CSMD1 Shows Complex Patterns of Somatic Copy Number Alterations and Expressions of mRNAs and Target Micro RNAs in Esophageal Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cases, Specimen Processing, and Arrays
2.1.1. Case Selection
2.1.2. Biological Specimen Collection and Processing
2.1.3. Target Preparation for GeneChip Human Mapping 500K Array Set
2.1.4. CSMD1 Gene Expression by Quantitative Reverse Transcription-PCR (qRT-PCR)
2.1.5. ABI miRNA Expression Array by RT-PCR
2.2. Data Analysis
2.2.1. GeneChip 500K and SNP5 Array Data Analysis
2.2.2. CSMD1 Gene Expression by qRT-PCR Data Analysis
2.2.3. CSMD1 Target miRNA Expression Array Analysis
2.2.4. Correlation Analyses
2.2.5. Survival Analysis
3. Results
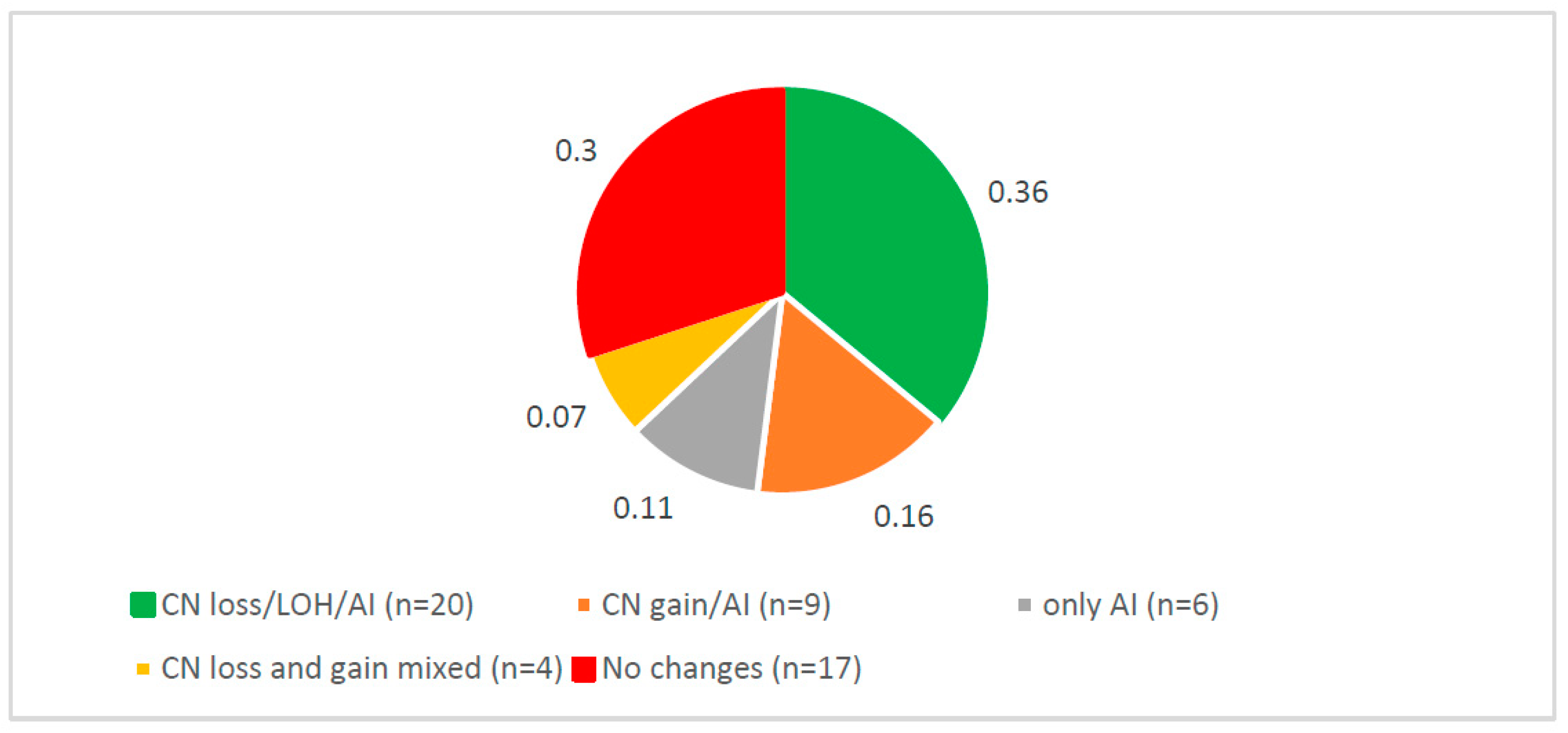
3.1. Complex Somatic DNA Alterations of CSMD1 in ESCC
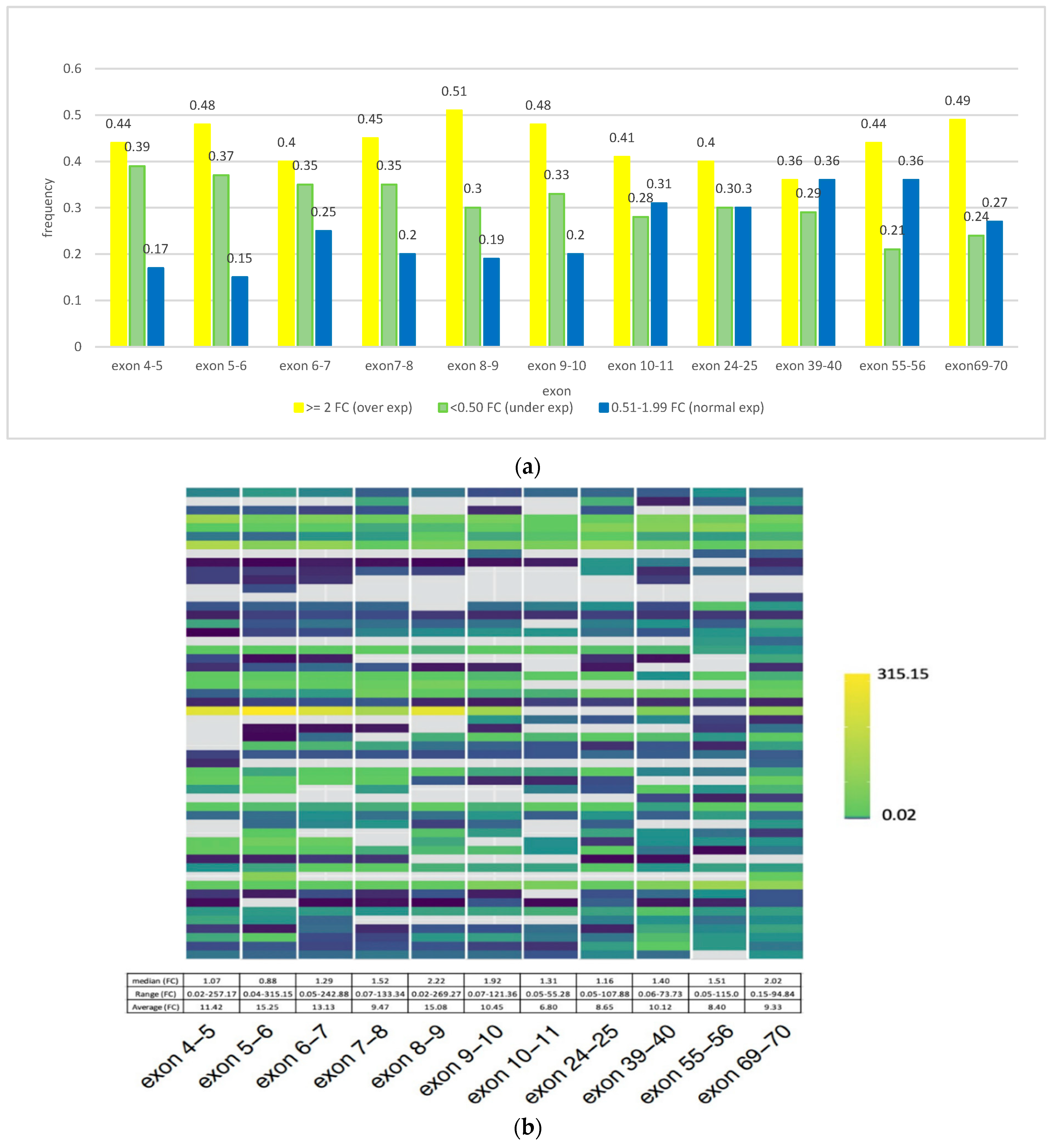
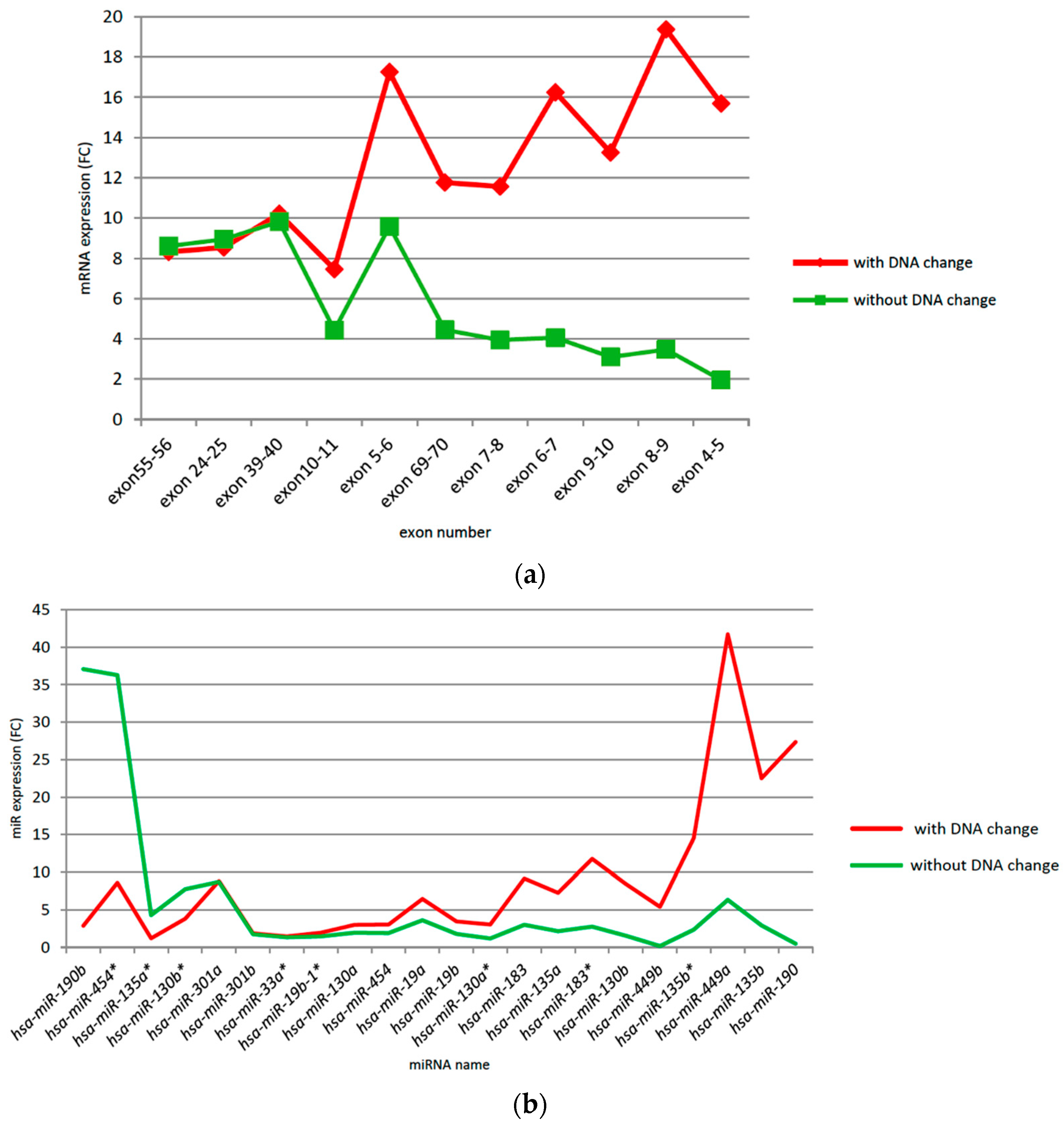
3.2. CSMD1 mRNA Expression Using qRT-PCR and Its Relation to Somatic DNA Alterations in ESCC Cases
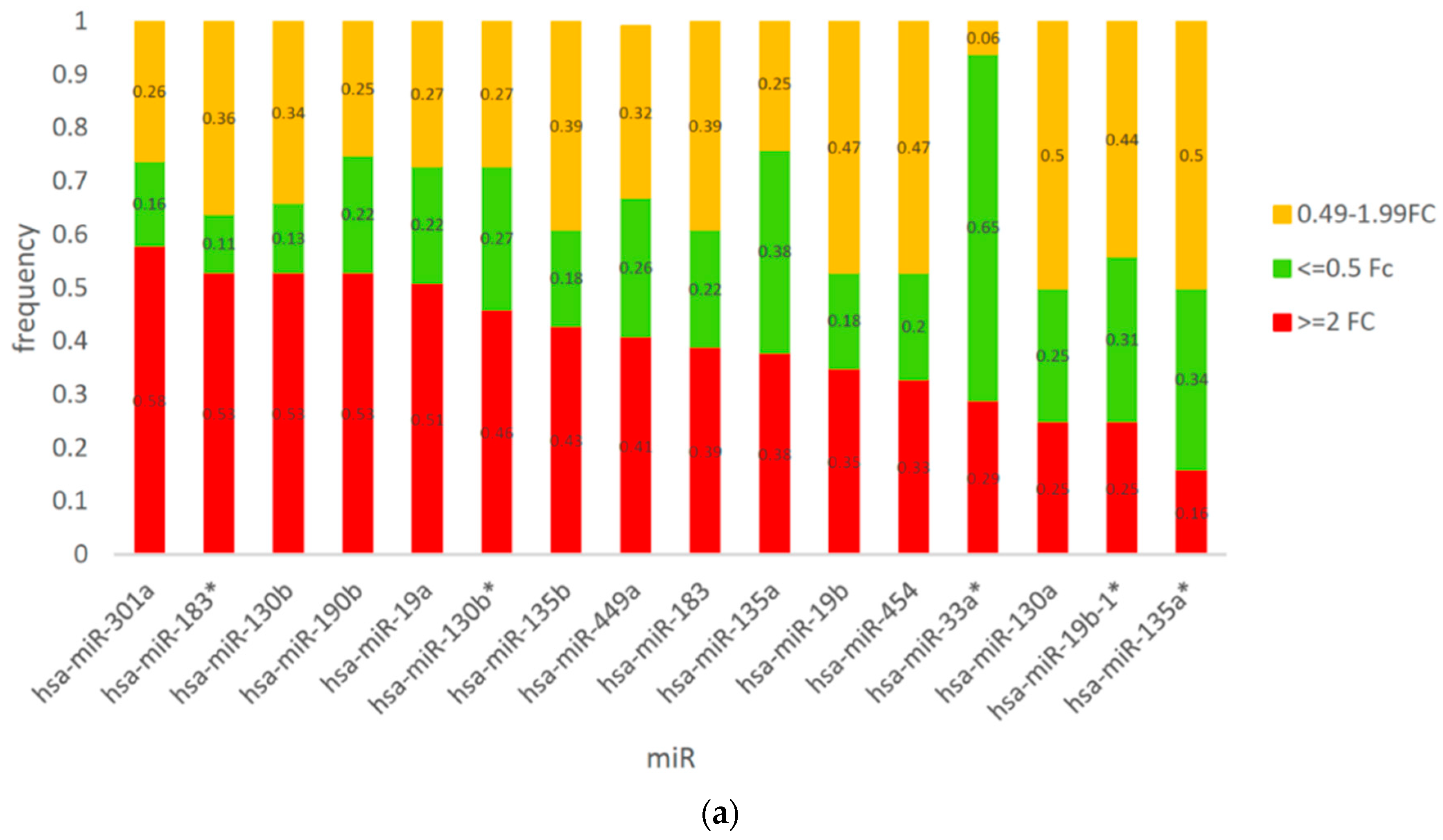
3.3. Expression of CSMD1 in Targeted miRNAs in ESCC
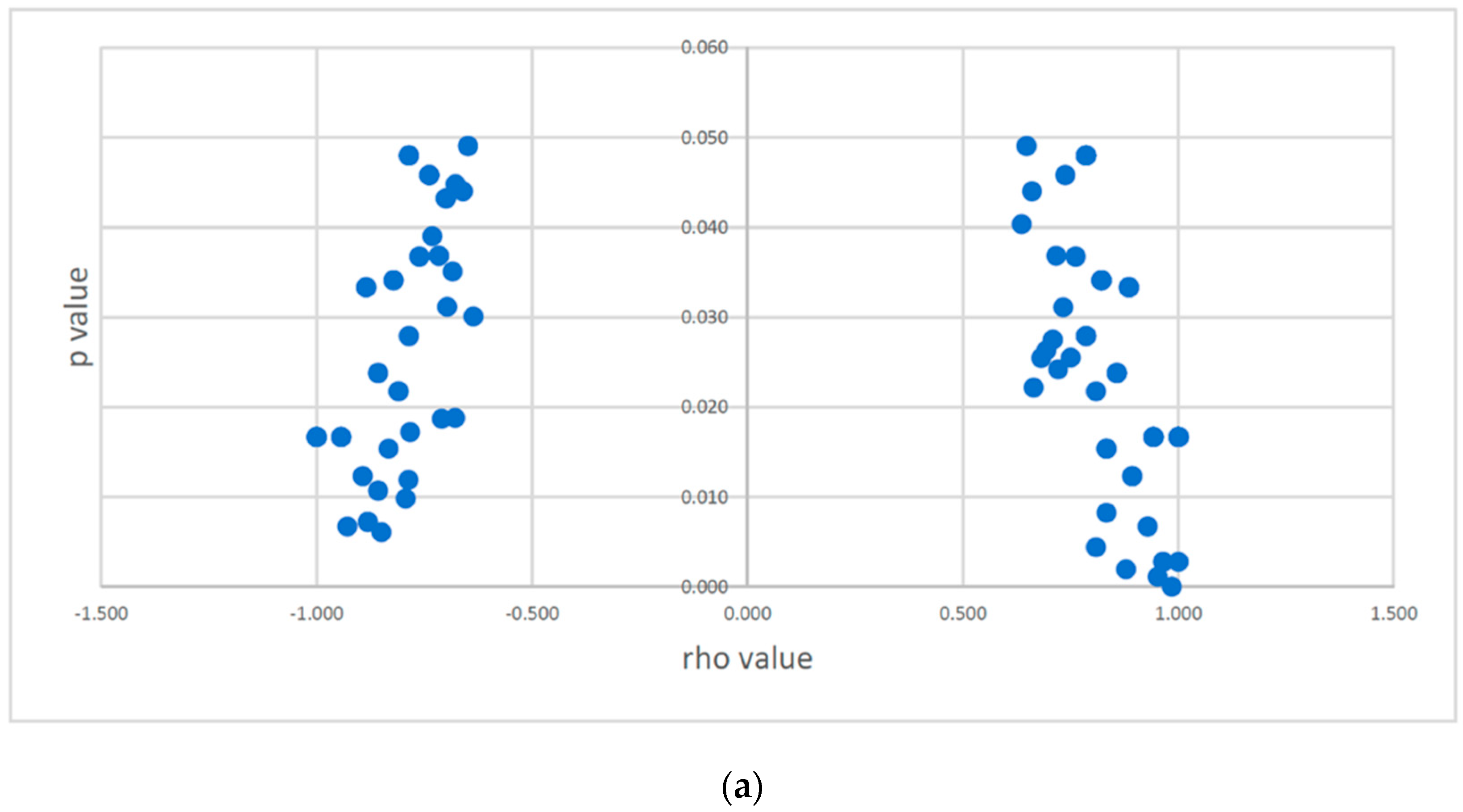
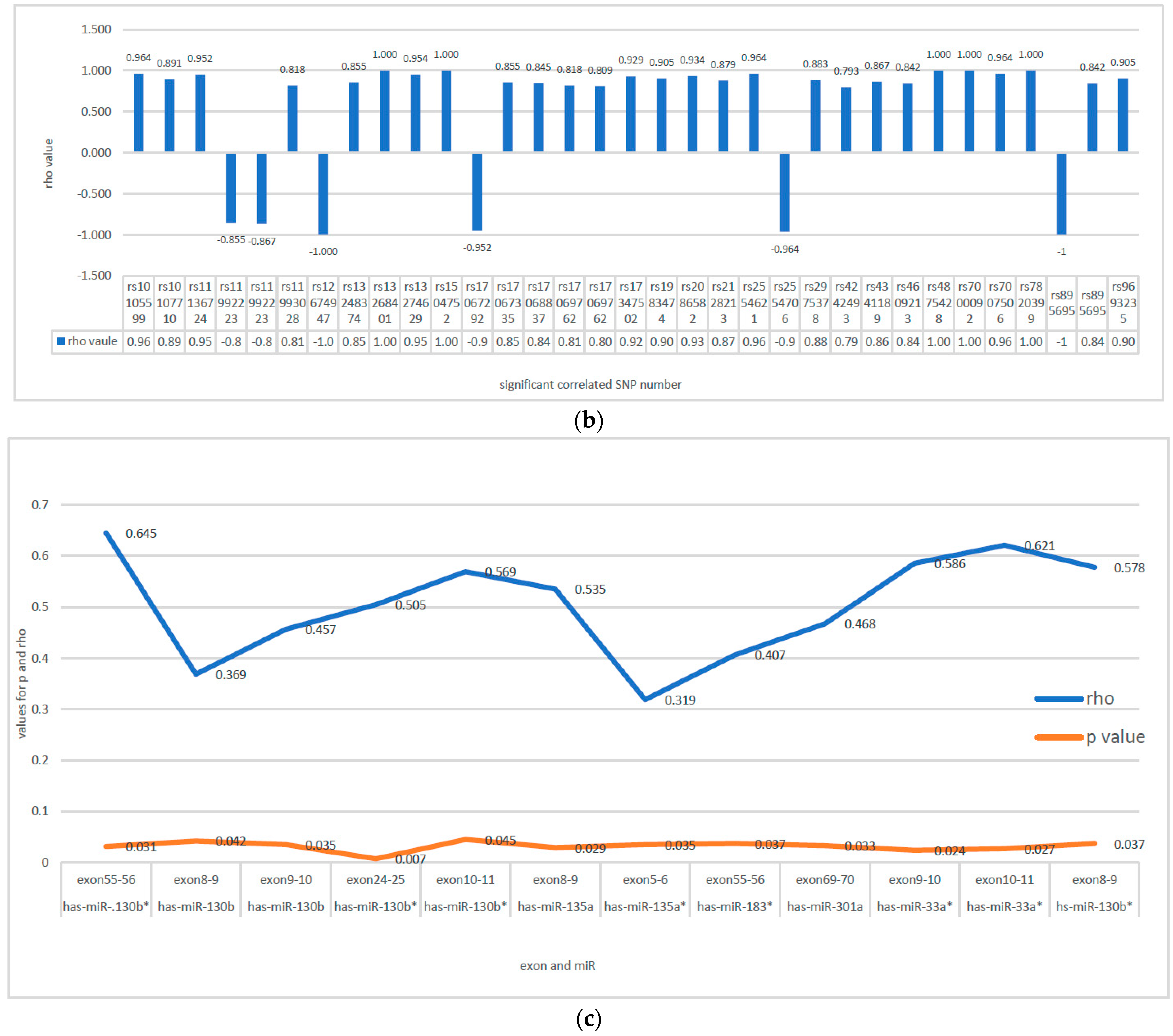
3.4. Correlation between SNPs with CNAs and Expression of CSMD1 and Its Target miRNAs in ESCC
Correlation between SNPs with CNAs and Expression of CSMD1
3.5. Correlation between SNPs with CNAs and Expression of CSMD1 Target miRNAs
3.6. Correlation between Expression of CSMD1 mRNA and Target miRNAs
3.7. CSMD1 Expression and Target miRNAs in ESCC Patients with and without Somatic DNA Alterations
3.8. Risk Factors and Clinical Factors and Somatic DNA Alterations in CSMD1
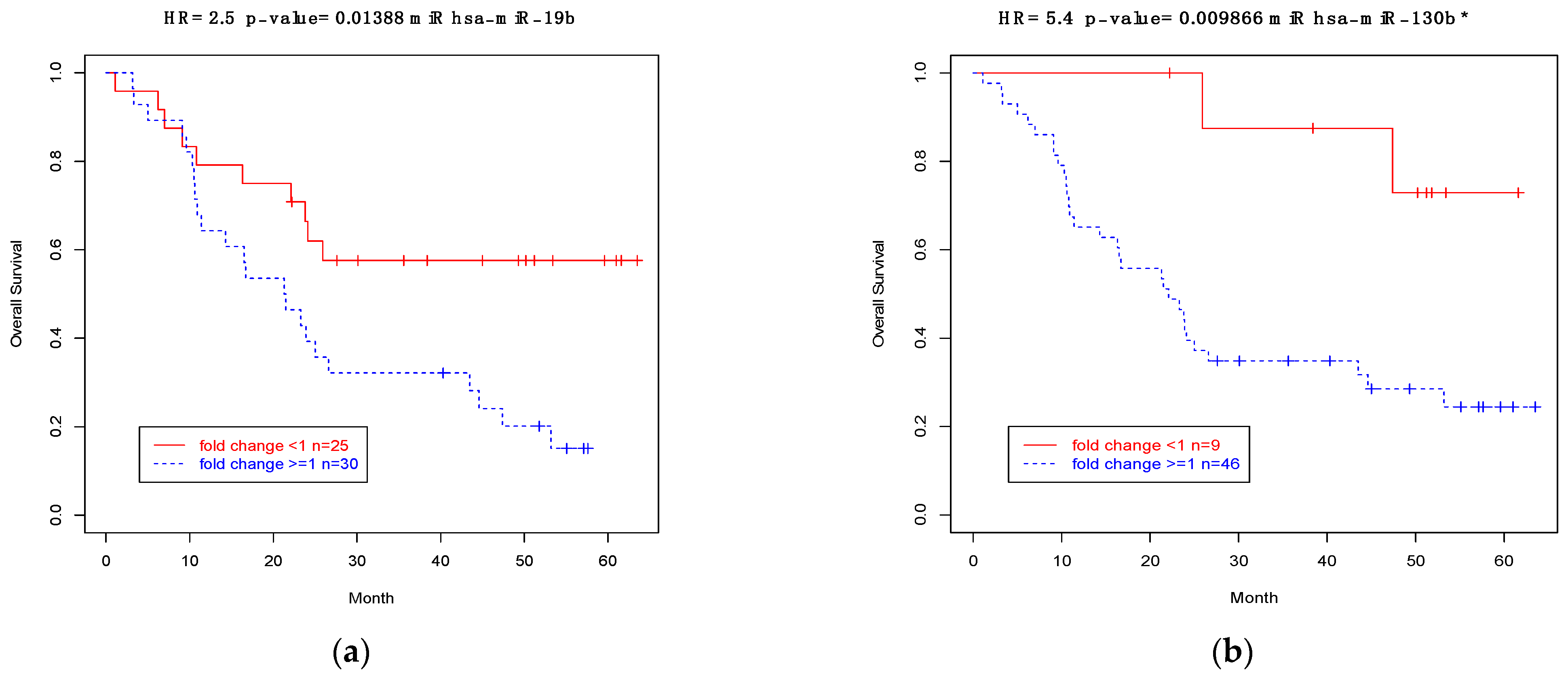
3.9. Association between Expressions of CSMD1 and Target miRNAs and Survival in ESCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, P.C.; Uppaluri, R.; Schmidt, A.P.; Pashia, M.E.; Quant, E.C.; Sunwoo, J.B.; Gollin, S.; Scholnick, S.B. Transcript map of the 8p23 putative tumor suppressor region. Genomics 2001, 75, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Kamal, M.; Shaaban, A.M.; Zhang, L.; Walker, C.; Gray, S.; Thakker, N.; Toomes, C.; Speirs, V.; Bell, S.M. Loss of CSMD1 expression is associated with high tumour grade and poor survival in invasive ductal breast carcinoma. Breast Cancer Res. Treat. 2010, 121, 555–563. [Google Scholar] [CrossRef]
- Escudero-Esparza, A.; Kalchishkova, N.; Kurbasic, E.; Jiang, W.G.; Blom, A.M. The novel complement inhibitor human CUB and Sushi multiple domains 1 (CSMD1) protein promotes factor I-mediated degradation of C4b and C3b and inhibits the membrane attack complex assembly. FASEB J. 2013, 27, 5083–5093. [Google Scholar] [CrossRef]
- Kraus, D.M.; Elliott, G.S.; Chute, H.; Horan, T.; Pfenninger, K.H.; Sanford, S.D.; Foster, S.; Scully, S.; Welcher, A.A.; Holers, V.M. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelia tissues. J. Immunol. 2006, 176, 4419–4430. [Google Scholar] [CrossRef]
- Ma, C.; Quesnelle, K.M.; Sparano, A.; Rao, S.; Park, M.S.; Cohen, M.A.; Wang, Y.; Samanta, M.; Kumar, M.S.; Aziz, M.U.; et al. Characterization CSMD1 in a large set of primary lung, head and neck, breast and skin cancer tissues. Cancer Biol. Ther. 2009, 8, 907–916. [Google Scholar] [CrossRef]
- Sheffer, M.; Bacolod, M.D.; Zuk, O.; Giardina, S.F.; Pincas, H.; Barany, F.; Paty, P.B.; Gerald, W.L.; Notterman, D.A.; Domany, E. Association of survival and disease progression with chromosomal instability: A genomic exploration of colorectal cancer. Proc. Natl. Acad. Sci. USA 2009, 106, 7131–7136. [Google Scholar] [CrossRef]
- Hassan, N.Z.A.; Mokhtar, N.M.; Sin, T.K.; Rose, I.M.; Sagap, I.; Harun, R.; Jamal, R. Integrated analysis of copy number Variation and genome-wide expression profiling in colorectal cancer tissues. PLoS ONE 2014, 9, e92553. [Google Scholar] [CrossRef]
- Deng, N.; Goh, L.K.; Wang, H.; Das, K.; Tao, J.; Tan, L.B.; Zhang, S.; Lee, M.; Wu, J.; Lim, K.H.; et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic target. Gut 2012, 61, 673–684. [Google Scholar] [CrossRef]
- Kuo, K.-T.; Guan, B.; Feng, Y.; Mao, T.-L.; Chen, X.; Jinawath, N.; Wang, Y.; Kurman, R.J.; Shih, I.-M.; Wang, T.-L. Analysis of DNA Copy Number Alterations in Ovarian Serous Tumors Identifies New Molecular Genetic Changes in Low-Grade and High-Grade Carcinomas. Cancer Res. 2009, 69, 4036–4042. [Google Scholar] [CrossRef]
- Farrell, C.L.; Crimm, H.; Meeh, P.; Croshaw, R.; Barbar, T.D.; Vandersteenhoven, J.J.; Butler, W.; Buckhaults, P. Somatic mutations to CSMD1 in colorectal adenocarcinomas. Cancer Biol. Ther. 2008, 7, 609–613. [Google Scholar] [CrossRef]
- Shull, A.Y.; Clendenning, M.L.; Ghoshal-Gupta, S.; Farrell, C.L.; Vangapandu, H.V.; Dudas, L.; Wilkerson, B.J.; Buckhaults, P.J. Somatic Mutations, Allele Loss, and DNA Methylation of the Cub and Sushi Multiple Domains 1 (CSMD1) Gene Reveals Association with Early Age of Diagnosis in Colorectal Cancer Patients. PLoS ONE 2013, 8, e58731. [Google Scholar] [CrossRef] [PubMed]
- Richter, T.M.; Tong, B.D.; Scholnick, S.B. Epigenetic inactivation and aberrant transcription of CSMD1 in squamous cell carcinoma cell lines. Cancer Cell Int. 2005, 5, 29. [Google Scholar] [CrossRef][Green Version]
- Hogan, L.E.; Meyer, J.A.; Yang, J.; Wang, J.; Wong, N.; Yang, W.; Condos, G.; Hunger, S.P.; Raetz, E.; Saffery, R.; et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood 2011, 118, 5128–5226. [Google Scholar] [CrossRef]
- Midorikawa, Y.; Yamamoto, S.; Tsuji, S.; Kamimura, N.; Ishikawa, S.; Igarashi, H.; Makuuchi, M.; Kokudo, N.; Sugimura, H.; Aburatani, H. Allelic imbalances and homozygous deletion on 8p23.2 for stepwise progression of hepatocarcinogenesis. Hepatology 2009, 49, 513–522. [Google Scholar] [CrossRef]
- Toomes, C.; Jackson, A.; Maguire, K.; Wood, J.; Gollin, S.; Ishwad, C.; Paterson, I.; Prime, S.; Parkinson, K.; Bell, S.; et al. The presence of multiple regions of homozygous deletion at the CSMD1 locus in oral squamous cell carcinoma question the role of CSMD1 in head and neck carcinogenesis. Genes Chromosomes Cancer 2003, 37, 132–140. [Google Scholar] [CrossRef]
- Tang, M.R.; Wang, Y.X.; Guo, S.; Han, S.Y.; Wang, D. CSMD1 exhibits antitumor activity in A375 melanoma cells through activation of the Smad pathway. Apoptosis 2012, 17, 927–937. [Google Scholar] [CrossRef]
- Hu, N.; Wang, C.; Clifford, R.J.; Yang, H.H.; Su, H.; Wang, L.; Wang, Y.; Xu, Y.; Tang, Z.-Z.; Ding, T.; et al. Integrative genomics analysis of genes with biallelic loss and its relation to the expression of mRNA and micro-RNA in esophageal squamous cell carcinoma. BMC Genom. 2015, 16, 732. [Google Scholar] [CrossRef]
- Hu, N.; Kadota, M.; Liu, H.; Abnet, C.C.; Su, H.; Wu, H.; Freedman, N.D.; Yang, H.H.; Wang, C.; Yan, C.; et al. Genomic Landscape of Somatic Alterations in Esophageal Squamous Cell Carcinoma and Gastric Cancer. Cancer Res. 2016, 7697, 1714–1723. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, Z.; Li, J.; Hu, X.-D.; Shi, X.-J.; Sun, Z.-M.; Zhang, F.; Zhao, Z.-R.; Li, Z.-T.; Liu, Z.-Y.; et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 1097–1102. [Google Scholar] [CrossRef]
- Moody, S.; Senkin, S.; Islam, S.M.A.; Wang, J.; Nasrollahzadeh, D.; Penha, R.C.C.; Fitzgerald, S.; Bergstrom, E.N.; Atkins, J.; He, Y.; et al. Mutational signatures in esophageal squamous cell carcinoma from 8 countries with varying incidence. Nat. Genet. 2021, 53, 1553–1563. [Google Scholar] [CrossRef]
- Lang, M.-F.; Yang, S.; Zhao, C.; Sun, G.; Murai, K.; Wu, X.; Wang, J.; Gao, H.; Brown, C.E.; Liu, X.; et al. Genome-wide profiling identified a set of miRNAs that are differentially expressed in glioblastoma stem cells and normal neural stem cells. PLoS ONE 2015, 7, e36248. [Google Scholar] [CrossRef]
- Zhu, Q.; Gong, L.; Wang, J.; Tu, Q.; Zhang, J.R.; Han, X.J.; Zhu, S.J.; Wang, S.M.; Li, Y.H.; Zhang, W. miR-10b experts oncogenic activity in human hepatocellular carcinoma cells by targeting expression of CUB and sushi multiple domains 1 (CSMD1). BMC Cancer 2016, 16, 806. [Google Scholar] [CrossRef]
- Yang, H.; Su, H.; Hu, N.; Wang, C.; Wang, L.; Giffen, C.; Goldstein, A.M.; Lee, M.P.; Taylor, P.R. Integrated analysis of genome-wide miRNAs and targeted gene expression in esophageal squamous cell carcinoma (ESCC) and relation to prognosis. BMC Cancer 2020, 20, 388. [Google Scholar] [CrossRef]
- Yang, H.H.; Liu, H.; Hu, N.; Su, H.; Wang, C.; Giffen, C.; Goldstein, A.M.; Taylor, P.R.; Lee, M.P. Modified eQTL and Somatic DNA Segment Alterations in Esophageal Squamous Cell Carcinoma for Genes Related to Immunity, DNA Repair, and Inflammation. Cancers 2022, 14, 1629. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef]
- ABI. User Bulletin No. 2ABI Prism 7700 Sequence Detection System. SUBJECT: Relative Quantitation of Gene Expression; Applied Biosystems: Waltham, MA, USA, 1997. [Google Scholar]
- Livak, M.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-CT) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Aranda, V.A.; Bardelli, A.; Blanpain, C.; Bock, C.; Borowski, C.; Caldas, C.; Califano, A.; Doherty, M.; Elsner, M.; et al. Toward understanding & exploiting tumor heterogeneity. Nat. Med. 2015, 2, 846–853. [Google Scholar]
- Fort, R.S.; Mathó, C.; Oliveira-Rizzo, C.; Garat, B.; Sotelo-Silveira, J.R.; Duhagon, M.A. An integrated view of the role of miR-130b/301b miRNA cluster in prostate cancer. Exp. Hematol. Oncol. 2018, 7, 10. [Google Scholar] [CrossRef]

| No | Case ID | DNA Somatic Alterations on CSMD1 | Gene Expression (FC) 1 | Target miRNAs (FC) 2 |
|---|---|---|---|---|
| onCSMD1 | onCSMD1 | |||
| 1 | E1450 | Allelic Imbalance | 3.67 | 1.77 |
| 2 | E1535 | Allelic Imbalance | 0.87 | 1.41 |
| 3 | E1566 | Allelic Imbalance | 25.25 | 26.53 |
| 4 | E1860 | Allelic Imbalance | 0.36 | 2.07 |
| 5 | E1866 | Allelic Imbalance | NA | 1 |
| 6 | E1885 | Allelic Imbalance | 41.10 | 72.57 |
| 7 | E0362 | Allelic Imbalance, CN Gain | 0.98 | 12.02 |
| 8 | E0844 | Allelic Imbalance, CN Gain | 48.01 | 15.25 |
| 9 | E1293 | Allelic Imbalance, CN Gain | 3.26 | 0.81 |
| 10 | E1546 | Allelic Imbalance, CN Gain | 0.25 | 2.51 |
| 11 | E1635 | Allelic Imbalance, CN Gain | 1.52 | 0.56 |
| 12 | E1862 | Allelic Imbalance, CN Gain | 0.24 | 6.02 |
| 13 | E1874 | Allelic Imbalance, CN Gain | 0.38 | 1.61 |
| 14 | E0796 | Allelic Imbalance, CN Gain, CN Loss | 0.20 | 3.08 |
| 15 | E1507 | Allelic Imbalance, CN Gain, CN Loss | 0.7 | 8.61 |
| 16 | E1575 | Allelic Imbalance, CN Gain, CN Loss | 0.77 | 0.44 |
| 17 | E0410 | Allelic Imbalance, CN Gain, LOH | 1.05 | 10.65 |
| 18 | E0387 | Allelic Imbalance, CN Loss | 2.77 | 4.36 |
| 19 | E1210 | Allelic Imbalance, CN Loss | 0.15 | 5.58 |
| 20 | E1242 | Allelic Imbalance, CN Loss | 5.18 | 2.85 |
| 21 | E1256 | Allelic Imbalance, CN Loss | 10.25 | 1.11 |
| 22 | E1520 | Allelic Imbalance, CN Loss | 2.08 | 1.4 |
| 23 | E1521 | Allelic Imbalance, CN Loss | 5.03 | 1.07 |
| 24 | E1532 | Allelic Imbalance, CN Loss | 0.27 | 3.15 |
| 25 | E1558 | Allelic Imbalance, CN Loss | 18.69 | 7.58 |
| 26 | E1572 | Allelic Imbalance, CN Loss | 9.38 | 3.75 |
| 27 | E1756 | Allelic Imbalance, CN Loss | 0.25 | 8.6 |
| 28 | E1782 | Allelic Imbalance, CN Loss | 1.63 | 9.28 |
| 29 | E1793 | Allelic Imbalance, CN Loss | NA | 3.34 |
| 30 | E1879 | Allelic Imbalance, CN Loss | 59.56 | 1.65 |
| 31 | E1897 | Allelic Imbalance, CN Loss | 0.42 | 1.83 |
| 32 | E1910 | Allelic Imbalance, CN Loss | 1.04 | 1.27 |
| 33 | E1542 | CN Gain | 186.84 | 13.01 |
| 34 | E1584 | CN Gain | 7.37 | 9.93 |
| 35 | E1416 | CN Loss | 7.70 | 2.7 |
| 36 | E1510 | CN Loss | 0.53 | 7.83 |
| 37 | E1610 | CN Loss | 1.11 | 38.59 |
| 38 | E1475 | CN Loss, LOH | 6.42 | 8.00 |
| 39 | E1864 | LOH | 0.52 | 1.47 |
| 40 | E0381 | not observed | 1.57 | 3.81 |
| 41 | E0742 | not observed | 1.65 | 2.35 |
| 42 | E0746 | not observed | 3.22 | 1.89 |
| 43 | E0822 | not observed | 0.45 | 3.00 |
| 44 | E1179 | not observed | 47.84 | 25.37 |
| 45 | E1195 | not observed | 4.81 | 1.55 |
| 46 | E1400 | not observed | 1.11 | 7.37 |
| 47 | E1415 | not observed | 1.25 | 2.8 |
| 48 | E1435 | not observed | 0.25 | 2.09 |
| 49 | E1451 | not observed | 8.54 | 0.76 |
| 50 | E1573 | not observed | 0.31 | 0.9 |
| 51 | E1589 | not observed | 1.62 | 7.6 |
| 52 | E1796 | not observed | 0.29 | 1.65 |
| 53 | E1877 | not observed | 0.72 | 0.71 |
| 54 | E1880 | not observed | 5.28 | 6.5 |
| 55 | E1882 | not observed | 28.97 | 60.39 |
| 56 | E1905 | not observed | 2.36 | 2.19 |
| CSMD1 | CSMD1 | |||||||
|---|---|---|---|---|---|---|---|---|
| No | rs # | mRNA Expression | rs # | Target miRNA Expression | ||||
| (Intron) | Region | rho Value | p Value | (Intron) | Name | rho Value | p Value | |
| 1 | rs12674947 (3) | exon 6–7 | 0.833 | 0.015 | rs12674947 (3) | Has-miR-130b | −1.000 | 0.000 |
| rs12674947 | exon 5–6 | 0.786 | 0.028 | |||||
| rs12674947 | exon 4–5 | 0.821 | 0.034 | |||||
| 2 | rs17068837 (3) | exon 10–11 | −0.881 | 0.007 | rs17068837 (3) | Has-miR-135b | 0.845 | 0.004 |
| 3 | rs2128213 (7) | exon 10–11 | −0.821 | 0.034 | rs2128213 (7) | has-miR-135b | 0.879 | 0.000 |
| 4 | rs2554706 (3) | exon 10–11 | −0.857 | 0.011 | rs2554706 (3) | has-miR-449a | −0.964 | 0.003 |
| rs2554706 | exon 24–25 | −0.648 | 0.049 | |||||
| 5 | rs7000092 | exon 69–70 | 1.000 | 0.017 | rs7000092 | has-miR-130a | 1.000 | 0.003 |
| 6 | rs7007506 (1) | exon 69–70 | 0.964 | 0.003 | rs7007506 (1) | has-miR-130a | 0.964 | 0.003 |
| rs7007506 | exon 39–40 | 0.929 | 0.007 | |||||
| 7 | rs9693235 (3) | exon 9–10 | 0.964 | 0.003 | rs9693235 (3) | has-miR-454 | 0.905 | 0.005 |
| rs9693235 | exon 4–5 | 0.786 | 0.048 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, N.; Wang, C.; Zhang, T.; Su, H.; Liu, H.; Yang, H.H.; Giffen, C.; Hu, Y.; Taylor, P.R.; Goldstein, A.M. CSMD1 Shows Complex Patterns of Somatic Copy Number Alterations and Expressions of mRNAs and Target Micro RNAs in Esophageal Squamous Cell Carcinoma. Cancers 2022, 14, 5001. https://doi.org/10.3390/cancers14205001
Hu N, Wang C, Zhang T, Su H, Liu H, Yang HH, Giffen C, Hu Y, Taylor PR, Goldstein AM. CSMD1 Shows Complex Patterns of Somatic Copy Number Alterations and Expressions of mRNAs and Target Micro RNAs in Esophageal Squamous Cell Carcinoma. Cancers. 2022; 14(20):5001. https://doi.org/10.3390/cancers14205001
Chicago/Turabian StyleHu, Nan, Chaoyu Wang, Tongwu Zhang, Hua Su, Huaitian Liu, Howard H. Yang, Carol Giffen, Ying Hu, Philip R. Taylor, and Alisa M. Goldstein. 2022. "CSMD1 Shows Complex Patterns of Somatic Copy Number Alterations and Expressions of mRNAs and Target Micro RNAs in Esophageal Squamous Cell Carcinoma" Cancers 14, no. 20: 5001. https://doi.org/10.3390/cancers14205001
APA StyleHu, N., Wang, C., Zhang, T., Su, H., Liu, H., Yang, H. H., Giffen, C., Hu, Y., Taylor, P. R., & Goldstein, A. M. (2022). CSMD1 Shows Complex Patterns of Somatic Copy Number Alterations and Expressions of mRNAs and Target Micro RNAs in Esophageal Squamous Cell Carcinoma. Cancers, 14(20), 5001. https://doi.org/10.3390/cancers14205001






