Metabolomics by NMR Combined with Machine Learning to Predict Neoadjuvant Chemotherapy Response for Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.1.1. Clinical and Histopathologic Diagnosis of Breast Cancer
2.1.2. Immunohistochemical Diagnosis
2.1.3. Response to NACT
2.2. Untargeted Nuclear Magnetic Resonance (NMR) Metabolomic Analysis of Serum Samples
2.2.1. NMR Analysis
2.2.2. Statistical Analysis
3. Results
3.1. Subjects and Clinical Features
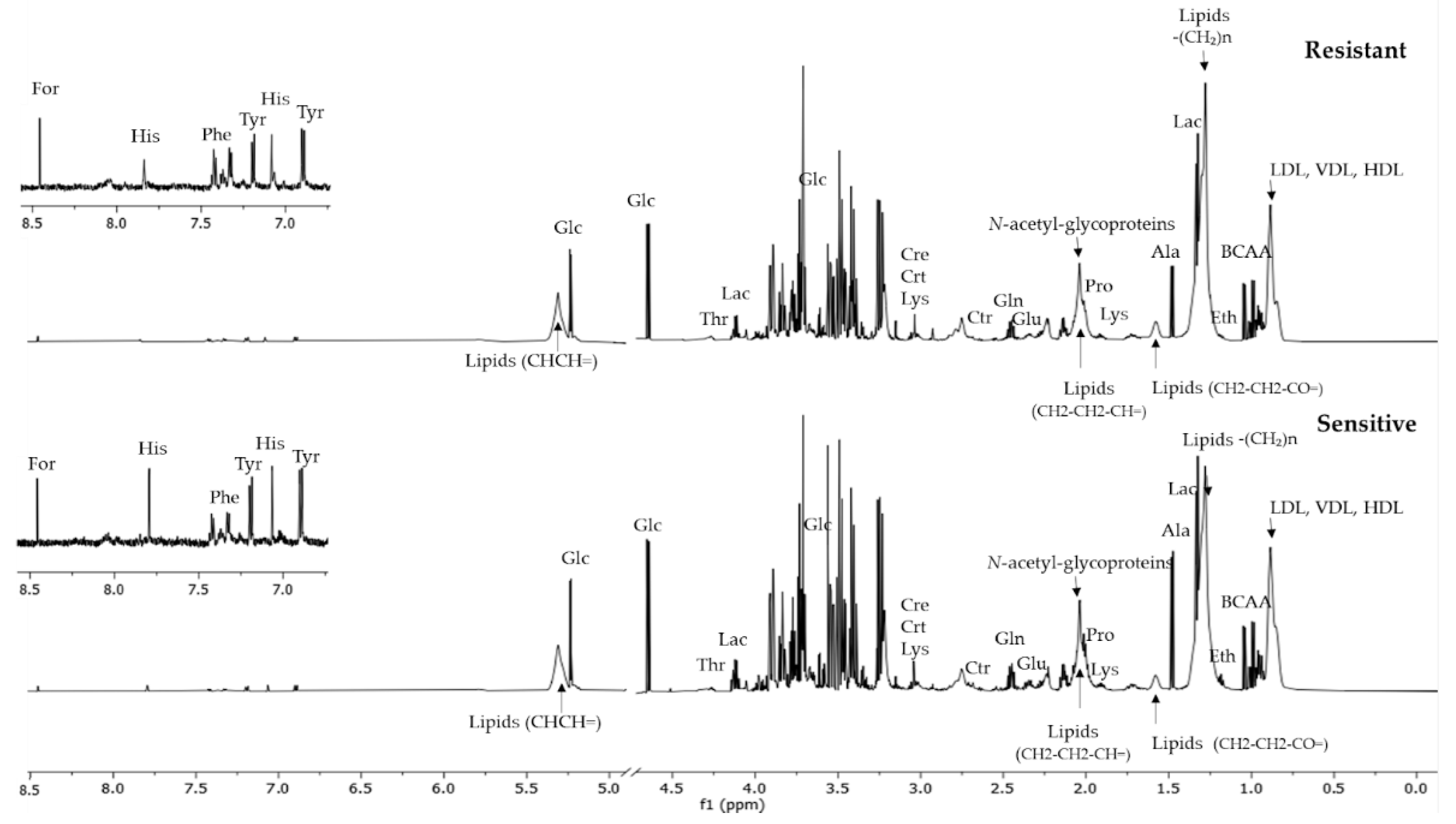
3.2. NMR-Based Metabolomic Analysis
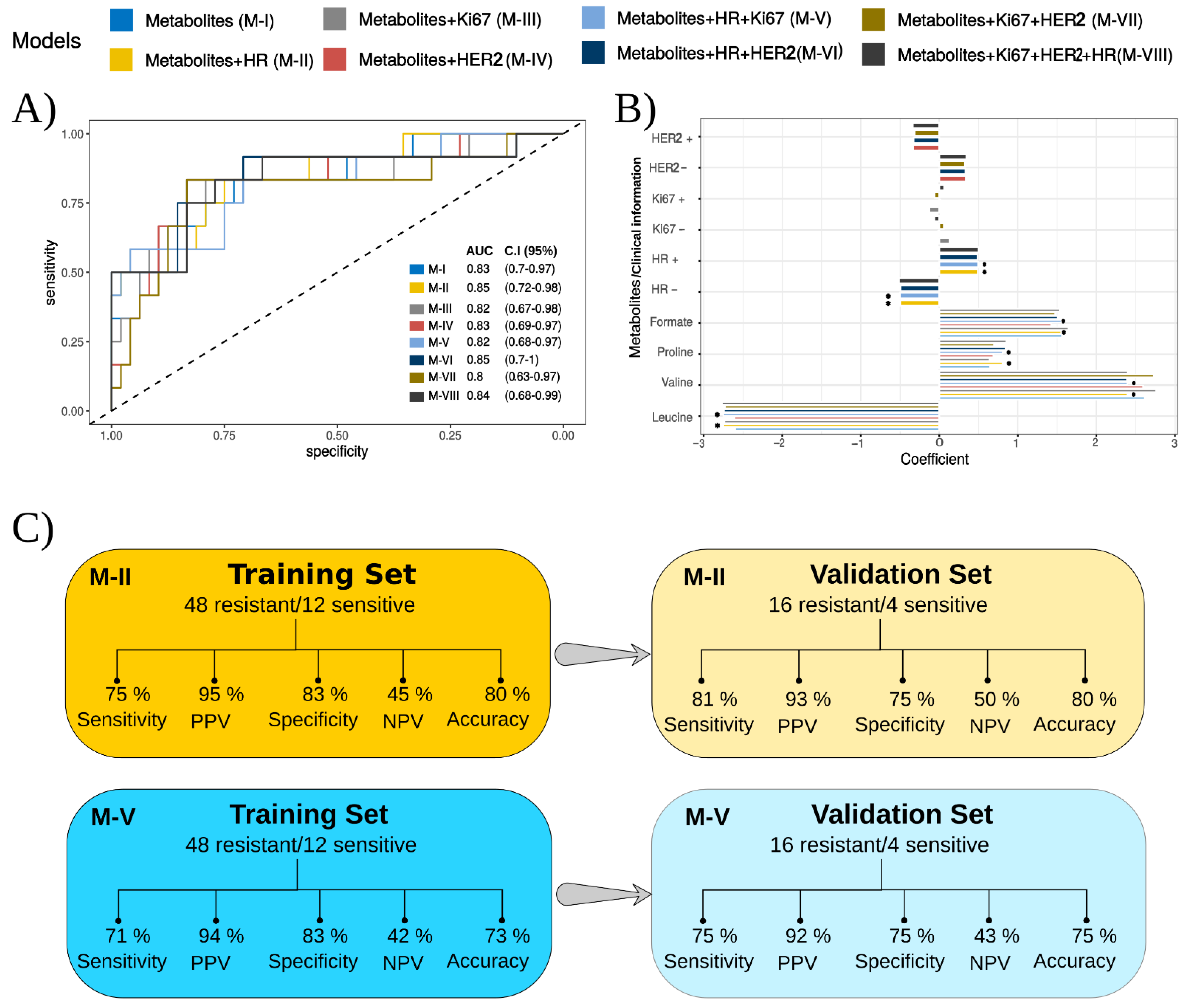
3.3. Classification Models for Predicting Response to NACT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- INCA/Estimativa de Câncer No Brasil. Available online: https://www.gov.br/inca/pt-br/assuntos/cancer/numeros/ (accessed on 26 July 2022).
- Esserman, L.J.; Berry, D.A.; DeMichele, A.; Carey, L.; Davis, S.E.; Buxton, M.; Hudis, C.; Gray, J.W.; Perou, C.; Yau, C.; et al. Pathologic Complete Response Predicts Recurrence-Free Survival More Effectively by Cancer Subset: Results from the I-SPY 1 TRIAL--CALGB 150007/150012, ACRIN 6657. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 3242–3249. [Google Scholar] [CrossRef] [PubMed]
- Cheang, M.C.U.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 Index, HER2 Status, and Prognosis of Patients with Luminal B Breast Cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.-J. Strategies for Subtypes—Dealing with the Diversity of Breast Cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef]
- Yardley, D.A. Drug Resistance and the Role of Combination Chemotherapy in Improving Patient Outcomes. Int. J. Breast Cancer 2013, 2013, 137414. [Google Scholar] [CrossRef] [PubMed]
- von Minckwitz, G.; Untch, M.; Blohmer, J.-U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and Impact of Pathologic Complete Response on Prognosis after Neoadjuvant Chemotherapy in Various Intrinsic Breast Cancer Subtypes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef]
- Haque, W.; Verma, V.; Hatch, S.; Klimberg, V.S.; Butler, E.B.; Teh, B.S. Response Rates and Pathologic Complete Response by Breast Cancer Molecular Subtype Following Neoadjuvant Chemotherapy. Breast Cancer Res. Treat. 2018, 170, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Masoud, V.; Pagès, G. Targeted Therapies in Breast Cancer: New Challenges to Fight against Resistance. World J. Clin. Oncol. 2017, 8, 120–134. [Google Scholar] [CrossRef]
- Bilkey, J.; Tata, A.; McKee, T.D.; Porcari, A.M.; Bluemke, E.; Woolman, M.; Ventura, M.; Eberlin, M.N.; Zarrine-Afsar, A. Variations in the Abundance of Lipid Biomarker Ions in Mass Spectrometry Images Correlate to Tissue Density. Anal. Chem. 2016, 88, 12099–12107. [Google Scholar] [CrossRef]
- Cardoso, M.R.; Santos, J.C.; Ribeiro, M.L.; Talarico, M.C.R.; Viana, L.R.; Derchain, S.F.M. A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response. Int. J. Mol. Sci. 2018, 19, 617. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.A.R.; Cardoso, M.R.; Rezende, L.M.; Lin, J.Q.; Guimaraes, F.; Silva, G.R.P.; Murgu, M.; Priolli, D.G.; Eberlin, M.N.; Tata, A.; et al. Multiplatform Investigation of Plasma and Tissue Lipid Signatures of Breast Cancer Using Mass Spectrometry Tools. Int. J. Mol. Sci. 2020, 21, 3611. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.L.; Pohl, U. Chapter 13-The Role of NMR-Based Metabolomics in Cancer. In The Handbook of Metabonomics and Metabolomics; Lindon, J.C., Nicholson, J.K., Holmes, E., Eds.; Elsevier Science B.V.: Amsterdam, The Netherland, 2007; pp. 345–374. ISBN 978-0-444-52841-4. [Google Scholar]
- Vandergrift, L.A.; Decelle, E.A.; Kurth, J.; Wu, S.; Fuss, T.L.; Defeo, E.M.; Halpern, E.F.; Taupitz, M.; McDougal, W.S.; Olumi, A.F.; et al. Metabolomic Prediction of Human Prostate Cancer Aggressiveness: Magnetic Resonance Spectroscopy of Histologically Benign Tissue. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Schult, T.A.; Lauer, M.J.; Berker, Y.; Cardoso, M.R.; Vandergrift, L.A.; Habbel, P.; Nowak, J.; Taupitz, M.; Aryee, M.; Mino-Kenudson, M.A.; et al. Screening Human Lung Cancer with Predictive Models of Serum Magnetic Resonance Spectroscopy Metabolomics. Proc. Natl. Acad. Sci. USA 2021, 118, e2110633118. [Google Scholar] [CrossRef] [PubMed]
- Kerbel, R.S.; Kobayashi, H.; Graham, C.H. Intrinsic or Acquired Drug Resistance and Metastasis: Are They Linked Phenotypes? J. Cell Biochem. 1994, 56, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass Spectrometry-Based Metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Ahmed, S. Emerging Field of Metabolomics: Big Promise for Cancer Biomarker Identification and Drug Discovery. J. Pharm. Biomed. Anal. 2015, 107, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Shajahan-Haq, A.N.; Cheema, M.S.; Clarke, R. Application of Metabolomics in Drug Resistant Breast Cancer Research. Metabolites 2015, 5, 100–118. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Bucher, E.; Hilvo, M.; Salek, R.; Orešič, M.; Griffin, J.; Brockmöller, S.; Klauschen, F.; Loibl, S.; Barupal, D.K.; et al. Metabolomics of Human Breast Cancer: New Approaches for Tumor Typing and Biomarker Discovery. Genome Med. 2012, 4, 37. [Google Scholar] [CrossRef]
- Debik, J.; Euceda, L.R.; Lundgren, S.; Gythfeldt, H.V.D.L.; Garred, Ø.; Borgen, E.; Engebraaten, O.; Bathen, T.F.; Giskeødegård, G.F. Assessing Treatment Response and Prognosis by Serum and Tissue Metabolomics in Breast Cancer Patients. J. Proteome Res. 2019, 18, 3649–3660. [Google Scholar] [CrossRef]
- Lin, X.; Xu, R.; Mao, S.; Zhang, Y.; Dai, Y.; Guo, Q.; Song, X.; Zhang, Q.; Li, L.; Chen, Q. Metabolic Biomarker Signature for Predicting the Effect of Neoadjuvant Chemotherapy of Breast Cancer. Ann. Transl. Med. 2019, 7, 670. [Google Scholar] [CrossRef] [PubMed]
- Vignoli, A.; Muraro, E.; Miolo, G.; Tenori, L.; Turano, P.; Gregorio, D.E.; Steffan, A.; Luchinat, C.; Corona, G. Effect of Estrogen Receptor Status on Circulatory Immune and Metabolomics Profiles of HER2-Positive Breast Cancer Patients Enrolled for Neoadjuvant Targeted Chemotherapy. Cancers 2020, 12, 314. [Google Scholar] [CrossRef] [PubMed]
- Shortliffe, E.H.; Blois, M.S. The Computer Meets Medicine and Biology: Emergence of a Discipline. In Biomedical Informatics; Springer: Berlin, Germany, 2006; pp. 3–45. [Google Scholar]
- Nattkemper, T.W.; Arnrich, B.; Lichte, O.; Timm, W.; Degenhard, A.; Pointon, L.; Hayes, C.; Leach, M.O. Evaluation of Radiological Features for Breast Tumour Classification in Clinical Screening with Machine Learning Methods. Artif. Intell. Med. 2005, 34, 129–139. [Google Scholar] [CrossRef]
- Wei, L.; Yang, Y.; Nishikawa, R.M.; Jiang, Y. A Study on Several Machine-Learning Methods for Classification of Malignant and Benign Clustered Microcalcifications. IEEE Trans. Med. Imaging 2005, 24, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Delen, D.; Walker, G.; Kadam, A. Predicting Breast Cancer Survivability: A Comparison of Three Data Mining Methods. Artif. Intell. Med. 2005, 34, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Antropova, N.; Huynh, B.Q.; Giger, M.L. A Deep Feature Fusion Methodology for Breast Cancer Diagnosis Demonstrated on Three Imaging Modality Datasets. Med. Phys. 2017, 44, 5162–5171. [Google Scholar] [CrossRef] [PubMed]
- Antropova, N.; Abe, H.; Giger, M.L. Use of Clinical MRI Maximum Intensity Projections for Improved Breast Lesion Classification with Deep Convolutional Neural Networks. J. Med. Imaging 2018, 5, 14503. [Google Scholar] [CrossRef] [PubMed]
- Lakhani, S.; Ellis, I.; Schnitt, S.; Tan, P.; van de Vijver, M. Breast Tumours, WHO Classification of Tumours, 5th ed.; WHO/IARC, Ed.; Publisher: Lyon, France, 2019; Volume 2, ISBN 9789283245001. [Google Scholar]
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Guideline Update. Arch. Pathol. Lab. Med. 2020, 144, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, M.; Nielsen, T.O.; A’Hern, R.; Bartlett, J.; Coombes, R.C.; Cuzick, J.; Ellis, M.; Henry, N.L.; Hugh, J.C.; Lively, T.; et al. Assessment of Ki67 in Breast Cancer: Recommendations from the International Ki67 in Breast Cancer Working Group. J. Natl. Cancer Inst. 2011, 103, 1656–1664. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; McShane, L.M.; Dowsett, M. HER2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update Summary. J. Oncol. Pract. 2018, 14, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.; Rönnlund, C.; de Boniface, J.; Hartman, J. Re-Testing of Predictive Biomarkers on Surgical Breast Cancer Specimens Is Clinically Relevant. Breast Cancer Res. Treat. 2019, 174, 795–805. [Google Scholar] [CrossRef]
- Bossuyt, V.; Provenzano, E.; Symmans, W.F.; Boughey, J.C.; Coles, C.; Curigliano, G.; Dixon, J.M.; Esserman, L.J.; Fastner, G.; Kuehn, T.; et al. Recommendations for Standardized Pathological Characterization of Residual Disease for Neoadjuvant Clinical Trials of Breast Cancer by the BIG-NABCG Collaboration. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 1280–1291. [Google Scholar] [CrossRef] [PubMed]
- Symmans, W.F.; Peintinger, F.; Hatzis, C.; Rajan, R.; Kuerer, H.; Valero, V.; Assad, L.; Poniecka, A.; Hennessy, B.; Green, M.; et al. Measurement of Residual Breast Cancer Burden to Predict Survival after Neoadjuvant Chemotherapy. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007, 25, 4414–4422. [Google Scholar] [CrossRef]
- Provenzano, E.; Bossuyt, V.; Viale, G.; Cameron, D.; Badve, S.; Denkert, C.; MacGrogan, G.; Penault-Llorca, F.; Boughey, J.; Curigliano, G.; et al. Standardization of Pathologic Evaluation and Reporting of Postneoadjuvant Specimens in Clinical Trials of Breast Cancer: Recommendations from an International Working Group. Mod. Pathol. Off. J. U.S. Can. Acad. Pathol. Inc. 2015, 28, 1185–1201. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.D.; Cai, T.T.; DasGupta, A. Interval Estimation for a Binomial Proportion. Stat. Sci. 2001, 16, 101–133. [Google Scholar] [CrossRef]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; Team, R.C.; et al. Classification and Regression Training. R Package Version 6.0–88. Available online: https://cran.r-project.org/package=caret (accessed on 20 November 2021).
- Guan, W.; Zhou, M.; Hampton, C.Y.; Benigno, B.B.; Walker, L.D.; Gray, A.; McDonald, J.F.; Fernández, F.M. Ovarian Cancer Detection from Metabolomic Liquid Chromatography/Mass Spectrometry Data by Support Vector Machines. BMC Bioinform. 2009, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification Using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Sheng, J.; Shao, M.; Zhang, Q.; Zhou, R.; Wang, L.; Xin, Y. Alzheimer’s Disease, Mild Cognitive Impairment, and Normal Aging Distinguished by Multi-Modal Parcellation and Machine Learning. Sci. Rep. 2020, 10, 5475. [Google Scholar] [CrossRef]
- Murata, T.; Yanagisawa, T.; Kurihara, T.; Kaneko, M.; Ota, S.; Enomoto, A.; Tomita, M.; Sugimoto, M.; Sunamura, M.; Hayashida, T.; et al. Salivary Metabolomics with Alternative Decision Tree-Based Machine Learning Methods for Breast Cancer Discrimination. Breast Cancer Res. Treat. 2019, 177, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; Morais, D.A.D.L.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Eberlin, L.S.; Tibshirani, R.J.; Zhang, J.; Longacre, T.A.; Berry, G.J.; Bingham, D.B.; Norton, J.A.; Zare, R.N.; Poultsides, G.A. Molecular Assessment of Surgical-Resection Margins of Gastric Cancer by Mass-Spectrometric Imaging. Proc. Natl. Acad. Sci. USA 2014, 111, 2436–2441. [Google Scholar] [CrossRef] [PubMed]
- Hosseinnataj, A.; RezaBaneshi, M.; Bahrampour, A. Mortality Risk Factors in Patients with Gastric Cancer Using Bayesian and Ordinary Lasso Logistic Models: A Study in the Southeast of Iran. Gastroenterol. Hepatol. Bed Bench 2020, 13, 31–36. [Google Scholar] [PubMed]
- Kim, S.M.; Kim, Y.; Jeong, K.; Jeong, H.; Kim, J. Logistic LASSO Regression for the Diagnosis of Breast Cancer Using Clinical Demographic Data and the BI-RADS Lexicon for Ultrasonography. Ultrasonography 2018, 37, 36–42. [Google Scholar] [CrossRef]
- Sans, M.; Gharpure, K.; Tibshirani, R.; Zhang, J.; Liang, L.; Liu, J.; Young, J.H.; Dood, R.L.; Sood, A.K.; Eberlin, L.S. Metabolic Markers and Statistical Prediction of Serous Ovarian Cancer Aggressiveness by Ambient Ionization Mass Spectrometry Imaging. Cancer Res. 2017, 77, 2903–2913. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Cai, H.; Wang, S.; Shen, Y.; Ke, C. Application of Metabolomics in the Diagnosis of Breast Cancer: A Systematic Review. J. Cancer 2020, 11, 2540–2551. [Google Scholar] [CrossRef]
- Ganapathy, V.; Thangaraju, M.; Prasad, P.D. Nutrient Transporters in Cancer: Relevance to Warburg Hypothesis and Beyond. Pharmacol. Ther. 2009, 121, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Bhutia, Y.D.; Babu, E.; Ramachandran, S.; Ganapathy, V. Amino Acid Transporters in Cancer and Their Relevance to “Glutamine Addiction”: Novel Targets for the Design of a New Class of Anticancer Drugs. Cancer Res. 2015, 75, 1782–1788. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.J.; Kim, E.-S.; Koo, J.S. Amino Acid Transporters and Glutamine Metabolism in Breast Cancer. Int. J. Mol. Sci. 2018, 19, 907. [Google Scholar] [CrossRef] [PubMed]
- Häfliger, P.; Graff, J.; Rubin, M.; Stooss, A.; Dettmer, M.S.; Altmann, K.-H.; Gertsch, J.; Charles, R.-P. The LAT1 Inhibitor JPH203 Reduces Growth of Thyroid Carcinoma in a Fully Immunocompetent Mouse Model. J. Exp. Clin. Cancer Res. 2018, 37, 234. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.; Kim, B.; Lee, H.; Lee, S.; Kang, H.-S.; Kim, S.J. Epigenetically Regulated Fibronectin Leucine Rich Transmembrane Protein 2 (FLRT2) Shows Tumor Suppressor Activity in Breast Cancer Cells. Sci. Rep. 2017, 7, 272. [Google Scholar] [CrossRef]
- Shennan, D.B.; Thomson, J.; Gow, I.F.; Travers, M.T.; Barber, M.C. L-Leucine Transport in Human Breast Cancer Cells (MCF-7 and MDA-MB-231): Kinetics, Regulation by Estrogen and Molecular Identity of the Transporter. Biochim. Biophys. Acta 2004, 1664, 206–216. [Google Scholar] [CrossRef]
- Speers, C.; Zhao, S.G.; Kothari, V.; Santola, A.; Liu, M.; Wilder-Romans, K.; Evans, J.; Batra, N.; Bartelink, H.; Hayes, D.F.; et al. Maternal Embryonic Leucine Zipper Kinase (MELK) as a Novel Mediator and Biomarker of Radioresistance in Human Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 5864–5875. [Google Scholar] [CrossRef]
- Saito, Y.; Li, L.; Coyaud, E.; Luna, A.; Sander, C.; Raught, B.; Asara, J.M.; Brown, M.; Muthuswamy, S.K. LLGL2 Rescues Nutrient Stress by Promoting Leucine Uptake in ER (+) Breast Cancer. Nature 2019, 569, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sui, C.; Yang, W.; Luo, Q. Amino Acid Transporters: Emerging Roles in Drug Delivery for Tumor-Targeting Therapy. Asian J. Pharm. Sci. 2020, 15, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Boutouja, F.; Stiehm, C.M.; Platta, H.W. MTOR: A Cellular Regulator Interface in Health and Disease. Cells 2019, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zhang, F.; Han, P.; Wang, Z.-Z.; Deng, K.; Zhang, Y.-Y.; Zhao, W.-W.; Song, W.; Cai, Y.-Q.; Li, K.; et al. Metabolomics Approach for Predicting Response to Neoadjuvant Chemotherapy for Colorectal Cancer. Metabolomics 2018, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Ananieva, E.A.; Wilkinson, A.C. Branched-Chain Amino Acid Metabolism in Cancer. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Lind, D.S. Arginine and Cancer. J. Nutr. 2004, 134, 2837S–2841S. [Google Scholar] [CrossRef] [PubMed]
- Albaugh, V.L.; Pinzon-Guzman, C.; Barbul, A. Arginine-Dual Roles as an Onconutrient and Immunonutrient. J. Surg. Oncol. 2017, 115, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Jayant, S.; Anant, A. The Relation of Serum Arginine Levels with Serum Arginase and Nitric Oxide Synthase Activity in Patients with Breast Cancer. J. Clin. Diagn. Res. 2017, 11, 11–14. [Google Scholar] [CrossRef]
- Pietzke, M.; Meiser, J.; Vazquez, A. Formate Metabolism in Health and Disease. Mol. Metab. 2020, 33, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Pietzke, M.; Arroyo, S.F.; Sumpton, D.; Mackay, G.M.; Martin-Castillo, B.; Camps, J.; Joven, J.; Menendez, J.A.; Vazquez, A. Stratification of Cancer and Diabetes Based on Circulating Levels of Formate and Glucose. Cancer Metab. 2019, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Delbrouck, C.; Pozdeev, V.I.; Oudin, A.; Grzyb, K.; Neises, L.; Kiweler, N.; Skupin, A.; Letellier, E.; Niclou, S.P.; Meiser, J. FSMP-09. Formate Promotes Cancer Cell Invasion and Metastasis Via Calcium Signaling. Neurocol. Adv. 2021, 3, i18. [Google Scholar] [CrossRef]
- Jiang, L.; Lee, S.C.; Ng, T.C. Pharmacometabonomics Analysis Reveals Serum Formate and Acetate Potentially Associated with Varying Response to Gemcitabine-Carboplatin Chemotherapy in Metastatic Breast Cancer Patients. J. Proteome. Res. 2018, 17, 1248–1257. [Google Scholar] [CrossRef]
- Chen, Z.; Dai, Y.; Huang, X.; Chen, K.; Gao, Y.; Li, N.; Wang, D.; Chen, A.; Yang, Q.; Hong, Y.; et al. Combined Metabolomic Analysis of Plasma and Tissue Reveals a Prognostic Risk Score System and Metabolic Dysregulation in Esophageal Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 1545. [Google Scholar] [CrossRef]
- Ge, C.; Luo, L.; Zhang, J.; Meng, X.; Chen, Y. FRL: An Integrative Feature Selection Algorithm Based on the Fisher Score, Recursive Feature Elimination, and Logistic Regression to Identify Potential Genomic Biomarkers. Biomed. Res. Int. 2021, 2021, 4312850. [Google Scholar] [CrossRef]
- Han, Y.; Huang, L.; Zhou, F. A Dynamic Recursive Feature Elimination Framework (DRFE) to Further Refine a Set of OMIC Biomarkers. Bioinformatics 2021, 37, 2183–2189. [Google Scholar] [CrossRef]
- Sutton, E.J.; Onishi, N.; Fehr, D.A.; Dashevsky, B.Z.; Sadinski, M.; Pinker, K.; Martinez, D.F.; Brogi, E.; Braunstein, L.; Razavi, P.; et al. A Machine Learning Model That Classifies Breast Cancer Pathologic Complete Response on MRI Post-Neoadjuvant Chemotherapy. Breast Cancer Res. 2020, 22, 57. [Google Scholar] [CrossRef]
- Lopez-Rincon, A.; Mendoza-Maldonado, L.; Martinez-Archundia, M.; Schönhuth, A.; Kraneveld, A.D.; Garssen, J.; Tonda, A. Machine Learning-Based Ensemble Recursive Feature Selection of Circulating MiRNAs for Cancer Tumor Classification. Cancers 2020, 12, 1785. [Google Scholar] [CrossRef]
- Liu, L. Research on Logistic Regression Algorithm of Breast Cancer Diagnose Data by Machine Learning. In Proceedings of the 2018 International Conference on Robots & Intelligent System (ICRIS), Amsterdam, The Netherlands, 21–23 February 2018; pp. 157–160. [Google Scholar]
- Pfob, A.; Sidey-Gibbons, C.; Lee, H.-B.; Tasoulis, M.K.; Koelbel, V.; Golatta, M.; Rauch, G.M.; Smith, B.D.; Valero, V.; Han, W.; et al. Identification of Breast Cancer Patients with Pathologic Complete Response in the Breast after Neoadjuvant Systemic Treatment by an Intelligent Vacuum-Assisted Biopsy. Eur. J. Cancer 2021, 143, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Tahmassebi, A.; Wengert, G.J.; Helbich, T.H.; Bago-Horvath, Z.; Alaei, S.; Bartsch, R.; Dubsky, P.; Baltzer, P.; Clauser, P.; Kapetas, P.; et al. Impact of Machine Learning With Multiparametric Magnetic Resonance Imaging of the Breast for Early Prediction of Response to Neoadjuvant Chemotherapy and Survival Outcomes in Breast Cancer Patients. Investig. Radiol. 2019, 54, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xiao, L.; Pu, S.; Liu, Y.; He, J.; Wang, K. Can We Reliably Identify the Pathological Outcomes of Neoadjuvant Chemotherapy in Patients with Breast Cancer? Development and Validation of a Logistic Regression Nomogram Based on Preoperative Factors. Ann. Surg. Oncol. 2021, 28, 2632–2645. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Lee, K.M.; Kim, S.H.; Kwon, Y.J.; Chun, Y.J.; Choi, H.K. Comparative Metabolic and Lipidomic Profiling of Human Breast Cancer Cells with Different Metastatic Potentials. Oncotarget 2016, 7, 67111–67128. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Wang, Y.; Luo, W. Multifaceted Role of Branched-Chain Amino Acid Metabolism in Cancer. Oncogene 2020, 39, 6747–6756. [Google Scholar] [CrossRef]

| Characteristic | n (%) | Sensitive | Resistant | OR | p-Value | |
|---|---|---|---|---|---|---|
| n = 16 (%) | n = 64 (%) | (95% CI) | ||||
| Age | ≥50 | 46 (57.5) | 8 (50.0) | 38 (59.4) | ref | |
| <50 | 34 (42.5) | 8 (50.0) | 26 (40.6) | 0.68 (0.23–2.05) | 0.499 | |
| Race | Caucasian | 69 (86.3) | 15 (93.8) | 54 (84.4) | ref | |
| Non-Caucasian | 11 (13.8) | 1 (6.2) | 10 (15.6) | 2.78 (0.33–23.45) | 0.293 | |
| Age of menarche | >12 | 42 (52.5) | 9 (56.2) | 33 (51.6) | ref | |
| ≤12 | 38 (47.5) | 7 (43.8) | 31 (48.4) | 1.21 (0.4–3.64) | 0.737 | |
| Menopause | No | 36 (45.0) | 9 (56.2) | 27 (42.2) | ref | |
| Yes | 44 (55.0) | 7 (43.8) | 37 (57.8) | 1.76 (0.58–5.32) | 0.313 | |
| Hormone therapy | No | 68 (85.0) | 15 (93.8) | 53 (82.8) | ref | |
| Yes | 12 (15.0) | 1 (6.2) | 11 (17.2) | 3.11 (0.37–26.09) | 0.233 | |
| Pregnancy (previous or current) | Yes | 73 (91.3) | 15 (93.8) | 58 (90.6) | ref | |
| No | 7 (8.7) | 1 (6.2) | 6 (9.4) | 1.55 (0.17–13.89) | 0.681 | |
| Lactation * | Yes | 63 (78.8) | 13 (81.2) | 50 (78.1) | ref | |
| No | 17 (21.2) | 3 (18.8) | 14 (21.9) | 1.21 (0.3–4.86) | 0.782 | |
| Smoking | Yes | 17 (21.2) | 4 (25.0) | 13 (20.3) | ref | |
| No | 63 (78.8) | 12 (75.0) | 51 (79.7) | 1.31 (0.36–4.73) | 0.686 | |
| Chronic alcoholism | No | 79 (98.8) | 16 (100.0) | 63 (98.4) | ref | |
| Yes | 1 (1.2) | 0 (0.0) | 1 (1.6) | NC | 0.503 | |
| BMI | Normal weight | 24 (30.0) | 6 (37.5) | 18 (28.1) | ref | |
| Overweight | 21 (26.3) | 2 (12.5) | 19 (29.7) | 3.17 (0.56–17.77) | 0.19 | |
| Obese | 35 (43.7) | 8 (50.0) | 27 (42.2) | 1.13 (0.33–3.79) | 0.849 | |
| Diabetes | No | 71 (88.7) | 15 (93.8) | 56 (87.5) | ref | |
| Yes | 9 (11.3) | 1 (6.2) | 8 (12.5) | 2.14 (0.25–18.5) | 0.452 | |
| Family history of breast or ovarian cancer | No | 59 (73.8) | 9 (56.2) | 50 (78.1) | ref | |
| Yes | 21 (26.2) | 7 (43.8) | 14 (21.9) | 0.36 (0.11–1.14) | 0.087 | |
| Characteristic | n (%) | Sensitive | Resistant | OR | p-Value | |
|---|---|---|---|---|---|---|
| n = 16 (%) | n = 64 (%) | (95% CI) | ||||
| Histological grade | 1/2 | 38 (48.75) | 3 (18.8) | 36 (56.2) | ref | |
| 3 | 41 (51.25) | 13 (81.2) | 28 (43.8) | 0.18 (0.05–0.69) | 0.006 | |
| Ki67 | Low | 34 (42.5) | 5 (31.2) | 29 (45.3) | ref | |
| High | 46 (57.5) | 11 (68.8) | 35 (54.7) | 0.55 (0.17–1.76) | 0.303 | |
| HER2 | Negative | 46 (57.5) | 6 (37.5) | 40 (62.5) | ref | |
| Positive * | 34 (42.5) | 10 (62.5) | 24 (37.5) | 0.36 (0.12–1.12) | 0.072 | |
| Tumor size ** | T1/T2 | 53 (66.25) | 12 (75.0) | 41 (64.1) | ref | |
| T3/T4 | 27 (33.75) | 4 (25.0) | 23 (35.9) | 1.68 (0.49–5.82) | 0.399 | |
| Regional lymph node | N0 | 33 (41.25) | 6 (37.5) | 27 (42.2) | ref | |
| N1 or higher | 47 (58.75) | 10 (62.5) | 37 (57.8) | 0.82 (0.27–2.54) | 0.732 | |
| Metastasis | M0 | 74 (92.5) | 16 (100.0) | 58 (90.6) | ref | |
| M1 | 6 (7.5) | 0 (0.0) | 6 (9.4) | NC | 0.094 | |
| Hormonal Receptor | Negative | 21 (26.25) | 9 (56.2) | 12 (18.8) | ref | |
| Positive | 59 (73.75) | 7 (43.8) | 52 (81.2) | 5.57 (1.73–17.96) | 0.004 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardoso, M.R.; Silva, A.A.R.; Talarico, M.C.R.; Sanches, P.H.G.; Sforça, M.L.; Rocco, S.A.; Rezende, L.M.; Quintero, M.; Costa, T.B.B.C.; Viana, L.R.; et al. Metabolomics by NMR Combined with Machine Learning to Predict Neoadjuvant Chemotherapy Response for Breast Cancer. Cancers 2022, 14, 5055. https://doi.org/10.3390/cancers14205055
Cardoso MR, Silva AAR, Talarico MCR, Sanches PHG, Sforça ML, Rocco SA, Rezende LM, Quintero M, Costa TBBC, Viana LR, et al. Metabolomics by NMR Combined with Machine Learning to Predict Neoadjuvant Chemotherapy Response for Breast Cancer. Cancers. 2022; 14(20):5055. https://doi.org/10.3390/cancers14205055
Chicago/Turabian StyleCardoso, Marcella R., Alex Ap. Rosini Silva, Maria Cecília R. Talarico, Pedro H. Godoy Sanches, Maurício L. Sforça, Silvana A. Rocco, Luciana M. Rezende, Melissa Quintero, Tassia B. B. C. Costa, Laís R. Viana, and et al. 2022. "Metabolomics by NMR Combined with Machine Learning to Predict Neoadjuvant Chemotherapy Response for Breast Cancer" Cancers 14, no. 20: 5055. https://doi.org/10.3390/cancers14205055
APA StyleCardoso, M. R., Silva, A. A. R., Talarico, M. C. R., Sanches, P. H. G., Sforça, M. L., Rocco, S. A., Rezende, L. M., Quintero, M., Costa, T. B. B. C., Viana, L. R., Canevarolo, R. R., Ferracini, A. C., Ramalho, S., Gutierrez, J. M., Guimarães, F., Tasic, L., Tata, A., Sarian, L. O., Cheng, L. L., ... Derchain, S. F. M. (2022). Metabolomics by NMR Combined with Machine Learning to Predict Neoadjuvant Chemotherapy Response for Breast Cancer. Cancers, 14(20), 5055. https://doi.org/10.3390/cancers14205055







