Claudins and Gastric Cancer: An Overview
Abstract
Simple Summary
Abstract
1. Introduction
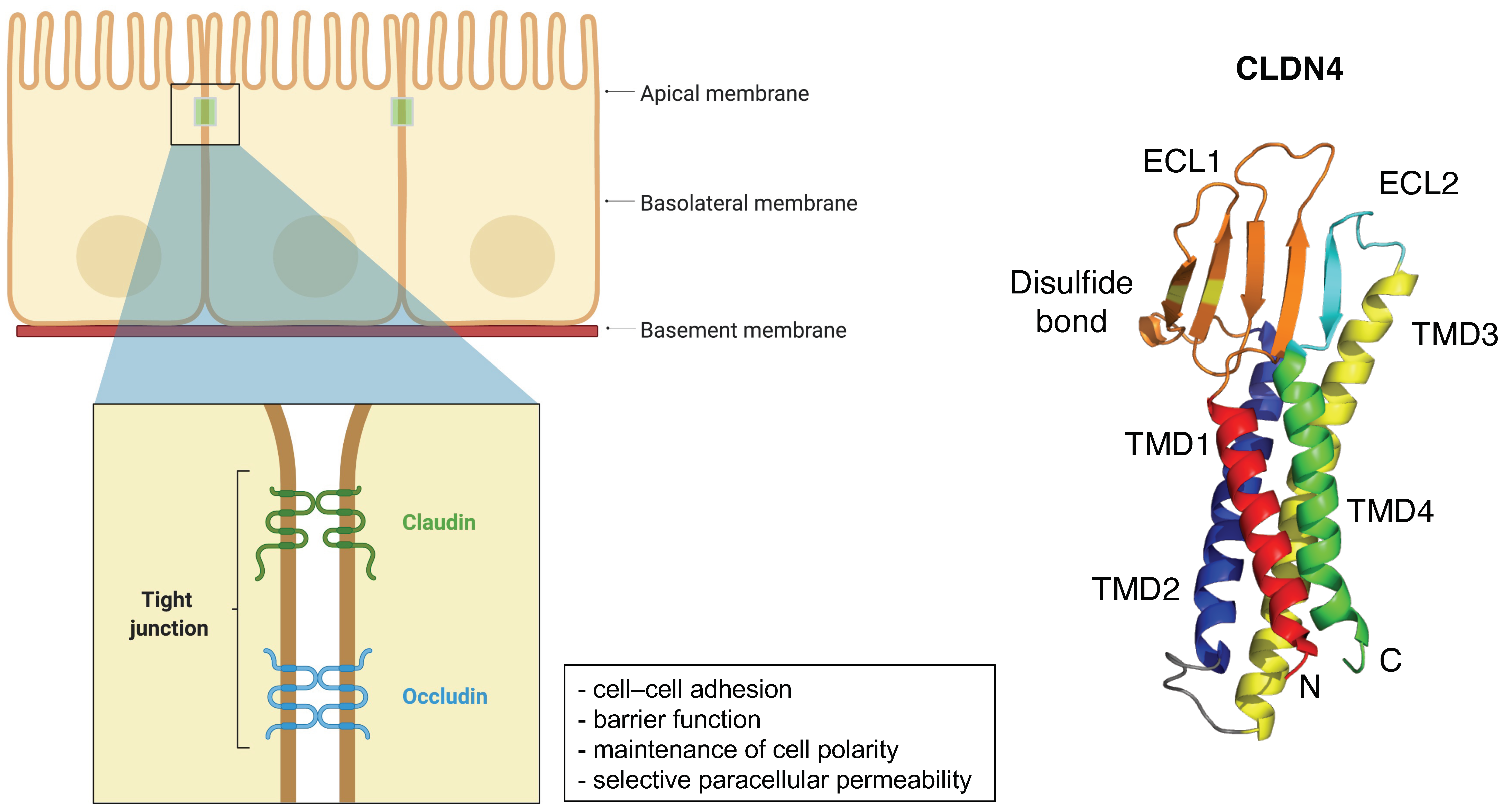
2. General Information of CLDNs
3. CLDN Expression in GC
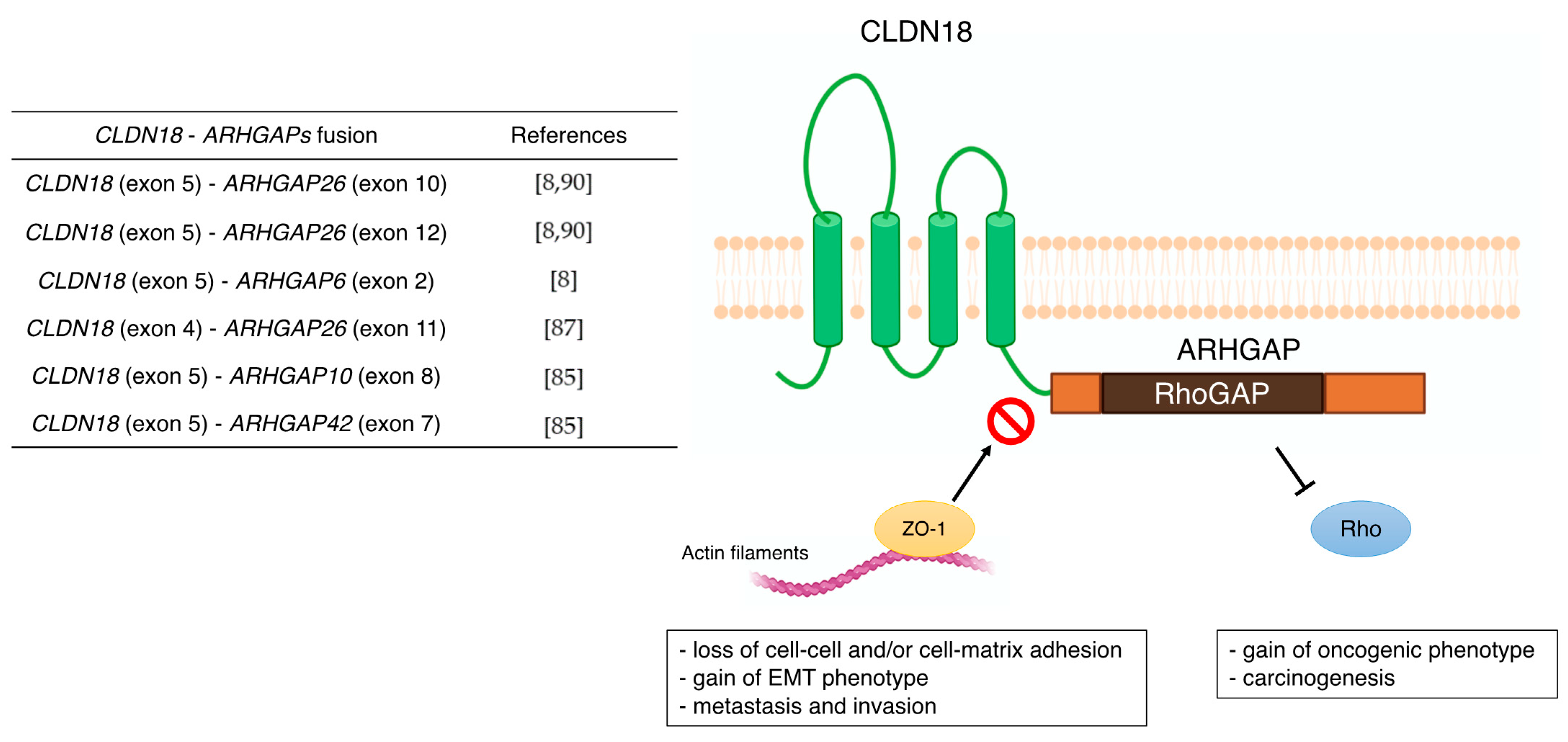
3.1. CLDN18
| Type of CLDN | Main Functions, Signaling Molecules Involved, and Clinical Significances in GC | References |
|---|---|---|
| CLDN1 | - correlation with tumor infiltration and metastasis - Akt, Src, and NF-κB signaling pathway - poor prognostic factor | [39,40] [41,42] [40,43] |
| CLDN2 | - CDX2-dependent targeting relationship with CagA produced from H. pylori | [44] |
| CLDN3 | - correlation with lymph node metastasis - important immunosuppressive regulator | [45] [46] |
| CLDN4 | - correlation with lymph node metastasis - promoting EMT and infiltration of MMP-2 and MMP-9 | [47] [48,49,50] |
| CLDN6 | - cell proliferation and migration/infiltration with YAP1 - high expression was positively correlated with decreased OS | [51] [51,52] |
| CLDN7 | - proliferation in a CagA-and β-catenin-dependent manner - poor prognostic factor | [53] [54] |
| CLDN10 | - association with metastasis and proliferation | [55,56] |
| CLDN11 | - correlation with H. pylori infection and Borrmann classification, not with lymph node metastasis and TNM stage | [57] |
| CLDN14 | - correlation with lymph node metastasis | [58] |
| CLDN17 | - correlation with lymph node metastasis | [58] |
| CLDN18 | - correlation with metastasis (lymph node, peritoneal, bone, and liver metastasis) - poor prognostic factor - Wnt, β-catenin, CD44, EFNB/ EPHB receptor signals, and HIPPO signals - CLDN18-ARHGAP fusion in genomically stable type - therapeutic target of Claudiximab (IMAB362, Zolbetuximab) | [59,60] [61,62,63,64,65,66,67,68] [54,69,70] [8] [10,71,72,73] |
| CLDN23 | - poor prognostic factor | [57] |
3.2. CLDN1
3.3. CLDN2
3.4. CLDN3
3.5. CLDN4
3.6. CLDN6
3.7. CLDN7
3.8. CLDN10
3.9. CLDN11
3.10. CLDN14
3.11. CLDN17
3.12. CLDN23
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Riihimäki, M.; Hemminki, A.; Sundquist, K.; Sundquist, J.; Hemminki, K. Metastatic Spread in Patients with Gastric Cancer. Oncotarget 2016, 7, 52307–52316. [Google Scholar] [CrossRef]
- Li, W.; Ng, J.M.-K.; Wong, C.C.; Ng, E.K.W.; Yu, J. Molecular Alterations of Cancer Cell and Tumour Microenvironment in Metastatic Gastric Cancer. Oncogene 2018, 37, 4903–4920. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular Mechanisms of Epithelial-Mesenchymal Transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Acloque, H.; Adams, M.S.; Fishwick, K.; Bronner-Fraser, M.; Nieto, M.A. Epithelial-Mesenchymal Transitions: The Importance of Changing Cell State in Development and Disease. J. Clin. Investig. 2009, 119, 1438–1449. [Google Scholar] [CrossRef]
- Milatz, S.; Piontek, J.; Hempel, C.; Meoli, L.; Grohe, C.; Fromm, A.; Lee, I.-F.M.; El-Athman, R.; Günzel, D. Tight Junction Strand Formation by Claudin-10 Isoforms and Claudin-10a/-10b Chimeras. Ann. N. Y. Acad. Sci. 2017, 1405, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Tabariès, S.; Siegel, P.M. The Role of Claudins in Cancer Metastasis. Oncogene 2017, 36, 1176–1190. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [CrossRef]
- Türeci, O.; Sahin, U.; Schulze-Bergkamen, H.; Zvirbule, Z.; Lordick, F.; Koeberle, D.; Thuss-Patience, P.; Ettrich, T.; Arnold, D.; Bassermann, F.; et al. A Multicentre, Phase II a Study of Zolbetuximab as a Single Agent in Patients with Recurrent or Refractory Advanced Adenocarcinoma of the Stomach or Lower Oesophagus: The MONO Study. Ann. Oncol. 2019, 30, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Türeci, Ö.; Manikhas, G.; Lordick, F.; Rusyn, A.; Vynnychenko, I.; Dudov, A.; Bazin, I.; Bondarenko, I.; Melichar, B.; et al. FAST: A Randomised Phase II Study of Zolbetuximab (IMAB362) plus EOX versus EOX Alone for First-Line Treatment of Advanced CLDN18.2-Positive Gastric and Gastro-Oesophageal Adenocarcinoma. Ann. Oncol. 2021, 32, 609–619. [Google Scholar] [CrossRef]
- Krause, G.; Winkler, L.; Mueller, S.L.; Haseloff, R.F.; Piontek, J.; Blasig, I.E. Structure and Function of Claudins. Biochim. Biophys. Acta 2008, 1778, 631–645. [Google Scholar] [CrossRef]
- Bhat, A.A.; Syed, N.; Therachiyil, L.; Nisar, S.; Hashem, S.; Macha, M.A.; Yadav, S.K.; Krishnankutty, R.; Muralitharan, S.; Al-Naemi, H.; et al. Claudin-1, A Double-Edged Sword in Cancer. Int. J. Mol. Sci. 2020, 21, 569. [Google Scholar] [CrossRef] [PubMed]
- Tsukita, S.; Furuse, M. The Structure and Function of Claudins, Cell Adhesion Molecules at Tight Junctions. Ann. N. Y. Acad. Sci. 2000, 915, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Lal-Nag, M.; Morin, P.J. The Claudins. Genome Biol. 2009, 10, 235. [Google Scholar] [CrossRef]
- Colegio, O.R.; Van Itallie, C.M.; McCrea, H.J.; Rahner, C.; Anderson, J.M. Claudins Create Charge-Selective Channels in the Paracellular Pathway between Epithelial Cells. Am. J. Physiol. Cell Physiol. 2002, 283, C142–C147. [Google Scholar] [CrossRef] [PubMed]
- Angelow, S.; Ahlstrom, R.; Yu, A.S.L. Biology of Claudins. Am. J. Physiol. Renal Physiol. 2008, 295, F867–F876. [Google Scholar] [CrossRef] [PubMed]
- Piontek, J.; Winkler, L.; Wolburg, H.; Müller, S.L.; Zuleger, N.; Piehl, C.; Wiesner, B.; Krause, G.; Blasig, I.E. Formation of Tight Junction: Determinants of Homophilic Interaction between Classic Claudins. FASEB J. 2008, 22, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Wen, W.; Wang, R.; Kam, C.; Xia, J.; Feng, W.; Zhang, M. Cdc42-Dependent Formation of the ZO-1/MRCKβ Complex at the Leading Edge Controls Cell Migration. EMBO J. 2011, 30, 665–678. [Google Scholar] [CrossRef]
- Kremerskothen, J.; Stölting, M.; Wiesner, C.; Korb-Pap, A.; van Vliet, V.; Linder, S.; Huber, T.B.; Rottiers, P.; Reuzeau, E.; Genot, E.; et al. Zona Occludens Proteins Modulate Podosome Formation and Function. FASEB J. 2011, 25, 505–514. [Google Scholar] [CrossRef]
- Capaldo, C.T.; Koch, S.; Kwon, M.; Laur, O.; Parkos, C.A.; Nusrat, A. Tight Function Zonula Occludens-3 Regulates Cyclin D1-Dependent Cell Proliferation. Mol. Biol. Cell 2011, 22, 1677–1685. [Google Scholar] [CrossRef]
- González-Mariscal, L.; Tapia, R.; Chamorro, D. Crosstalk of Tight Junction Components with Signaling Pathways. Biochim. Biophys. Acta 2008, 1778, 729–756. [Google Scholar] [CrossRef] [PubMed]
- Tsukita, S.; Furuse, M.; Itoh, M. Multifunctional Strands in Tight Junctions. Nat. Rev. Mol. Cell Biol. 2001, 2, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Van Itallie, C.M.; Gambling, T.M.; Carson, J.L.; Anderson, J.M. Palmitoylation of Claudins Is Required for Efficient Tight-Junction Localization. J. Cell Sci. 2005, 118, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Van Itallie, C.M.; Mitic, L.L.; Anderson, J.M. SUMOylation of Claudin-2. Ann. N. Y. Acad. Sci. 2012, 1258, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Van Itallie, C.M.; Tietgens, A.J.; LoGrande, K.; Aponte, A.; Gucek, M.; Anderson, J.M. Phosphorylation of Claudin-2 on Serine 208 Promotes Membrane Retention and Reduces Trafficking to Lysosomes. J. Cell Sci. 2012, 125, 4902–4912. [Google Scholar] [CrossRef]
- D’Souza, T.; Agarwal, R.; Morin, P.J. Phosphorylation of Claudin-3 at Threonine 192 by CAMP-Dependent Protein Kinase Regulates Tight Junction Barrier Function in Ovarian Cancer Cells. J. Biol. Chem. 2005, 280, 26233–26240. [Google Scholar] [CrossRef] [PubMed]
- Stamatovic, S.M.; Dimitrijevic, O.B.; Keep, R.F.; Andjelkovic, A.V. Protein Kinase Calpha-RhoA Cross-Talk in CCL2-Induced Alterations in Brain Endothelial Permeability. J. Biol. Chem. 2006, 281, 8379–8388. [Google Scholar] [CrossRef]
- Yamauchi, K.; Rai, T.; Kobayashi, K.; Sohara, E.; Suzuki, T.; Itoh, T.; Suda, S.; Hayama, A.; Sasaki, S.; Uchida, S. Disease-Causing Mutant WNK4 Increases Paracellular Chloride Permeability and Phosphorylates Claudins. Proc. Natl. Acad. Sci. USA 2004, 101, 4690–4694. [Google Scholar] [CrossRef]
- Tanaka, M.; Kamata, R.; Sakai, R. EphA2 Phosphorylates the Cytoplasmic Tail of Claudin-4 and Mediates Paracellular Permeability. J. Biol. Chem. 2005, 280, 42375–42382. [Google Scholar] [CrossRef]
- Soma, T.; Chiba, H.; Kato-Mori, Y.; Wada, T.; Yamashita, T.; Kojima, T.; Sawada, N. Thr(207) of Claudin-5 Is Involved in Size-Selective Loosening of the Endothelial Barrier by Cyclic AMP. Exp. Cell Res. 2004, 300, 202–212. [Google Scholar] [CrossRef]
- Gyõrffy, H.; Holczbauer, A.; Nagy, P.; Szabó, Z.; Kupcsulik, P.; Páska, C.; Papp, J.; Schaff, Z.; Kiss, A. Claudin Expression in Barrett’s Esophagus and Adenocarcinoma. Virchows Arch. 2005, 447, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Toom, S.; Huang, Y. Anti-Claudin 18.2 Antibody as New Targeted Therapy for Advanced Gastric Cancer. J. Hematol. Oncol. 2017, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Lu, Z.; Lu, Q.; Chen, Y.-H. The Claudin Family of Proteins in Human Malignancy: A Clinical Perspective. Cancer Manag. Res. 2013, 5, 367–375. [Google Scholar] [CrossRef]
- Hu, Y.-J.; Wang, Y.-D.; Tan, F.-Q.; Yang, W.-X. Regulation of Paracellular Permeability: Factors and Mechanisms. Mol. Biol. Rep. 2013, 40, 6123–6142. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.R.; Buschmann, M.M.; Romero-Calvo, I.; Sailer, A.; Shen, L. The Role of Molecular Remodeling in Differential Regulation of Tight Junction Permeability. Semin. Cell Dev. Biol. 2014, 36, 204–212. [Google Scholar] [CrossRef]
- Niimi, T.; Nagashima, K.; Ward, J.M.; Minoo, P.; Zimonjic, D.B.; Popescu, N.C.; Kimura, S. Claudin-18, a Novel Downstream Target Gene for the T/EBP/NKX2.1 Homeodomain Transcription Factor, Encodes Lung- and Stomach-Specific Isoforms through Alternative Splicing. Mol. Cell Biol. 2001, 21, 7380–7390. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Koslowski, M.; Dhaene, K.; Usener, D.; Brandenburg, G.; Seitz, G.; Huber, C.; Türeci, O. Claudin-18 Splice Variant 2 Is a Pan-Cancer Target Suitable for Therapeutic Antibody Development. Clin. Cancer Res. 2008, 14, 7624–7634. [Google Scholar] [CrossRef]
- Shinozaki, A.; Ushiku, T.; Morikawa, T.; Hino, R.; Sakatani, T.; Uozaki, H.; Fukayama, M. Epstein-Barr Virus-Associated Gastric Carcinoma: A Distinct Carcinoma of Gastric Phenotype by Claudin Expression Profiling. J. Histochem. Cytochem. 2009, 57, 775–785. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Zhang, S.; Wang, G.-R.; Chen, Y.-P. Expression Transformation of Claudin-1 in the Process of Gastric Adenocarcinoma Invasion. World J. Gastroenterol. 2008, 14, 4943–4948. [Google Scholar] [CrossRef]
- Huang, J.; Li, J.; Qu, Y.; Zhang, J.; Zhang, L.; Chen, X.; Liu, B.; Zhu, Z. The Expression of Claudin 1 Correlates with β-Catenin and Is a Prognostic Factor of Poor Outcome in Gastric Cancer. Int. J. Oncol. 2014, 44, 1293–1301. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, L.; He, C.; Qu, Y.; Li, J.; Zhang, J.; Du, T.; Chen, X.; Yu, Y.; Liu, B.; et al. Claudin-1 Enhances Tumor Proliferation and Metastasis by Regulating Cell Anoikis in Gastric Cancer. Oncotarget 2015, 6, 1652–1665. [Google Scholar] [CrossRef] [PubMed]
- Shiozaki, A.; Shimizu, H.; Ichikawa, D.; Konishi, H.; Komatsu, S.; Kubota, T.; Fujiwara, H.; Okamoto, K.; Iitaka, D.; Nakashima, S.; et al. Claudin 1 Mediates Tumor Necrosis Factor Alpha-Induced Cell Migration in Human Gastric Cancer Cells. World J. Gastroenterol. 2014, 20, 17863–17876. [Google Scholar] [CrossRef] [PubMed]
- Eftang, L.L.; Esbensen, Y.; Tannæs, T.M.; Blom, G.P.; Bukholm, I.R.K.; Bukholm, G. Up-Regulation of CLDN1 in Gastric Cancer Is Correlated with Reduced Survival. BMC Cancer 2013, 13, 586. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Chen, H.-X.; Wang, X.-Y.; Deng, X.-Y.; Xi, Y.-X.; He, Q.; Peng, T.-L.; Chen, J.; Chen, W.; Wong, B.C.-Y.; et al. H. Pylori-Encoded CagA Disrupts Tight Junctions and Induces Invasiveness of AGS Gastric Carcinoma Cells via Cdx2-Dependent Targeting of Claudin-2. Cell Immunol. 2013, 286, 22–30. [Google Scholar] [CrossRef]
- Wang, H.; Yang, X. The Expression Patterns of Tight Junction Protein Claudin-1, -3, and -4 in Human Gastric Neoplasms and Adjacent Non-Neoplastic Tissues. Int. J. Clin. Exp. Pathol. 2015, 8, 881–887. [Google Scholar]
- Ren, F.; Zhao, Q.; Zhao, M.; Zhu, S.; Liu, B.; Bukhari, I.; Zhang, K.; Wu, W.; Fu, Y.; Yu, Y.; et al. Immune Infiltration Profiling in Gastric Cancer and Their Clinical Implications. Cancer Sci. 2021, 112, 3569–3584. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhao, J.; Li, A.; Gao, P.; Sun, J.; Song, Y.; Liu, J.; Chen, P.; Wang, Z. Clinicopathological Significance of Claudin 4 Expression in Gastric Carcinoma: A Systematic Review and Meta-Analysis. OncoTargets Ther. 2016, 9, 3205–3212. [Google Scholar] [CrossRef]
- Agarwal, R.; D’Souza, T.; Morin, P.J. Claudin-3 and Claudin-4 Expression in Ovarian Epithelial Cells Enhances Invasion and Is Associated with Increased Matrix Metalloproteinase-2 Activity. Cancer Res. 2005, 65, 7378–7385. [Google Scholar] [CrossRef]
- Lin, X.; Shang, X.; Manorek, G.; Howell, S.B. Regulation of the Epithelial-Mesenchymal Transition by Claudin-3 and Claudin-4. PLoS ONE 2013, 8, e67496. [Google Scholar] [CrossRef]
- Hwang, T.-L.; Lee, L.-Y.; Wang, C.-C.; Liang, Y.; Huang, S.-F.; Wu, C.-M. Claudin-4 Expression Is Associated with Tumor Invasion, MMP-2 and MMP-9 Expression in Gastric Cancer. Exp. Ther. Med. 2010, 1, 789–797. [Google Scholar] [CrossRef]
- Kohmoto, T.; Masuda, K.; Shoda, K.; Takahashi, R.; Ujiro, S.; Tange, S.; Ichikawa, D.; Otsuji, E.; Imoto, I. Claudin-6 Is a Single Prognostic Marker and Functions as a Tumor-Promoting Gene in a Subgroup of Intestinal Type Gastric Cancer. Gastric Cancer 2020, 23, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhang, Y.; Li, Q.; Zhang, Z.; Zhao, G.; Xu, J. CLDN6 Promotes Tumor Progression through the YAP1-Snail1 Axis in Gastric Cancer. Cell Death Dis. 2019, 10, 949. [Google Scholar] [CrossRef] [PubMed]
- Wroblewski, L.E.; Piazuelo, M.B.; Chaturvedi, R.; Schumacher, M.; Aihara, E.; Feng, R.; Noto, J.M.; Delgado, A.; Israel, D.A.; Zavros, Y.; et al. Helicobacter Pylori Targets Cancer-Associated Apical-Junctional Constituents in Gastroids and Gastric Epithelial Cells. Gut 2015, 64, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Jun, K.-H.; Kim, J.-H.; Jung, J.-H.; Choi, H.-J.; Chin, H.-M. Expression of Claudin-7 and Loss of Claudin-18 Correlate with Poor Prognosis in Gastric Cancer. Int. J. Surg. 2014, 12, 156–162. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, X.; Wang, A.; Li, Q.; Zhou, M.; Li, T. CLDN10 Promotes a Malignant Phenotype of Osteosarcoma Cells via JAK1/Stat1 Signaling. J. Cell Commun. Signal 2019, 13, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Cui, C.; Xiao, F.; Wang, H.; Xu, J.; Shi, X.; Yang, Y.; Zhang, Q.; Zheng, X.; Yang, X.; et al. MiR-486 Regulates Metastasis and Chemosensitivity in Hepatocellular Carcinoma by Targeting CLDN10 and CITRON. Hepatol. Res. 2015, 45, 1312–1322. [Google Scholar] [CrossRef]
- Lu, Y.; Jing, J.; Sun, L.; Gong, Y.; Chen, M.; Wang, Z.; Sun, M.; Yuan, Y. Expression of Claudin-11, -23 in Different Gastric Tissues and Its Relationship with the Risk and Prognosis of Gastric Cancer. PLoS ONE 2017, 12, e0174476. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Li, W.; Wang, H.; Wang, G. The Distinct Expression Patterns of Claudin-10, -14, -17 and E-Cadherin between Adjacent Non-Neoplastic Tissues and Gastric Cancer Tissues. Diagn. Pathol. 2013, 8, 205. [Google Scholar] [CrossRef]
- Kim, S.R.; Shin, K.; Park, J.M.; Lee, H.H.; Song, K.Y.; Lee, S.H.; Kim, B.; Kim, S.-Y.; Seo, J.; Kim, J.-O.; et al. Clinical Significance of CLDN18.2 Expression in Metastatic Diffuse-Type Gastric Cancer. J. Gastric. Cancer 2020, 20, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Pellino, A.; Brignola, S.; Riello, E.; Niero, M.; Murgioni, S.; Guido, M.; Nappo, F.; Businello, G.; Sbaraglia, M.; Bergamo, F.; et al. Association of CLDN18 Protein Expression with Clinicopathological Features and Prognosis in Advanced Gastric and Gastroesophageal Junction Adenocarcinomas. J. Pers. Med. 2021, 11, 1095. [Google Scholar] [CrossRef]
- Franco, A.T.; Israel, D.A.; Washington, M.K.; Krishna, U.; Fox, J.G.; Rogers, A.B.; Neish, A.S.; Collier-Hyams, L.; Perez-Perez, G.I.; Hatakeyama, M.; et al. Activation of Beta-Catenin by Carcinogenic Helicobacter Pylori. Proc. Natl. Acad. Sci. USA 2005, 102, 10646–10651. [Google Scholar] [CrossRef] [PubMed]
- Nagy, T.A.; Wroblewski, L.E.; Wang, D.; Piazuelo, M.B.; Delgado, A.; Romero-Gallo, J.; Noto, J.; Israel, D.A.; Ogden, S.R.; Correa, P.; et al. β-Catenin and P120 Mediate PPARδ-Dependent Proliferation Induced by Helicobacter Pylori in Human and Rodent Epithelia. Gastroenterology 2011, 141, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.S.; Riehl, T.E.; Moore, B.D.; Fassan, M.; Rugge, M.; Romero-Gallo, J.; Noto, J.; Peek, R.M.; Stenson, W.F.; Mills, J.C. The Hyaluronic Acid Receptor CD44 Coordinates Normal and Metaplastic Gastric Epithelial Progenitor Cell Proliferation. J. Biol. Chem. 2013, 288, 16085–16097. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, S.; Saeki, N.; Ogawa, K. Aberrant EphB/Ephrin-B Expression in Experimental Gastric Lesions and Tumor Cells. World J. Gastroenterol. 2015, 21, 453–464. [Google Scholar] [CrossRef]
- Wong, S.S.; Kim, K.-M.; Ting, J.C.; Yu, K.; Fu, J.; Liu, S.; Cristescu, R.; Nebozhyn, M.; Gong, L.; Yue, Y.G.; et al. Genomic Landscape and Genetic Heterogeneity in Gastric Adenocarcinoma Revealed by Whole-Genome Sequencing. Nat. Commun. 2014, 5, 5477. [Google Scholar] [CrossRef]
- Ma, L.-G.; Bian, S.-B.; Cui, J.-X.; Xi, H.-Q.; Zhang, K.-C.; Qin, H.-Z.; Zhu, X.-M.; Chen, L. LKB1 Inhibits the Proliferation of Gastric Cancer Cells by Suppressing the Nuclear Translocation of Yap and β-Catenin. Int. J. Mol. Med. 2016, 37, 1039–1048. [Google Scholar] [CrossRef]
- Jiao, S.; Wang, H.; Shi, Z.; Dong, A.; Zhang, W.; Song, X.; He, F.; Wang, Y.; Zhang, Z.; Wang, W.; et al. A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer. Cancer Cell 2014, 25, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Sentani, K.; Tanaka, H.; Yano, T.; Suzuki, K.; Oshima, M.; Yasui, W.; Tamura, A.; Tsukita, S. Deficiency of Stomach-Type Claudin-18 in Mice Induces Gastric Tumor Formation Independent of H Pylori Infection. Cell Mol. Gastroenterol. Hepatol. 2019, 8, 119–142. [Google Scholar] [CrossRef] [PubMed]
- Sanada, Y.; Oue, N.; Mitani, Y.; Yoshida, K.; Nakayama, H.; Yasui, W. Down-Regulation of the Claudin-18 Gene, Identified through Serial Analysis of Gene Expression Data Analysis, in Gastric Cancer with an Intestinal Phenotype. J. Pathol. 2006, 208, 633–642. [Google Scholar] [CrossRef]
- Matsuda, Y.; Semba, S.; Ueda, J.; Fuku, T.; Hasuo, T.; Chiba, H.; Sawada, N.; Kuroda, Y.; Yokozaki, H. Gastric and Intestinal Claudin Expression at the Invasive Front of Gastric Carcinoma. Cancer Sci. 2007, 98, 1014–1019. [Google Scholar] [CrossRef]
- Sahin, U.; Schuler, M.; Richly, H.; Bauer, S.; Krilova, A.; Dechow, T.; Jerling, M.; Utsch, M.; Rohde, C.; Dhaene, K.; et al. A Phase I Dose-Escalation Study of IMAB362 (Zolbetuximab) in Patients with Advanced Gastric and Gastro-Oesophageal Junction Cancer. Eur. J. Cancer 2018, 100, 17–26. [Google Scholar] [CrossRef]
- Sahin, U.; Al-Batran, S.-E.; Hozaeel, W.; Zvirbule, Z.; Freiberg-Richter, J.; Lordick, F.; Just, M.; Bitzer, M.; Thuss-Patience, P.C.; Krilova, A.; et al. IMAB362 plus Zoledronic Acid (ZA) and Interleukin-2 (IL-2) in Patients (Pts) with Advanced Gastroesophageal Cancer (GEC): Clinical Activity and Safety Data from the PILOT Phase I Trial. JCO 2015, 33, e15079. [Google Scholar] [CrossRef]
- Astellas Pharma Global Development, Inc. A Phase 3, Global, Multi-Center, Double-Blind, Randomized, Efficacy Study of Zolbetuximab (IMAB362) Plus CAPOX Compared With Placebo Plus CAPOX as First-Line Treatment of Subjects with Claudin (CLDN) 18.2-Positive, HER2-Negative, Locally Advanced Unresectable or Metastatic Gastric or Gastroesophageal Junction (GEJ) Adenocarcinoma. 2021. Available online: https://clinicaltrials.gov (accessed on 30 September 2021).
- Correa, P. Human Gastric Carcinogenesis: A Multistep and Multifactorial Process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992, 52, 6735–6740. [Google Scholar] [PubMed]
- Correa, P.; Houghton, J. Carcinogenesis of Helicobacter Pylori. Gastroenterology 2007, 133, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Shan, J.; Okugawa, T.; Chen, X.; Hori, K.; Tomita, T.; Fukui, H.; Watari, J.; Miwa, H. Down-Regulation of Claudin-18 Is Associated with the Proliferative and Invasive Potential of Gastric Cancer at the Invasive Front. PLoS ONE 2013, 8, e74757. [Google Scholar] [CrossRef]
- Hagen, S.J.; Ang, L.-H.; Zheng, Y.; Karahan, S.N.; Wu, J.; Wang, Y.E.; Caron, T.J.; Gad, A.P.; Muthupalani, S.; Fox, J.G. Loss of Tight Junction Protein Claudin 18 Promotes Progressive Neoplasia Development in Mouse Stomach. Gastroenterology 2018, 155, 1852–1867. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.P.; Mills, J.C.; Goldenring, J.R. Murine Models of Gastric Corpus Preneoplasia. Cell Mol. Gastroenterol. Hepatol. 2017, 3, 11–26. [Google Scholar] [CrossRef]
- Hayashi, D.; Tamura, A.; Tanaka, H.; Yamazaki, Y.; Watanabe, S.; Suzuki, K.; Suzuki, K.; Sentani, K.; Yasui, W.; Rakugi, H.; et al. Deficiency of Claudin-18 Causes Paracellular H+ Leakage, up-Regulation of Interleukin-1β, and Atrophic Gastritis in Mice. Gastroenterology 2012, 142, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Dottermusch, M.; Krüger, S.; Behrens, H.-M.; Halske, C.; Röcken, C. Expression of the Potential Therapeutic Target Claudin-18.2 Is Frequently Decreased in Gastric Cancer: Results from a Large Caucasian Cohort Study. Virchows Arch. 2019, 475, 563–571. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Hu, D.; Gong, T.; Xu, R.; Gao, J. Analysis of the Expression and Genetic Alteration of CLDN18 in Gastric Cancer. Aging 2020, 12, 14271–14284. [Google Scholar] [CrossRef]
- Coati, I.; Lotz, G.; Fanelli, G.N.; Brignola, S.; Lanza, C.; Cappellesso, R.; Pellino, A.; Pucciarelli, S.; Spolverato, G.; Guzzardo, V.; et al. Claudin-18 Expression in Oesophagogastric Adenocarcinomas: A Tissue Microarray Study of 523 Molecularly Profiled Cases. Br. J. Cancer 2019, 121, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-T.; Jeng, Y.-M.; Yang, C.-Y. Claudin-18 as a Marker for Identifying the Stomach and Pancreatobiliary Tract as the Primary Sites of Metastatic Adenocarcinoma. Am. J. Surg. Pathol. 2020, 44, 1643–1648. [Google Scholar] [CrossRef] [PubMed]
- Kakiuchi, M.; Nishizawa, T.; Ueda, H.; Gotoh, K.; Tanaka, A.; Hayashi, A.; Yamamoto, S.; Tatsuno, K.; Katoh, H.; Watanabe, Y.; et al. Recurrent Gain-of-Function Mutations of RHOA in Diffuse-Type Gastric Carcinoma. Nat. Genet. 2014, 46, 583–587. [Google Scholar] [CrossRef]
- Yao, F.; Kausalya, J.P.; Sia, Y.Y.; Teo, A.S.M.; Lee, W.H.; Ong, A.G.M.; Zhang, Z.; Tan, J.H.J.; Li, G.; Bertrand, D.; et al. Recurrent Fusion Genes in Gastric Cancer: CLDN18-ARHGAP26 Induces Loss of Epithelial Integrity. Cell Rep. 2015, 12, 272–285. [Google Scholar] [CrossRef]
- Tanaka, A.; Ishikawa, S.; Ushiku, T.; Yamazawa, S.; Katoh, H.; Hayashi, A.; Kunita, A.; Fukayama, M. Frequent CLDN18-ARHGAP Fusion in Highly Metastatic Diffuse-Type Gastric Cancer with Relatively Early Onset. Oncotarget 2018, 9, 29336–29350. [Google Scholar] [CrossRef]
- Nakayama, I.; Shinozaki, E.; Sakata, S.; Yamamoto, N.; Fujisaki, J.; Muramatsu, Y.; Hirota, T.; Takeuchi, K.; Takahashi, S.; Yamaguchi, K.; et al. Enrichment of CLDN18-ARHGAP Fusion Gene in Gastric Cancers in Young Adults. Cancer Sci. 2019, 110, 1352–1363. [Google Scholar] [CrossRef] [PubMed]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Shu, Y.; Zhang, W.; Hou, Q.; Zhao, L.; Zhang, S.; Zhou, J.; Song, X.; Zhang, Y.; Jiang, D.; Chen, X.; et al. Prognostic Significance of Frequent CLDN18-ARHGAP26/6 Fusion in Gastric Signet-Ring Cell Cancer. Nat. Commun. 2018, 9, 2447. [Google Scholar] [CrossRef]
- Ushiku, T.; Arnason, T.; Ban, S.; Hishima, T.; Shimizu, M.; Fukayama, M.; Lauwers, G.Y. Very Well-Differentiated Gastric Carcinoma of Intestinal Type: Analysis of Diagnostic Criteria. Mod. Pathol. 2013, 26, 1620–1631. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, N.; Kawachi, H.; Yoshida, T.; Kitagaki, K.; Sekine, M.; Kojima, K.; Kawano, T.; Eishi, Y. “Crawling-Type” Adenocarcinoma of the Stomach: A Distinct Entity Preceding Poorly Differentiated Adenocarcinoma. Gastric. Cancer 2013, 16, 220–232. [Google Scholar] [CrossRef]
- Hashimoto, T.; Ogawa, R.; Tang, T.-Y.; Yoshida, H.; Taniguchi, H.; Katai, H.; Oda, I.; Sekine, S. RHOA Mutations and CLDN18-ARHGAP Fusions in Intestinal-Type Adenocarcinoma with Anastomosing Glands of the Stomach. Mod. Pathol. 2019, 32, 568–575. [Google Scholar] [CrossRef]
- Fukata, M.; Kaibuchi, K. Rho-Family GTPases in Cadherin-Mediated Cell-Cell Adhesion. Nat. Rev. Mol. Cell Biol. 2001, 2, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.N.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-Genome Sequencing and Comprehensive Molecular Profiling Identify New Driver Mutations in Gastric Cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef]
- Kataoka, K.; Ogawa, S. Variegated RHOA Mutations in Human Cancers. Exp. Hematol. 2016, 44, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Schaefer, A.; Wang, Y.; Hodge, R.G.; Blake, D.R.; Diehl, J.N.; Papageorge, A.G.; Stachler, M.D.; Liao, J.; Zhou, J.; et al. Gain-of-Function RHOA Mutations Promote Focal Adhesion Kinase Activation and Dependency in Diffuse Gastric Cancer. Cancer Discov. 2020, 10, 288–305. [Google Scholar] [CrossRef] [PubMed]
- Klamp, T.; Schumacher, J.; Huber, G.; Kühne, C.; Meissner, U.; Selmi, A.; Hiller, T.; Kreiter, S.; Markl, J.; Türeci, Ö.; et al. Highly Specific Auto-Antibodies against Claudin-18 Isoform 2 Induced by a Chimeric HBcAg Virus-like Particle Vaccine Kill Tumor Cells and Inhibit the Growth of Lung Metastases. Cancer Res. 2011, 71, 516–527. [Google Scholar] [CrossRef]
- Micke, P.; Mattsson, J.S.M.; Edlund, K.; Lohr, M.; Jirström, K.; Berglund, A.; Botling, J.; Rahnenfuehrer, J.; Marincevic, M.; Pontén, F.; et al. Aberrantly Activated Claudin 6 and 18.2 as Potential Therapy Targets in Non-Small-Cell Lung Cancer. Int. J. Cancer 2014, 135, 2206–2214. [Google Scholar] [CrossRef] [PubMed]
- Wöll, S.; Schlitter, A.M.; Dhaene, K.; Roller, M.; Esposito, I.; Sahin, U.; Türeci, Ö. Claudin 18.2 Is a Target for IMAB362 Antibody in Pancreatic Neoplasms. Int. J. Cancer 2014, 134, 731–739. [Google Scholar] [CrossRef]
- Türeci, O.; Koslowski, M.; Helftenbein, G.; Castle, J.; Rohde, C.; Dhaene, K.; Seitz, G.; Sahin, U. Claudin-18 Gene Structure, Regulation, and Expression Is Evolutionary Conserved in Mammals. Gene 2011, 481, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Sentani, K.; Oue, N.; Tashiro, T.; Sakamoto, N.; Nishisaka, T.; Fukuhara, T.; Taniyama, K.; Matsuura, H.; Arihiro, K.; Ochiai, A.; et al. Immunohistochemical Staining of Reg IV and Claudin-18 Is Useful in the Diagnosis of Gastrointestinal Signet Ring Cell Carcinoma. Am. J. Surg. Pathol. 2008, 32, 1182–1189. [Google Scholar] [CrossRef]
- Jiang, H.; Shi, Z.; Wang, P.; Wang, C.; Yang, L.; Du, G.; Zhang, H.; Shi, B.; Jia, J.; Li, Q.; et al. Claudin18.2-Specific Chimeric Antigen Receptor Engineered T Cells for the Treatment of Gastric Cancer. J. Natl. Cancer Inst. 2019, 111, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Foletti, D.; Liu, X.; Ding, S.; Melton Witt, J.; Hasa-Moreno, A.; Rickert, M.; Holz, C.; Aschenbrenner, L.; Yang, A.H.; et al. Targeting CLDN18.2 by CD3 Bispecific and ADC Modalities for the Treatments of Gastric and Pancreatic Cancer. Sci. Rep. 2019, 9, 8420. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Chong, X.; Zhao, M.; Jia, F.; Wang, Z.; Zhou, Y.; Lu, X.; Huang, Q.; Li, P.; Yang, Y.; et al. Ultrasensitive Gastric Cancer Circulating Tumor Cellular CLDN18.2 RNA Detection Based on a Molecular Beacon. Anal. Chem. 2021, 93, 665–670. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 30 September 2021).
- Resnick, M.B.; Gavilanez, M.; Newton, E.; Konkin, T.; Bhattacharya, B.; Britt, D.E.; Sabo, E.; Moss, S.F. Claudin Expression in Gastric Adenocarcinomas: A Tissue Microarray Study with Prognostic Correlation. Hum. Pathol. 2005, 36, 886–892. [Google Scholar] [CrossRef] [PubMed]
- Soini, Y.; Tommola, S.; Helin, H.; Martikainen, P. Claudins 1, 3, 4 and 5 in Gastric Carcinoma, Loss of Claudin Expression Associates with the Diffuse Subtype. Virchows Arch. 2006, 448, 52–58. [Google Scholar] [CrossRef]
- Kinugasa, T.; Huo, Q.; Higashi, D.; Shibaguchi, H.; Kuroki, M.; Tanaka, T.; Futami, K.; Yamashita, Y.; Hachimine, K.; Maekawa, S.; et al. Selective Up-Regulation of Claudin-1 and Claudin-2 in Colorectal Cancer. Anticancer Res. 2007, 27, 3729–3734. [Google Scholar] [CrossRef]
- Kim, T.H.; Huh, J.H.; Lee, S.; Kang, H.; Kim, G.I.; An, H.J. Down-Regulation of Claudin-2 in Breast Carcinomas Is Associated with Advanced Disease. Histopathology 2008, 53, 48–55. [Google Scholar] [CrossRef]
- Song, X.; Li, X.; Tang, Y.; Chen, H.; Wong, B.; Wang, J.; Chen, M. Expression of Claudin-2 in the Multistage Process of Gastric Carcinogenesis. Histol. Histopathol. 2008, 23, 673–682. [Google Scholar] [CrossRef]
- Xin, S.; Huixin, C.; Benchang, S.; Aiping, B.; Jinhui, W.; Xiaoyan, L.; Yu, W.B.C.; Minhu, C. Expression of Cdx2 and Claudin-2 in the Multistage Tissue of Gastric Carcinogenesis. Oncology 2007, 73, 357–365. [Google Scholar] [CrossRef]
- Aung, P.P.; Mitani, Y.; Sanada, Y.; Nakayama, H.; Matsusaki, K.; Yasui, W. Differential Expression of Claudin-2 in Normal Human Tissues and Gastrointestinal Carcinomas. Virchows Arch. 2006, 448, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhang, X.; Liu, Z.; Liu, Q.; Wang, L.; Lu, Y.; Liu, Y.; Wang, M.; Yang, M.; Jin, X.; et al. The Distinct Expression Patterns of Claudin-2, -6, and -11 between Human Gastric Neoplasms and Adjacent Non-Neoplastic Tissues. Diagn. Pathol. 2013, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Okugawa, T.; Oshima, T.; Chen, X.; Hori, K.; Tomita, T.; Fukui, H.; Watari, J.; Matsumoto, T.; Miwa, H. Down-Regulation of Claudin-3 Is Associated with Proliferative Potential in Early Gastric Cancers. Dig. Dis. Sci. 2012, 57, 1562–1567. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Jun, K.H.; Jung, J.H.; Chin, H.M.; Park, W.B. The Expression of Claudin-1, Claudin-2, Claudin-3, and Claudin-4 in Gastric Cancer Tissue. J. Surg. Res. 2011, 167, e185–e191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yu, W.; Chen, S.; Chen, Y.; Chen, L.; Zhang, S. Methylation of the Claudin-3 Promoter Predicts the Prognosis of Advanced Gastric Adenocarcinoma. Oncol. Rep. 2018, 40, 49–60. [Google Scholar] [CrossRef]
- Zhu, J.-L.; Gao, P.; Wang, Z.-N.; Song, Y.-X.; Li, A.-L.; Xu, Y.-Y.; Wang, M.-X.; Xu, H.-M. Clinicopathological Significance of Claudin-4 in Gastric Carcinoma. World J. Surg. Oncol. 2013, 11, 150. [Google Scholar] [CrossRef]
- Tokuhara, Y.; Morinishi, T.; Matsunaga, T.; Ohsaki, H.; Kushida, Y.; Haba, R.; Hirakawa, E. Claudin-1, but Not Claudin-4, Exhibits Differential Expression Patterns between Well- to Moderately-Differentiated and Poorly-Differentiated Gastric Adenocarcinoma. Oncol. Lett. 2015, 10, 93–98. [Google Scholar] [CrossRef][Green Version]
- Kwon, M.J.; Kim, S.-H.; Jeong, H.M.; Jung, H.S.; Kim, S.-S.; Lee, J.E.; Gye, M.C.; Erkin, O.C.; Koh, S.S.; Choi, Y.-L.; et al. Claudin-4 Overexpression Is Associated with Epigenetic Derepression in Gastric Carcinoma. Lab. Investig. 2011, 91, 1652–1667. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-T.; He, J.; Liang, Q.; Ren, L.-L.; Yan, T.-T.; Yu, T.-C.; Tang, J.-Y.; Bao, Y.-J.; Hu, Y.; Lin, Y.; et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016, 6, 784–801. [Google Scholar] [CrossRef]
- Han, T.-S.; Hur, K.; Xu, G.; Choi, B.; Okugawa, Y.; Toiyama, Y.; Oshima, H.; Oshima, M.; Lee, H.-J.; Kim, V.N.; et al. MicroRNA-29c Mediates Initiation of Gastric Carcinogenesis by Directly Targeting ITGB1. Gut 2015, 64, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-X.; Sun, J.-X.; Zhao, J.-H.; Yang, Y.-C.; Shi, J.-X.; Wu, Z.-H.; Chen, X.-W.; Gao, P.; Miao, Z.-F.; Wang, Z.-N. Non-Coding RNAs Participate in the Regulatory Network of CLDN4 via CeRNA Mediated MiRNA Evasion. Nat Commun 2017, 8, 289. [Google Scholar] [CrossRef]
- Ma, Z.-H.; Shuai, Y.; Gao, X.-Y.; Yan, Y.; Wang, K.-M.; Wen, X.-Z.; Ji, J.-F. BTEB2-Activated LncRNA TSPEAR-AS2 Drives GC Progression through Suppressing GJA1 Expression and Upregulating CLDN4 Expression. Mol. Ther. Nucleic Acids 2020, 22, 1129–1141. [Google Scholar] [CrossRef]
- Kuwada, M.; Chihara, Y.; Luo, Y.; Li, X.; Nishiguchi, Y.; Fujiwara, R.; Sasaki, T.; Fujii, K.; Ohmori, H.; Fujimoto, K.; et al. Pro-Chemotherapeutic Effects of Antibody against Extracellular Domain of Claudin-4 in Bladder Cancer. Cancer Lett. 2015, 369, 212–221. [Google Scholar] [CrossRef]
- Nishiguchi, Y.; Fujiwara-Tani, R.; Sasaki, T.; Luo, Y.; Ohmori, H.; Kishi, S.; Mori, S.; Goto, K.; Yasui, W.; Sho, M.; et al. Targeting Claudin-4 Enhances CDDP-Chemosensitivity in Gastric Cancer. Oncotarget 2019, 10, 2189–2202. [Google Scholar] [CrossRef]
- Gao, F.; Li, M.; Xiang, R.; Zhou, X.; Zhu, L.; Zhai, Y. Expression of CLDN6 in Tissues of Gastric Cancer Patients: Association with Clinical Pathology and Prognosis. Oncol. Lett. 2019, 17, 4621–4625. [Google Scholar] [CrossRef] [PubMed]
- Ushiku, T.; Shinozaki-Ushiku, A.; Maeda, D.; Morita, S.; Fukayama, M. Distinct Expression Pattern of Claudin-6, a Primitive Phenotypic Tight Junction Molecule, in Germ Cell Tumours and Visceral Carcinomas. Histopathology 2012, 61, 1043–1056. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Piao, X.; Wang, C.; Wang, R.; Song, Z. Identification of Claudin-1, -3, -7 and -8 as Prognostic Markers in Human Laryngeal Carcinoma. Mol. Med. Rep. 2019, 20, 393–400. [Google Scholar] [CrossRef]
- Tiwari-Woodruff, S.K.; Buznikov, A.G.; Vu, T.Q.; Micevych, P.E.; Chen, K.; Kornblum, H.I.; Bronstein, J.M. OSP/Claudin-11 Forms a Complex with a Novel Member of the Tetraspanin Super Family and Beta1 Integrin and Regulates Proliferation and Migration of Oligodendrocytes. J. Cell Biol. 2001, 153, 295–305. [Google Scholar] [CrossRef]
- Soini, Y.; Rauramaa, T.; Alafuzoff, I.; Sandell, P.-J.; Kärjä, V. Claudins 1, 11 and Twist in Meningiomas. Histopathology 2010, 56, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Awsare, N.S.; Martin, T.A.; Haynes, M.D.; Matthews, P.N.; Jiang, W.G. Claudin-11 Decreases the Invasiveness of Bladder Cancer Cells. Oncol. Rep. 2011, 25, 1503–1509. [Google Scholar] [CrossRef]
- Li, C.-P.; Cai, M.-Y.; Jiang, L.-J.; Mai, S.-J.; Chen, J.-W.; Wang, F.-W.; Liao, Y.-J.; Chen, W.-H.; Jin, X.-H.; Pei, X.-Q.; et al. CLDN14 Is Epigenetically Silenced by EZH2-Mediated H3K27ME3 and Is a Novel Prognostic Biomarker in Hepatocellular Carcinoma. Carcinogenesis 2016, 37, 557–566. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. CLDN23 Gene, Frequently down-Regulated in Intestinal-Type Gastric Cancer, Is a Novel Member of CLAUDIN Gene Family. Int. J. Mol. Med. 2003, 11, 683–689. [Google Scholar] [CrossRef] [PubMed]
| Study Name | NCT Number | Phase | Number of Participants | Design | Response Rate | OS | PFS | Adverse Effects |
|---|---|---|---|---|---|---|---|---|
| - | NCT00909025 | I | 15 | Single-dose escalation study evaluating safety and tolerability | - | - | - | Vomiting |
| PILOT | NCT01671774 | I | 32 | Multiple dose study of IMAB362 with immunomodulation (Zoledronic acid and IL-2) | 11 patients had disease control | 40 weeks | 12.7 weeks | Nausea and vomiting |
| MONO | NCT01197885 | IIa | 54 | Multiple dose study of IMAB362 as monotherapy | Clinical benefit rate: 23% | - | - | Nausea, vomiting, and fatigue |
| FAST | NCT01630083 | IIb | 246 | Randomized EOX vs. IMAB362 + EOX, extended with high-dose IMAB362 + EOX | Objective response rate: 25 vs. 39% | 8.3 vs. 13.0 months | 5.3 vs. 7.5 months | Neutropenia, anemia, weight loss, and vomiting |
| GLOW | NCT03653507 | III | 500 (estimated) | Double-blinded, Randomized, IMAB362 plus CAPOX compared with placebo plus CAPOX as first-line treatment of subjects with CLDN 18.2-positive, HER2-negative locally advanced unresectable or metastatic gastric or gastroesophageal junction adenocarcinoma | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashimoto, I.; Oshima, T. Claudins and Gastric Cancer: An Overview. Cancers 2022, 14, 290. https://doi.org/10.3390/cancers14020290
Hashimoto I, Oshima T. Claudins and Gastric Cancer: An Overview. Cancers. 2022; 14(2):290. https://doi.org/10.3390/cancers14020290
Chicago/Turabian StyleHashimoto, Itaru, and Takashi Oshima. 2022. "Claudins and Gastric Cancer: An Overview" Cancers 14, no. 2: 290. https://doi.org/10.3390/cancers14020290
APA StyleHashimoto, I., & Oshima, T. (2022). Claudins and Gastric Cancer: An Overview. Cancers, 14(2), 290. https://doi.org/10.3390/cancers14020290






