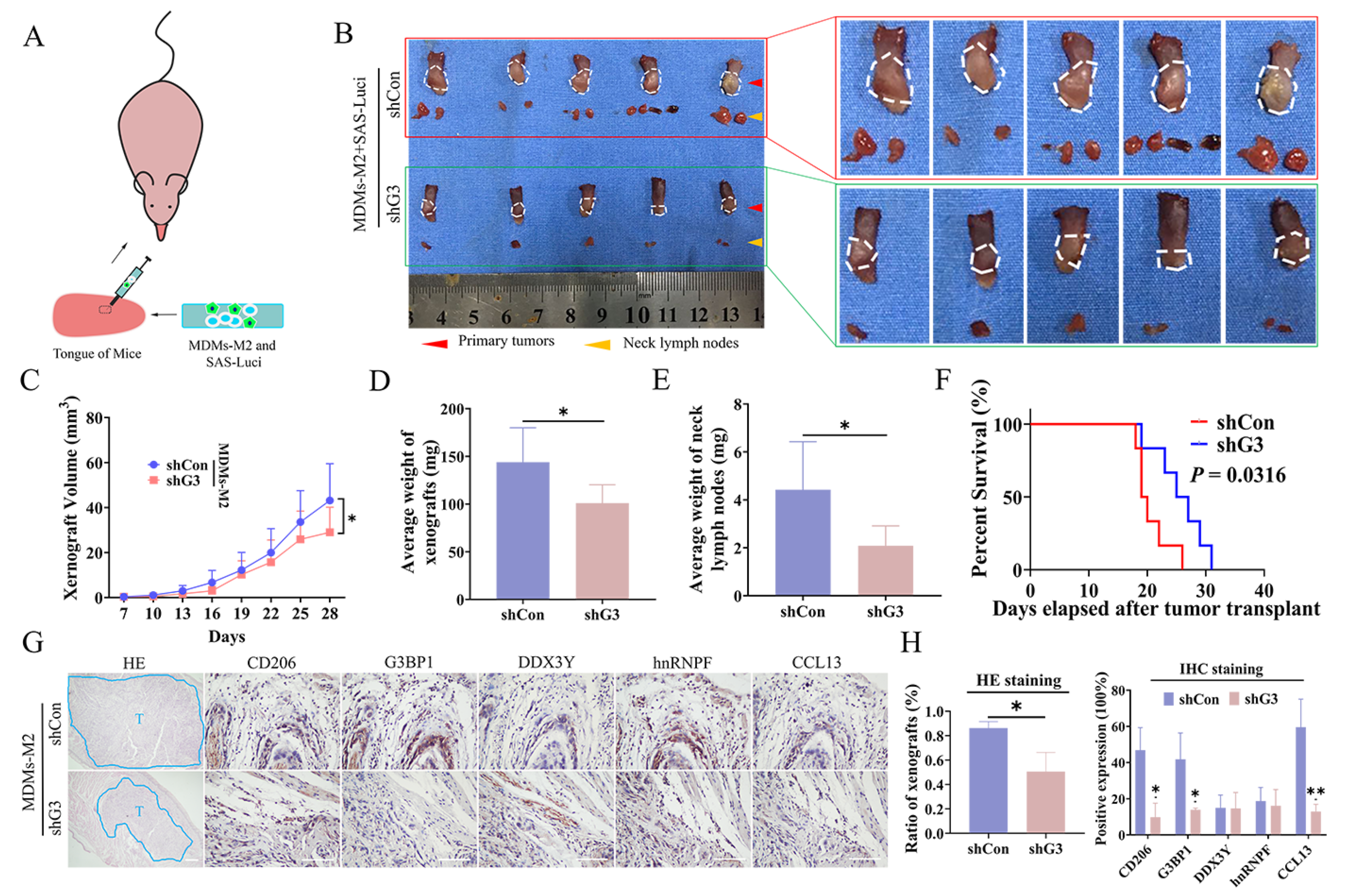
3.6. SG Formation in M2 TAMs Promoted OSCC Metastasis In Vivo
3.7. High Expression of CCL13 Predicts Shorter Overall Survival of OSCC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Koyfman, S.A.; Ismaila, N.; Crook, D.; D’Cruz, A.; Rodriguez, C.P.; Sher, D.J.; Silbermins, D.; Sturgis, E.M.; Tsue, T.T.; Weiss, J.; et al. Management of the Neck in Squamous Cell Carcinoma of the Oral Cavity and Oropharynx: ASCO Clinical Pr actice Guideline. J. Clin. Oncol. 2019, 37, 1753–1774. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zhuang, X.M.; Wang, Y.Y.; Lin, Z.Y.; Zhang, D.M.; Fan, S.; Li, J.S.; Chen, W.L. Tumor necrosis factor α induces myofibroblast differentiation in human tongue cancer and promotes inv asiveness and angiogenesis via secretion of stromal cell-derived factor-1. Oral Oncol. 2015, 51, 1095–1102. [Google Scholar] [CrossRef]
- Lippitz, B.E. Cytokine patterns in patients with cancer: A systematic review. Lancet Oncol. 2013, 14, e218–e228. [Google Scholar] [CrossRef]
- Wellenstein, M.D.; Coffelt, S.B.; Duits, D.E.M.; van Miltenburg, M.H.; Slagter, M.; de Rink, I.; Henneman, L.; Kas, S.M.; Prekovic, S.; Hau, C.S.; et al. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature 2019, 572, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Brüne, B.; Weigert, A.; Dehne, N. Macrophage Polarization in The Tumor Microenvironment. Redox Biol. 2015, 5, 419. [Google Scholar]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Weigert, A.; Reu, S.; Guenther, S.; Mansouri, S.; Bassaly, B.; Gattenlohner, S.; Grimminger, F.; Pullamsetti, S.; Seeger, W.; et al. Spatial Density and Distribution of Tumor-Associated Macrophages Predict Survival in Non-Small Cell Lung Carcinoma. Cancer Res. 2020, 80, 4414–4425. [Google Scholar] [CrossRef]
- Zhu, Y.; Herndon, J.M.; Sojka, D.K.; Kim, K.W.; Knolhoff, B.L.; Zuo, C.; Cullinan, D.R.; Luo, J.; Bearden, A.R.; Lavine, K.J.; et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity 2017, 47, 323–338.e326. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.K.; Wang, M.; Sun, Y.; Di Costanzo, N.; Mitchell, C.; Achuthan, A.; Hamilton, J.A.; Busuttil, R.A.; Boussioutas, A. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat. Commun. 2019, 10, 3928. [Google Scholar] [CrossRef] [PubMed]
- Boussiotis, V.A. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N. Engl. J. Med. 2016, 375, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.F.; Liu, H.; Gao, R.; Zhou, M.; Ma, J.; Zhang, Y.; Zhao, J.; Chen, Y.; Zhang, T.; Huang, F.; et al. Tumor cell-released autophagosomes (TRAPs) promote immunosuppression through induction of M2-like macrophages with increased expression of PD-L1. J. Immunother. Cancer 2018, 6, 151. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; He, H.; Liu, H.; Li, R.; Chen, Y.; Qi, Y.; Jiang, Q.; Chen, L.; Zhang, P.; Zhang, H.; et al. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut 2019, 68, 1764–1773. [Google Scholar] [CrossRef]
- Feng, M.; Jiang, W.; Kim, B.Y.S.; Zhang, C.C.; Fu, Y.X.; Weissman, I.L. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat. Rev. Cancer 2019, 19, 568–586. [Google Scholar] [CrossRef]
- Gordon, S.R.; Maute, R.L.; Dulken, B.W.; Hutter, G.; George, B.M.; McCracken, M.N.; Gupta, R.; Tsai, J.M.; Sinha, R.; Corey, D.; et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017, 545, 495–499. [Google Scholar] [CrossRef]
- Barkal, A.A.; Brewer, R.E.; Markovic, M.; Kowarsky, M.; Barkal, S.A.; Zaro, B.W.; Krishnan, V.; Hatakeyama, J.; Dorigo, O.; Barkal, L.J.; et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 2019, 572, 392–396. [Google Scholar] [CrossRef]
- Dong, L.; Chen, C.; Zhang, Y.; Guo, P.; Wang, Z.; Li, J.; Liu, Y.; Liu, J.; Chang, R.; Li, Y.; et al. The loss of RNA N6-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8+ T?cell dysfunction and tumor growth. Cancer Cell 2021, 39, 945–957.e910. [Google Scholar] [CrossRef]
- Chen, J.; Yao, Y.; Gong, C.; Yu, F.; Su, S.; Chen, J.; Liu, B.; Deng, H.; Wang, F.; Lin, L.; et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell 2011, 19, 541–555. [Google Scholar] [CrossRef]
- Ono, K.; Sogawa, C.; Kawai, H.; Tran, M.T.; Taha, E.A.; Lu, Y.; Oo, M.W.; Okusha, Y.; Okamura, H.; Ibaragi, S.; et al. Triple knockdown of CDC37, HSP90-alpha and HSP90-beta diminishes extracellular vesicles-driven malignancy events and macrophage M2 polarization in oral cancer. J. Extracell. Vesicles 2020, 9, 1769373. [Google Scholar] [CrossRef] [PubMed]
- Gomez, K.E.; Wu, F.; Keysar, S.B.; Morton, J.J.; Miller, B.; Chimed, T.S.; Le, P.N.; Nieto, C.; Chowdhury, F.N.; Tyagi, A.; et al. Cancer Cell CD44 Mediates Macrophage/Monocyte-Driven Regulation of Head and Neck Cancer Stem Cells. Cancer Res. 2020, 80, 4185–4198. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Hiroi, M.; Shimada, J.; Ohmori, Y. Infiltration of m2 tumor-associated macrophages in oral squamous cell carcinoma correlates with tumor malignancy. Cancers 2011, 3, 3726–3739. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Sun, M.; Gu, C.; Wang, X.; Chen, D.; Zhao, E.; Jiao, X.; Zheng, J. Expression of CD163, interleukin-10, and interferon-gamma in oral squamous cell carcinoma: Mutual relationships and prognostic implications. Eur. J. Oral. Sci. 2014, 122, 202–209. [Google Scholar] [CrossRef]
- Haque, A.; Moriyama, M.; Kubota, K.; Ishiguro, N.; Sakamoto, M.; Chinju, A.; Mochizuki, K.; Sakamoto, T.; Kaneko, N.; Munemura, R.; et al. CD206(+) tumor-associated macrophages promote proliferation and invasion in oral squamous cell carcinoma via EGF production. Sci. Rep. 2019, 9, 14611. [Google Scholar] [CrossRef]
- Anderson, P.; Kedersha, N. RNA granules. J. Cell Biol. 2006, 172, 803–808. [Google Scholar] [CrossRef]
- Chevet, E.; Hetz, C.; Samali, A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov. 2015, 5, 586–597. [Google Scholar] [CrossRef]
- Kedersha, N.; Ivanov, P.; Anderson, P. Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 2013, 38, 494–506. [Google Scholar] [CrossRef]
- Somasekharan, S.P.; El-Naggar, A.; Leprivier, G.; Cheng, H.; Hajee, S.; Grunewald, T.G.; Zhang, F.; Ng, T.; Delattre, O.; Evdokimova, V.; et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J. Cell Biol. 2015, 208, 913–929. [Google Scholar] [CrossRef]
- Rai, A.K.; Chen, J.X.; Selbach, M.; Pelkmans, L. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 2018, 559, 211–216. [Google Scholar] [CrossRef]
- Yoon, J.H.; Choi, E.J.; Parker, R. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 2010, 189, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.J.; Chambers, J.E.; Liniker, E.; Marciniak, S.J. Endoplasmic reticulum stress in malignancy. Cancer Cell 2014, 25, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Badeaux, M.; Liu, Y.; Naxerova, K.; Sgroi, D.; Munn, L.L.; Jain, R.K.; Garkavtsev, I. Stress granule-associated protein G3BP2 regulates breast tumor initiation. Proc. Natl. Acad. Sci. USA 2017, 114, 1033–1038. [Google Scholar] [CrossRef]
- Shi, Q.; Zhu, Y.; Ma, J.; Chang, K.; Ding, D.; Bai, Y.; Gao, K.; Zhang, P.; Mo, R.; Feng, K.; et al. Prostate Cancer-associated SPOP mutations enhance cancer cell survival and docetaxel resistance by up regulating Caprin1-dependent stress granule assembly. Mol. Cancer 2019, 18, 170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, H.; Zhang, D.; Xie, S.; Wang, W.; Li, Q.; Lin, Z.; Wang, Y. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018, 9, 742. [Google Scholar] [CrossRef]
- Udagawa, T.; Fujioka, Y.; Tanaka, M.; Honda, D.; Yokoi, S.; Riku, Y.; Ibi, D.; Nagai, T.; Yamada, K.; Watanabe, H.; et al. FUS regulates AMPA receptor function and FTLD/ALS-associated behaviour via GluA1 mRNA stabilization. Nat. Commun. 2015, 6, 7098. [Google Scholar] [CrossRef]
- Naz, S.; Battu, S.; Khan, R.A.; Afroz, S.; Giddaluru, J.; Vishwakarma, S.K.; Satti, V.; Habeeb, M.A.; Khan, A.A.; Khan, N. Activation of integrated stress response pathway regulates IL-1¶¬ production through posttranscription al and translational reprogramming in macrophages. Eur. J. Immunol. 2019, 49, 277–289. [Google Scholar] [CrossRef]
- Guillén-Boixet, J.; Kopach, A.; Holehouse, A.S.; Wittmann, S.; Jahnel, M.; Schlüßler, R.; Kim, K.; Trussina, I.; Wang, J.; Mateju, D.; et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181, 346–361.e317. [Google Scholar] [CrossRef]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e328. [Google Scholar] [CrossRef]
- Loughlin, F.E.; West, D.L.; Gunzburg, M.J.; Waris, S.; Crawford, S.A.; Wilce, M.C.J.; Wilce, J.A. Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1. Nucleic Acids Res. 2021, 49, 2403–2417. [Google Scholar] [CrossRef]
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef] [PubMed]
- Aulas, A.; Lyons, S.M.; Fay, M.M.; Anderson, P.; Ivanov, P. Nitric oxide triggers the assembly of “type II” stress granules linked to decreased cell viability. Cell Death Dis. 2018, 9, 1129. [Google Scholar] [CrossRef] [PubMed]
- Namkoong, S.; Ho, A.; Woo, Y.M.; Kwak, H.; Lee, J.H. Systematic Characterization of Stress-Induced RNA Granulation. Mol. Cell 2018, 70, 175–187.e178. [Google Scholar] [CrossRef] [PubMed]
- Mateju, D.; Eichenberger, B.; Voigt, F.; Eglinger, J.; Roth, G.; Chao, J.A. Single-Molecule Imaging Reveals Translation of mRNAs Localized to Stress Granules. Cell 2020, 183, 1801–1812.e1813. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhao, H.; Su, M.; Xie, W.; Fang, Y.; Du, Y.; Yu, Z.; Hou, L.; Tan, W. HnRNP-F regulates EMT in bladder cancer by mediating the stabilization of Snail1 mRNA by binding to i ts 3’ UTR. EBioMedicine 2019, 45, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhang, J.; Harvey, S.E.; Hu, X.; Cheng, C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes. Dev. 2017, 31, 2296–2309. [Google Scholar] [CrossRef]
- Mills, C.D.; Lenz, L.L.; Harris, R.A. A Breakthrough: Macrophage-Directed Cancer Immunotherapy. Cancer Res. 2016, 76, 513–516. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- Yamagata, Y.; Tomioka, H.; Sakamoto, K.; Sato, K.; Harada, H.; Ikeda, T.; Kayamori, K. CD163-Positive Macrophages Within the Tumor Stroma Are Associated with Lymphangiogenesis and Lymph Node Metastasis in Oral Squamous Cell Carcinoma. J. Oral Maxillofac. Surg. 2017, 75, 2144–2153. [Google Scholar] [CrossRef]
- He, K.F.; Zhang, L.; Huang, C.F.; Ma, S.R.; Wang, Y.F.; Wang, W.M.; Zhao, Z.L.; Liu, B.; Zhao, Y.F.; Zhang, W.F.; et al. CD163+ tumor-associated macrophages correlated with poor prognosis and cancer stem cells in oral squamous cell carcinoma. Biomed Res. Int. 2014, 2014, 838632. [Google Scholar] [CrossRef]
- Kubota, K.; Moriyama, M.; Furukawa, S.; Rafiul, H.; Maruse, Y.; Jinno, T.; Tanaka, A.; Ohta, M.; Ishiguro, N.; Yamauchi, M.; et al. CD163 (+) CD204 (+) tumor-associated macrophages contribute to T cell regulation via interleukin-10 and PD-L1 production in oral squamous cell carcinoma. Sci. Rep. 2017, 7, 1755. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Cathey, P.I.; Wu, H.; Parker, R.; Voeltz, G.K. Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science 2020, 367, 6477. [Google Scholar] [CrossRef] [PubMed]
- Franchini, D.M.; Lanvin, O.; Tosolini, M.; Patras de Campaigno, E.; Cammas, A.; Péricart, S.; Scarlata, C.M.; Lebras, M.; Rossi, C.; Ligat, L.; et al. Microtubule-Driven Stress Granule Dynamics Regulate Inhibitory Immune Checkpoint Expression in T Cell s. Cell Rep. 2019, 26, 94–107.e107. [Google Scholar] [CrossRef]
- Chen, H.H.; Yu, H.I.; Yang, M.H.; Tarn, W.Y. DDX3 Activates CBC-eIF3-Mediated Translation of uORF-Containing Oncogenic mRNAs to Promote Metastasis in HNSCC. Cancer Res. 2018, 78, 4512–4523. [Google Scholar] [CrossRef]
- Raman, D.; Baugher, P.J.; Thu, Y.M.; Richmond, A. Role of chemokines in tumor growth. Cancer Lett. 2007, 256, 137–165. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Y.; Jin, S.; Wang, M.; Jiao, Y.; Yang, B.; Lu, X.; Ji, X.; Fei, Y.; Yang, H.; et al. Single-cell sequencing of immune cells from anticitrullinated peptide antibody positive and negative rheumatoid arthritis. Nat. Commun. 2021, 12, 4977. [Google Scholar] [CrossRef]
- Okugawa, Y.; Toiyama, Y.; Mohri, Y.; Tanaka, K.; Kawamura, M.; Hiro, J.; Araki, T.; Inoue, Y.; Miki, C.; Kusunoki, M. Elevated serum concentration of monocyte chemotactic protein 4 (MCP-4) as a novel non-invasive progno stic and predictive biomarker for detection of metastasis in colorectal cancer. J. Surg. Oncol. 2016, 114, 483–489. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, D.; Liu, C.; Li, G.; Chen, H.; Ling, H.; Zhang, F.; Huang, D.; Wang, X.; Liu, Y.; et al. Comprehensive Analysis of Myeloid Signature Genes in Head and Neck Squamous Cell Carcinoma to Predict the Prognosis and Immune Infiltration. Front. Immunol. 2021, 12, 659184. [Google Scholar] [CrossRef]
- Pérez-Ortín, J.E.; Alepuz, P.; Chávez, S.; Choder, M. Eukaryotic mRNA decay: Methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 2013, 425, 3750–3775. [Google Scholar] [CrossRef]
- Xu, S.H.; Zhu, S.; Wang, Y.; Huang, J.Z.; Chen, M.; Wu, Q.X.; He, Y.T.; Yan, G.R. ECD promotes gastric cancer metastasis by blocking E3 ligase ZFP91-mediated hnRNP F ubiquitination and degradation. Cell Death Dis. 2018, 9, 479. [Google Scholar] [CrossRef]
- Zhang, F.L.; Cao, J.L.; Xie, H.Y.; Sun, R.; Yang, L.F.; Shao, Z.M.; Li, D.Q. Cancer-Associated MORC2-Mutant M276I Regulates an hnRNPM-Mediated CD44 Splicing Switch to Promote Inv asion and Metastasis in Triple-Negative Breast Cancer. Cancer Res. 2018, 78, 5780–5792. [Google Scholar] [CrossRef]
- Dominguez, C.; Fisette, J.F.; Chabot, B.; Allain, F.H. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat. Struct. Mol. Biol. 2010, 17, 853–861. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, Z.; Sun, L.; Fan, S.; Huang, Z.; Zhang, D.; Yang, Z.; Li, J.; Chen, W. Akt/Ezrin Tyr353/NF-κB pathway regulates EGF-induced EMT and metastasis in tongue squamous cell carcinoma. Br. J. Cancer 2014, 110, 695–705. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Rui, T.; Lin, Z.; Xie, S.; Zhou, B.; Fu, M.; Mai, L.; Zhu, C.; Wu, G.; Wang, Y. Tumor-Associated Macrophages Promote Metastasis of Oral Squamous Cell Carcinoma via CCL13 Regulated by Stress Granule. Cancers 2022, 14, 5081. https://doi.org/10.3390/cancers14205081
Liu Z, Rui T, Lin Z, Xie S, Zhou B, Fu M, Mai L, Zhu C, Wu G, Wang Y. Tumor-Associated Macrophages Promote Metastasis of Oral Squamous Cell Carcinoma via CCL13 Regulated by Stress Granule. Cancers. 2022; 14(20):5081. https://doi.org/10.3390/cancers14205081
Chicago/Turabian StyleLiu, Zhixin, Tao Rui, Zhaoyu Lin, Shule Xie, Bin Zhou, Min Fu, Lianxi Mai, Chuandong Zhu, Guotao Wu, and Youyuan Wang. 2022. "Tumor-Associated Macrophages Promote Metastasis of Oral Squamous Cell Carcinoma via CCL13 Regulated by Stress Granule" Cancers 14, no. 20: 5081. https://doi.org/10.3390/cancers14205081
APA StyleLiu, Z., Rui, T., Lin, Z., Xie, S., Zhou, B., Fu, M., Mai, L., Zhu, C., Wu, G., & Wang, Y. (2022). Tumor-Associated Macrophages Promote Metastasis of Oral Squamous Cell Carcinoma via CCL13 Regulated by Stress Granule. Cancers, 14(20), 5081. https://doi.org/10.3390/cancers14205081





