Prognostic and Predictive Significance of Stromal Tumor-Infiltrating Lymphocytes (sTILs) in ER-Positive/HER2−Negative Postmenopausal Breast Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
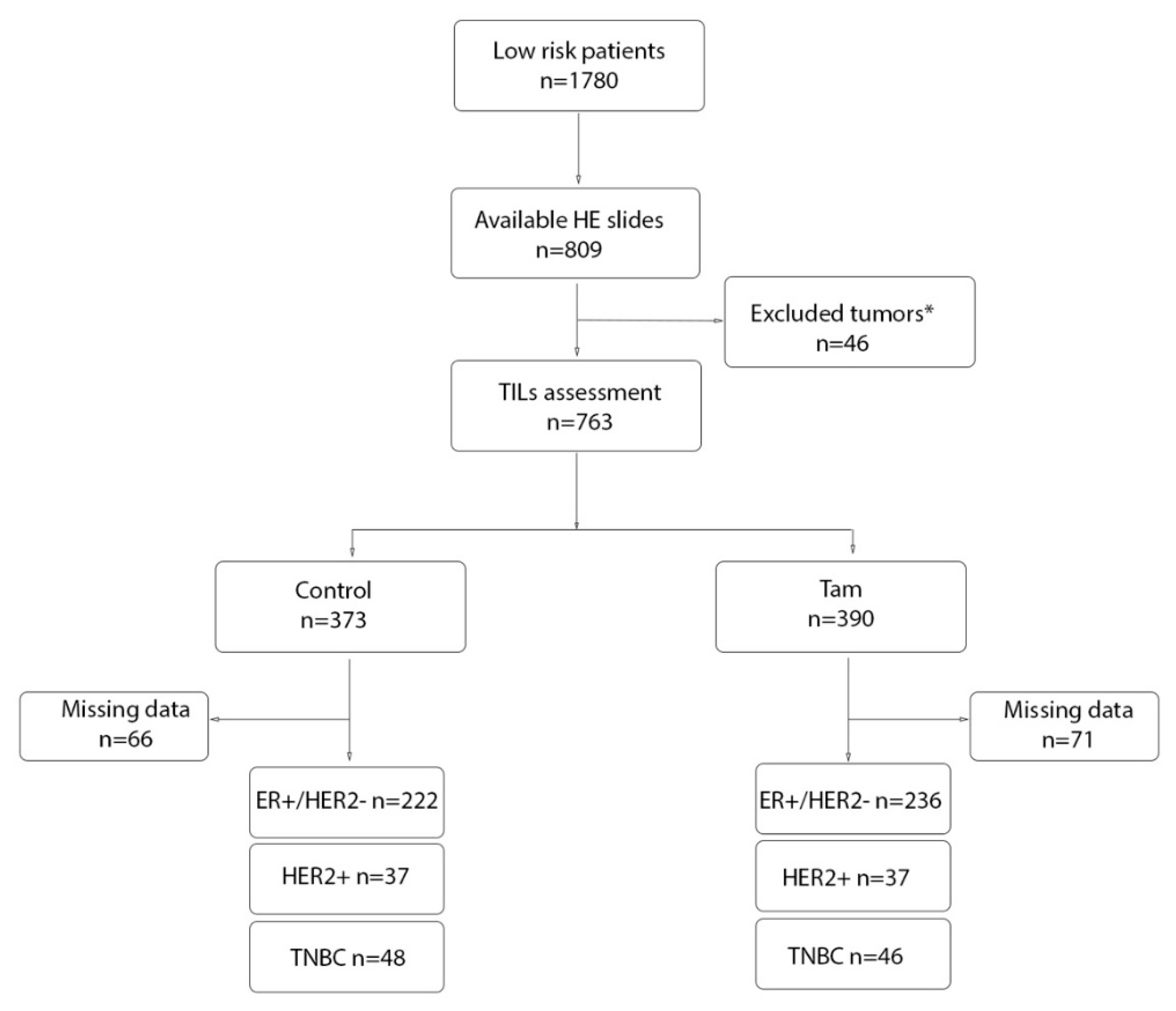
2. Materials and Methods
2.1. Definition of Clinical Variables and Tumor Characteristics
2.2. Immunohistochemical Detection of ER, PR, HER2, and Ki67
2.3. TILs Scoring
2.4. Gene Expression Data and Normalization
2.5. Statistical Analysis
3. Results
3.1. TILs Association with Clinicopathological Characteristics
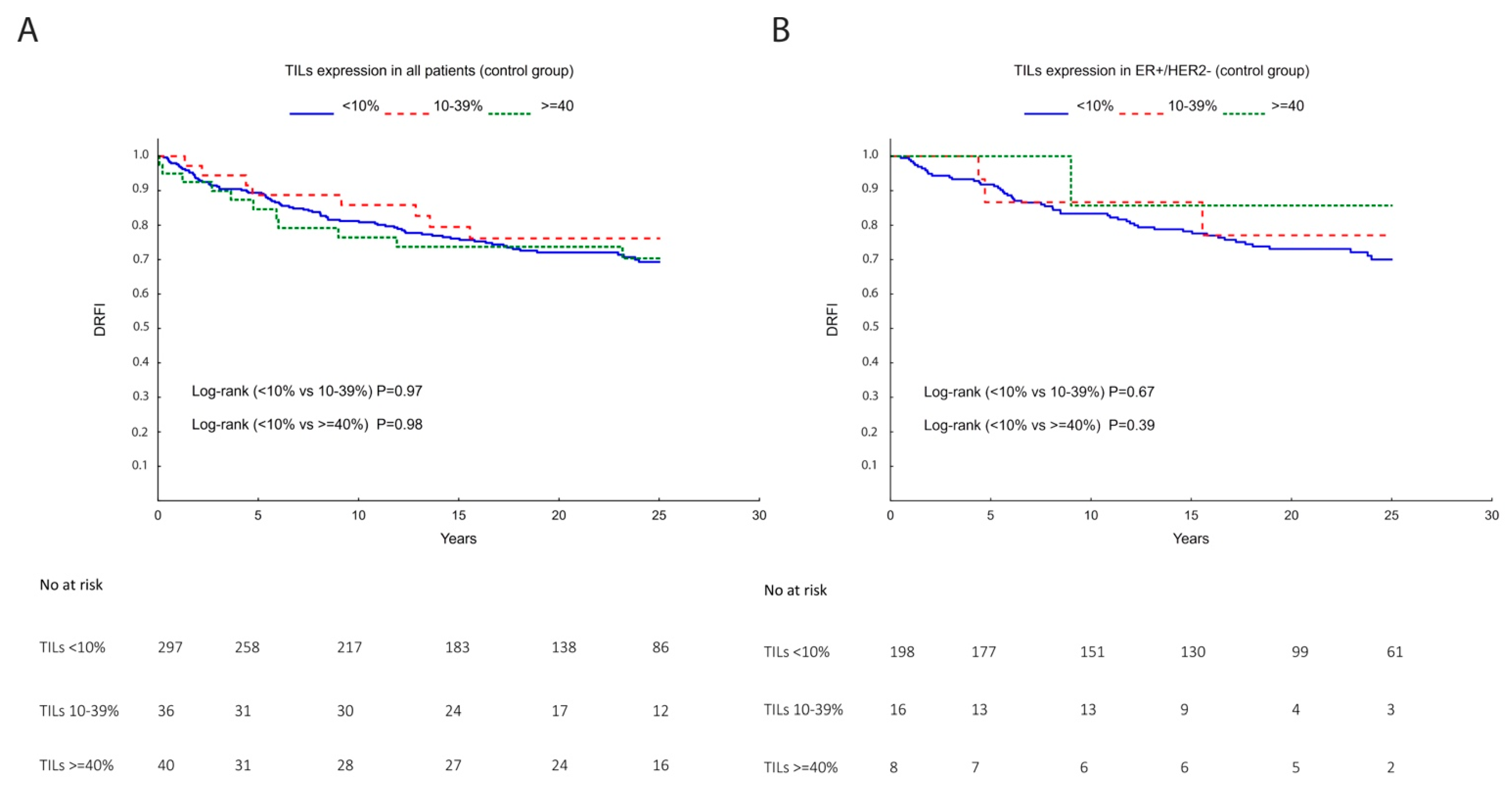
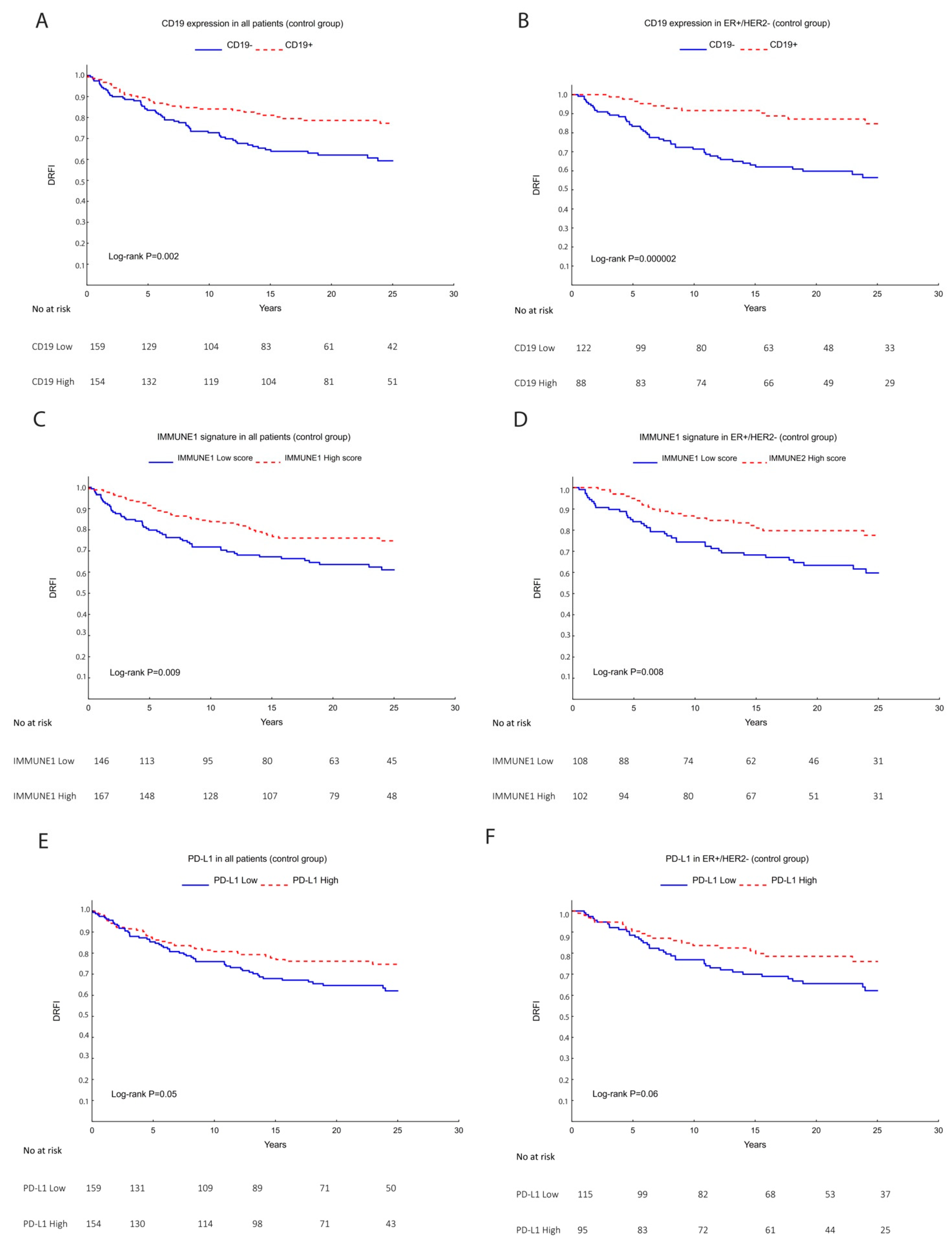
3.2. TILs, Lymphocytic Markers, and Gene Signatures as a Prognostic Marker
3.3. Predictive Role of TILs, Lymphocytic Gene Expression Markers, and Gene Signatures for TAM Efficacy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Gnant, M.; Harbeck, N.; Thomssen, C. St. Gallen 2011: Summary of the Consensus Discussion. Breast Care 2011, 6, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, N.; Yamamoto, S.; Gornbein, J.; Kuo, M.D. Receptor-based Surrogate Subtypes and Discrepancies with Breast Cancer Intrinsic Subtypes: Implications for Image Biomarker Development. Radiology 2018, 289, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, C.; Bendahl, P.O.; Borg, A.; Ehinger, A.; Hegardt, C.; Larsson, C.; Loman, N.; Malmberg, M.; Olofsson, H.; Saal, L.H.; et al. Agreement between molecular subtyping and surrogate subtype classification: A contemporary population-based study of ER-positive/HER2-negative primary breast cancer. Breast Cancer Res. Treat 2019, 178, 459–467. [Google Scholar] [CrossRef]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Pan, H.C.; Gray, R.; Braybrooke, J.; Davies, C.; Taylor, C.; McGale, P.; Peto, R.; Pritchard, K.I.; Bergh, J.; Dowsett, M.; et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N. Engl. J. Med. 2017, 377, 1836–1846. [Google Scholar] [CrossRef]
- Richman, J.; Dowsett, M. Beyond 5 years: Enduring risk of recurrence in oestrogen receptor-positive breast cancer. Nat. Rev. Clin. Oncol. 2019, 16, 296–311. [Google Scholar] [CrossRef]
- Goldberg, J.; Pastorello, R.G.; Vallius, T.; Davis, J.; Cui, Y.X.; Agudo, J.; Waks, A.G.; Keenan, T.; McAllister, S.S.; Tolaney, S.M.; et al. The Immunology of Hormone Receptor Positive Breast Cancer. Front. Immunol. 2021, 12, 674192. [Google Scholar] [CrossRef]
- Stanton, S.E.; Adams, S.; Disis, M.L. Variation in the Incidence and Magnitude of Tumor-Infiltrating Lymphocytes in Breast Cancer Subtypes: A Systematic Review. JAMA Oncol. 2016, 2, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Muller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Hendry, S.; Salgado, R.; Gevaert, T.; Russell, P.A.; John, T.; Thapa, B.; Christie, M.; van de Vijver, K.; Estrada, M.V.; Gonzalez-Ericsson, P.I.; et al. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method From the International Immunooncology Biomarkers Working Group: Part 1: Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Adv. Anat. Pathol. 2017, 24, 235–251. [Google Scholar] [CrossRef] [PubMed]
- Gnant, M.; Harbeck, N.; Thomssen, C. St. Gallen/Vienna 2017: A Brief Summary of the Consensus Discussion about Escalation and De-Escalation of Primary Breast Cancer Treatment. Breast Care 2017, 12, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Radosevic-Robin, N.; Fineberg, S.; van den Eynden, G.; Ternes, N.; Penault-Llorca, F.; Pruneri, G.; D’Alfonso, T.M.; Demaria, S.; Castaneda, C.; et al. Update on tumor-infiltrating lymphocytes (TILs) in breast cancer, including recommendations to assess TILs in residual disease after neoadjuvant therapy and in carcinoma in situ: A report of the International Immuno-Oncology Biomarker Working Group on Breast Cancer. Semin. Cancer Biol. 2018, 52, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Gray, R.J.; Demaria, S.; Goldstein, L.; Perez, E.A.; Shulman, L.N.; Martino, S.; Wang, M.; Jones, V.E.; Saphner, T.J.; et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J. Clin. Oncol. 2014, 32, 2959–2966. [Google Scholar] [CrossRef]
- Loi, S.; Michiels, S.; Salgado, R.; Sirtaine, N.; Jose, V.; Fumagalli, D.; Kellokumpu-Lehtinen, P.L.; Bono, P.; Kataja, V.; Desmedt, C.; et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. 2014, 25, 1544–1550. [Google Scholar] [CrossRef]
- Guarneri, V.; Dieci, M.V.; Bisagni, G.; Brandes, A.A.; Frassoldati, A.; Cavanna, L.; Musolino, A.; Giotta, F.; Rimanti, A.; Garrone, O.; et al. PIK3CA Mutation in the ShortHER Randomized Adjuvant Trial for Patients with Early HER2(+) Breast Cancer: Association with Prognosis and Integration with PAM50 Subtype. Clin. Cancer Res. 2020, 26, 5843–5851. [Google Scholar] [CrossRef]
- Sobral-Leite, M.; Salomon, I.; Opdam, M.; Kruger, D.T.; Beelen, K.J.; van der Noort, V.; van Vlierberghe, R.L.P.; Blok, E.J.; Giardiello, D.; Sanders, J.; et al. Cancer-immune interactions in ER-positive breast cancers: PI3K pathway alterations and tumor-infiltrating lymphocytes. Breast Cancer Res. 2019, 21, 90. [Google Scholar] [CrossRef]
- Blok, E.J.; Engels, C.C.; Dekker-Ensink, G.; Kranenbarg, E.M.K.; Putter, H.; Smit, V.T.H.B.M.; Liefers, G.J.; Morden, J.P.; Bliss, J.M.; Coombes, R.C.; et al. Exploration of tumour-infiltrating lymphocytes as a predictive biomarker for adjuvant endocrine therapy in early breast cancer. Breast Cancer Res. Treat. 2018, 171, 65–74. [Google Scholar] [CrossRef]
- Wu, D.; Tang, S.; Ye, R.; Li, D.; Gu, D.; Chen, R.; Zhang, H.; Sun, J.; Chen, Z. Case Report: Long-Term Response to Pembrolizumab Combined With Endocrine Therapy in Metastatic Breast Cancer Patients With Hormone Receptor Expression. Front. Immunol. 2021, 12, 610149. [Google Scholar] [CrossRef] [PubMed]
- Ruffell, B.; Au, A.; Rugo, H.S.; Esserman, L.J.; Hwang, E.S.; Coussens, L.M. Leukocyte composition of human breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 2796–2801. [Google Scholar] [CrossRef] [PubMed]
- Qian, F.; Qingping, Y.; Linquan, W.; Xiaojin, H.; Rongshou, W.; Shanshan, R.; Wenjun, L.; Yong, H.; Enliang, L. High tumor-infiltrating FoxP3(+) T cells predict poor survival in estrogen receptor-positive breast cancer: A meta-analysis. Eur. J. Surg. Oncol. 2017, 43, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Bates, G.J.; Fox, S.B.; Han, C.; Leek, R.D.; Garcia, J.F.; Harris, A.L.; Banham, A.H. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J. Clin. Oncol. 2006, 24, 5373–5380. [Google Scholar] [CrossRef] [PubMed]
- Garaud, S.; Buisseret, L.; Solinas, C.; Gu-Trantien, C.; de Wind, A.; Van den Eynden, G.; Naveaux, C.; Lodewyckx, J.N.; Boisson, A.; Duvillier, H.; et al. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight 2019, 5, 129641. [Google Scholar] [CrossRef]
- Desmedt, C.; Haibe-Kains, B.; Wirapati, P.; Buyse, M.; Larsimont, D.; Bontempi, G.; Delorenzi, M.; Piccart, M.; Sotiriou, C. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin. Cancer Res. 2008, 14, 5158–5165. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Miremadi, A.; Pinder, S.E.; Ellis, I.O.; Caldas, C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007, 8, R157. [Google Scholar] [CrossRef]
- Rutqvist, L.E.; Johansson, H.; Grp, S.B.C.S. Long-term follow-up of the randomized Stockholm trial on adjuvant tamoxifen among postmenopausal patients with early stage breast cancer. Acta Oncol. 2007, 46, 133–145. [Google Scholar] [CrossRef]
- Barlow, L.; Westergren, K.; Holmberg, L.; Talback, M. The completeness of the Swedish Cancer Register: A sample survey for year 1998. Acta Oncol. 2009, 48, 27–33. [Google Scholar] [CrossRef]
- Brooke, H.L.; Talback, M.; Hornblad, J.; Johansson, L.A.; Ludvigsson, J.F.; Druid, H.; Feychting, M.; Ljung, R. The Swedish cause of death register. Eur. J. Epidemiol. 2017, 32, 765–773. [Google Scholar] [CrossRef]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Jerevall, P.L.; Ma, X.J.; Li, H.; Salunga, R.; Kesty, N.C.; Erlander, M.G.; Sgroi, D.C.; Holmlund, B.; Skoog, L.; Fornander, T.; et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br. J. Cancer 2011, 104, 1762–1769. [Google Scholar] [CrossRef] [PubMed]
- Esserman, L.J.; Yau, C.; Thompson, C.K.; van ‘t Veer, L.J.; Borowsky, A.D.; Hoadley, K.A.; Tobin, N.P.; Nordenskjold, B.; Fornander, T.; Stal, O.; et al. Use of Molecular Tools to Identify Patients With Indolent Breast Cancers With Ultralow Risk Over 2 Decades. Jama Oncol. 2017, 3, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- van ‘t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef]
- Jerevall, P.L.; Jansson, A.; Fornander, T.; Skoog, L.; Nordenskjold, B.; Stal, O. Predictive relevance of HOXB13 protein expression for tamoxifen benefit in breast cancer. Breast Cancer Res. 2010, 12, R53. [Google Scholar] [CrossRef]
- Yu, N.Y.; Iftimi, A.; Yau, C.; Tobin, N.P.; van ‘t Veer, L.; Hoadley, K.A.; Benz, C.C.; Nordenskjold, B.; Fornander, T.; Stal, O.; et al. Assessment of Long-term Distant Recurrence-Free Survival Associated With Tamoxifen Therapy in Postmenopausal Patients With Luminal A or Luminal B Breast Cancer. JAMA Oncol. 2019, 5, 1304–1309. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef]
- Hudis, C.A.; Barlow, W.E.; Costantino, J.P.; Gray, R.J.; Pritchard, K.I.; Chapman, J.A.; Sparano, J.A.; Hunsberger, S.; Enos, R.A.; Gelber, R.D.; et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: The STEEP system. J. Clin. Oncol. 2007, 25, 2127–2132. [Google Scholar] [CrossRef]
- Tolaney, S.M.; Garrett-Mayer, E.; White, J.; Blinder, V.S.; Foster, J.C.; Amiri-Kordestani, L.; Hwang, E.S.; Bliss, J.M.; Rakovitch, E.; Perlmutter, J.; et al. Updated Standardized Definitions for Efficacy End Points (STEEP) in Adjuvant Breast Cancer Clinical Trials: STEEP Version 2.0. J. Clin. Oncol. 2021, 39, 2720–2731. [Google Scholar] [CrossRef]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Loi, S.; Sirtaine, N.; Piette, F.; Salgado, R.; Viale, G.; Van Eenoo, F.; Rouas, G.; Francis, P.; Crown, J.P.; Hitre, E.; et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 2013, 31, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Vihervuori, H.; Autere, T.A.; Repo, H.; Kurki, S.; Kallio, L.; Lintunen, M.M.; Talvinen, K.; Kronqvist, P. Tumor-infiltrating lymphocytes and CD8(+) T cells predict survival of triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 3105–3114. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Dieras, V.; Hegg, R.; Im, S.A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Paijens, S.T.; Vledder, A.; de Bruyn, M.; Nijman, H.W. Tumor-infiltrating lymphocytes in the immunotherapy era. Cell Mol. Immunol. 2021, 18, 842–859. [Google Scholar] [CrossRef]
- Lundgren, C.; Bendahl, P.O.; Ekholm, M.; Ferno, M.; Forsare, C.; Kruger, U.; Nordenskjold, B.; Stal, O.; Ryden, L. Tumour-infiltrating lymphocytes as a prognostic and tamoxifen predictive marker in premenopausal breast cancer: Data from a randomised trial with long-term follow-up. Breast Cancer Res. 2020, 22, 140. [Google Scholar] [CrossRef]
- Seo, A.N.; Lee, H.J.; Kim, E.J.; Kim, H.J.; Jang, M.H.; Lee, H.E.; Kim, Y.J.; Kim, J.H.; Park, S.Y. Tumour-infiltrating CD8+lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br. J. Cancer 2013, 109, 2705–2713. [Google Scholar] [CrossRef]
- Criscitiello, C.; Vingiani, A.; Maisonneuve, P.; Viale, G.; Viale, G.; Curigliano, G. Tumor-infiltrating lymphocytes (TILs) in ER+/HER2- breast cancer. Breast Cancer Res. Treat. 2020, 183, 347–354. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Watanabe, T.; Hida, A.I.; Higuchi, T.; Miyagawa, Y.; Ozawa, H.; Bun, A.; Fukui, R.; Sata, A.; Imamura, M.; et al. Prognostic significance of tumor-infiltrating lymphocytes may differ depending on Ki67 expression levels in estrogen receptor-positive/HER2-negative operated breast cancers. Breast Cancer 2019, 26, 738–747. [Google Scholar] [CrossRef]
- Joffroy, C.M.; Buck, M.B.; Stope, M.B.; Popp, S.L.; Pfizenmaier, K.; Knabbe, C. Antiestrogens induce transforming growth factor beta-mediated immunosuppression in breast cancer. Cancer Res. 2010, 70, 1314–1322. [Google Scholar] [CrossRef]
- Olkhanud, P.B.; Damdinsuren, B.; Bodogai, M.; Gress, R.E.; Sen, R.; Wejksza, K.; Malchinkhuu, E.; Wersto, R.P.; Biragyn, A. Tumor-evoked regulatory B cells promote breast cancer metastasis by converting resting CD4(+) T cells to T-regulatory cells. Cancer Res. 2011, 71, 3505–3515. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, T.; Li, M.; Yin, L.; Xue, J. Immunosuppressive B cells expressing PD-1/PD-L1 in solid tumors: A mini review. QJM Int. J. Med. 2022, 115, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Morgan, R.; Chen, C.; Cai, Y.; Clark, E.; Khan, W.N.; Shin, S.U.; Cho, H.M.; Al Bayati, A.; Pimentel, A.; et al. Mammary-tumor-educated B cells acquire LAP/TGF-beta and PD-L1 expression and suppress anti-tumor immune responses. Int. Immunol. 2016, 28, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Huhn, D.; Marti-Rodrigo, P.; Mouron, S.; Hansel, C.; Tschapalda, K.; Porebski, B.; Haggblad, M.; Lidemalm, L.; Quintela-Fandino, M.; Carreras-Puigvert, J.; et al. Prolonged estrogen deprivation triggers a broad immunosuppressive phenotype in breast cancer cells. Mol. Oncol. 2022, 16, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Zerdes, I.; Sifakis, E.G.; Matikas, A.; Chretien, S.; Tobin, N.P.; Hartman, J.; Rassidakis, G.Z.; Bergh, J.; Foukakis, T. Programmed death-ligand 1 gene expression is a prognostic marker in early breast cancer and provides additional prognostic value to 21-gene and 70-gene signatures in estrogen receptor-positive disease. Mol. Oncol. 2020, 14, 951–963. [Google Scholar] [CrossRef] [PubMed]
| Variables | Low TILs <10% n (%) | Intermediate TILs 10–39% n (%) | High TILs ≥40% n (%) | p Value |
|---|---|---|---|---|
| Tumor size | N = 450 | 0.07 | ||
| ≤20 mm | 337 (90) | 24 (6) | 14 (4) | |
| >20 mm | 62 (83) | 8 (10) | 5 (7) | |
| Tumor grade (NHG) | N = 394 | 0.001 | ||
| 1 | 83 (95) | 4 (5) | 0 (0) | |
| 2 | 223 (88) | 16 (6) | 14 (6) | |
| 3 | 42 (78) | 7 (13) | 5 (9) | |
| Ki67 | N = 355 | <0.0001 | ||
| <15% | 252 (92) | 17 (6) | 5 (2) | |
| ≥15% | 60 (74) | 8 (10) | 13 (16) | |
| 70-gene signature | N = 351 | <0.0001 | ||
| Ultra-low risk | 65 (100) | 0 (0) | 0 (0) | |
| Low risk | 163 (90) | 13 (7) | 5 (3) | |
| High risk | 82 (78) | 11 (11) | 12 (11) | |
| CD4 | N = 351 | 0.99 | ||
| Low | 145 (88) | 10 (6) | 9 (5) | |
| High | 165 (88) | 14 (7) | 8 (4) | |
| CD8A | N = 351 | <0.0001 | ||
| Low | 176 (96) | 8 (4) | 0 (0) | |
| High | 134 (80) | 16 (10) | 17 (10) | |
| CD19 | N = 351 | <0.0001 | ||
| Low | 199 (96) | 6 (3) | 1 (1) | |
| High | 111 (77) | 18 (12) | 16 (11) | |
| FOXP3 | N = 351 | 0.45 | ||
| Low | 155 (90) | 11 (6) | 7 (4) | |
| High | 155 (87) | 13 (7) | 10 (6) | |
| PD-L1 | N = 351 | 0.005 | ||
| Low | 181 (92) | 13 (7) | 2 (1) | |
| High | 129 (83) | 11 (7) | 15 (10) | |
| PD1 | N = 351 | <0.0001 | ||
| Low | 194 (97) | 3 (2) | 2 (1) | |
| High | 116 (76) | 21 (14) | 15 (10) | |
| IMMUNE1 | N = 351 | <0.0001 | ||
| Low | 178 (96) | 4 (2) | 3 (2) | |
| High | 132 (80) | 20 (12) | 14 (8) | |
| IMMUNE2 | N = 351 | <0.0001 | ||
| Low | 191 (98) | 2 (1) | 2 (1) | |
| High | 119 (76) | 22 (14) | 15 (10) |
| TILs | Univariate DRFI a HR (95% CI) | Multivariable DRFI b HR (95% CI) |
|---|---|---|
| All patients | (n = 373) | (n = 456) |
| <10% (Ref) | 1.00 | 1.00 |
| 10–39% | 0.77 (0.37–1.60) | 0.85 (0.45–1.60) |
| ≥40% | 1.01 (0.54–1.90) | 0.47 (0.24–0.95) |
| ER+/HER2− | (n = 222) | (n = 348) |
| <10% (Ref) | 1.00 | 1.00 |
| 10–39% | 0.77 (0.24–2.46) | 0.99 (0.39–2.47) |
| ≥40% | 0.44 (0.06–3.28) | 0.37 (0.09–1.64) |
| TILs | Univariate BCSS a HR (95% CI) | Multivariable BCSS b HR (95% CI) |
|---|---|---|
| All patients | (n = 373) | (n = 456) |
| <10% (ref) | 1.00 | 1.00 |
| 10–39% | 0.82 (0.37–1.78) | 0.84 (0.43–1.64) |
| ≥40% | 1.09 (0.56–2.11) | 0.45 (0.22–0.93) |
| ER+/HER2− | (n = 222) | (n = 348) |
| <10% (ref) | 1.00 | 1.00 |
| 10–39% | 0.94 (0.29–3.02) | 1.09 (0.43–2.76) |
| ≥40% | 0.54 (0.07–3.95) | 0.42 (0.10–1.92) |
| CD8 a | CD4 | FOXP3 | CD19 | IMMUNE1 | IMMUNE2 | PD1 | PD-L1 | |
|---|---|---|---|---|---|---|---|---|
| Univariate DRFI HR (95% CI) | ||||||||
| All patients (n = 313) | ||||||||
| Low | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| High | 0.74 (0.44–1.22) | 0.92 (0.61–1.40) | 0.74 (0.49–1.12) | 0.51 (0.33–0.79) | 0.58 (0.38–0.87) | 1.21 (0.80–1.83) | 1.21 (0.80–1.83) | 0.66 (0.43–0.99) |
| ER+/HER2− (n = 210) | ||||||||
| Low | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| High | 0.45 (0.20–0.98) | 0.85 (0.51–1.41) | 0.67 (0.40–1.13) | 0.26 (0.14–0.51) | 0.49 (0.29–0.84) | 0.87 (0.52–1.47) | 1.36 (0.81–2.26) | 0.60 (0.35–1.03) |
| CD8 a | CD4 | FOXP3 | CD19 | IMMUNE1 | IMMUNE2 | PD1 | PD-L1 | |
| Multivariable DRFI b HR (95% CI) | ||||||||
| All patients (n = 524) | ||||||||
| Low | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| High | 0.79 (0.52–1.20) | 0.83 (0.58–1.19) | 0.84 (0.58–1.19) | 0.63 (0.43–0.93) | 0.57 (0.39–0.83) | 1.11 (0.75–1.64) | 1.36 (0.92–2.01) | 0.60 (0.41–0.89) |
| ER+/HER2− (n = 418) | ||||||||
| Low | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| High | 0.71 (0.41–1.24) | 0.79 (0.51–1.21) | 0.84 (0.55–1.29) | 0.46 (0.29–0.74) | 0.63 (0.41–0.98) | 0.88 (0.57–1.37) | 1.46 (0.95–2.26) | 0.54 (0.34–0.85) |
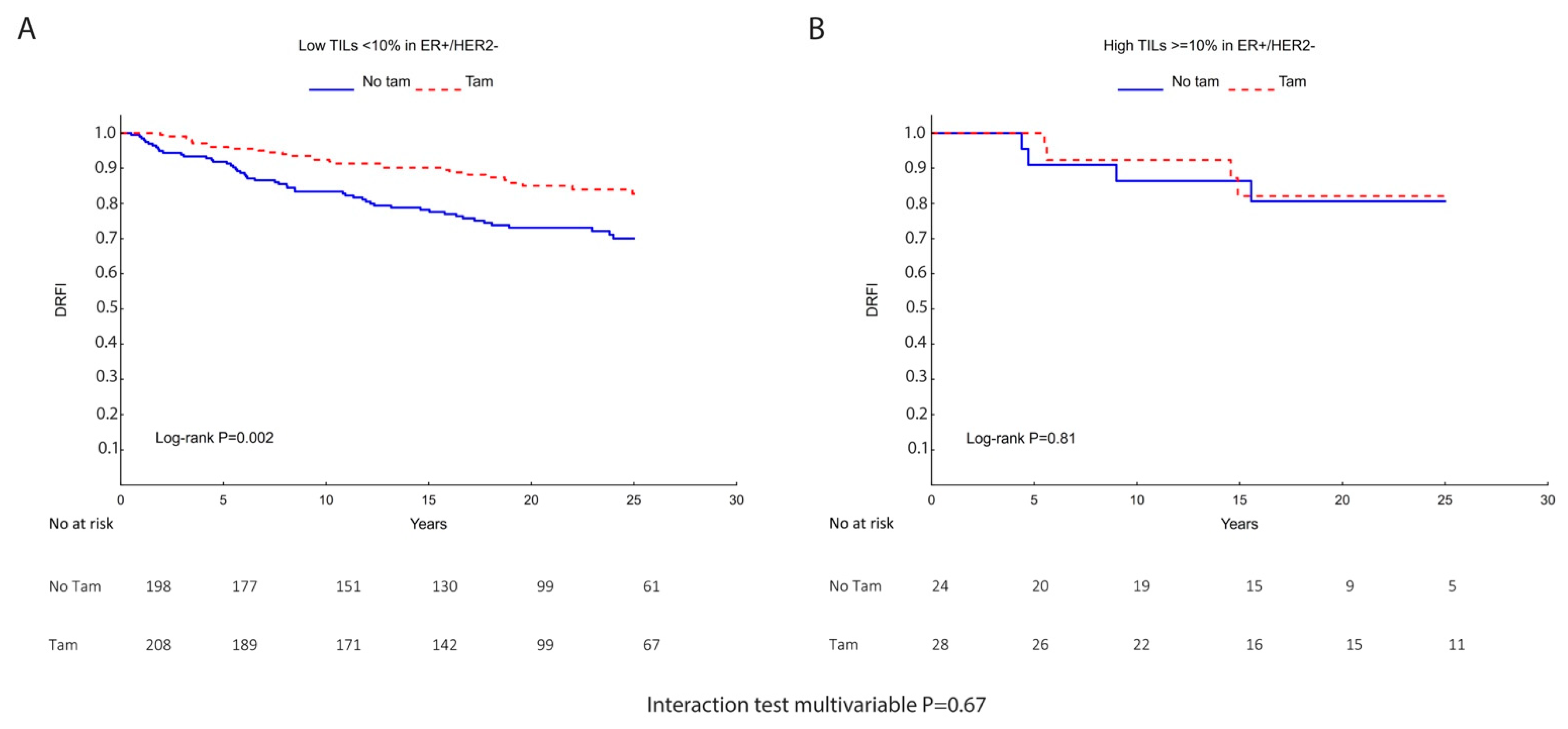
| TILs | N | Tamoxifen | Univariate DRFI HR (95% CI) | Interaction | N | Tamoxifen | Multivariable DRFI a HR (95% CI) | Interaction |
|---|---|---|---|---|---|---|---|---|
| Low <10% | 406 | − | 1.00 | 306 | − | 1.00 | ||
| + | 0.49 (0.31–0.78) | + | 0.46 (0.27–0.79) | |||||
| p = 0.50 | p = 0.67 | |||||||
| High ≥10% | 52 | − | 1.00 | 42 | − | 1.00 | ||
| + | 0.85 (0.21–3.39) | + | 0.38 (0.06–2.28) |
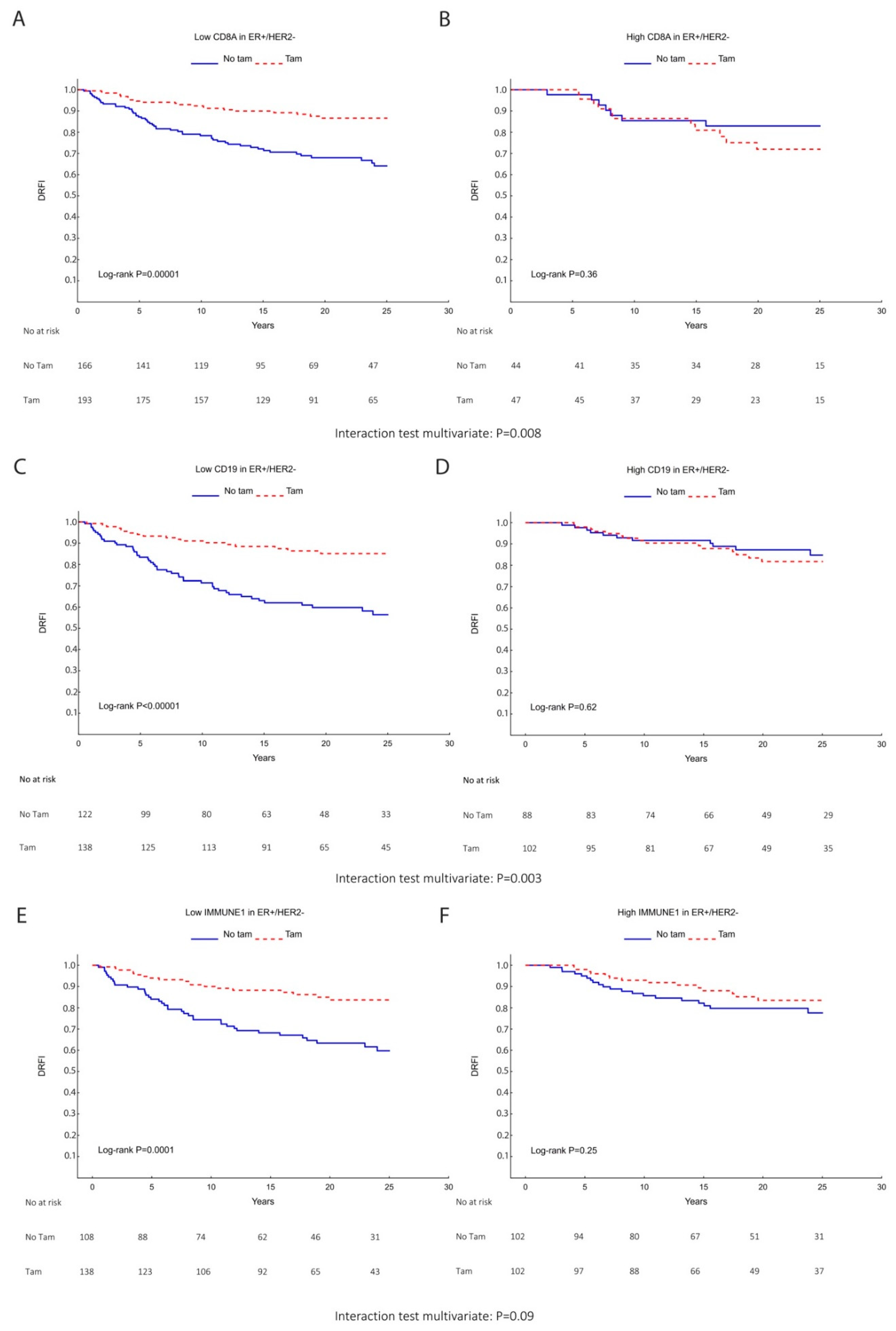
| TIL Markers | N | Tamoxifen | Univariate DRFI HR (95% CI) | Interaction | N | Tamoxifen | Multivariable DRFI a HR (95% CI) | Interaction |
|---|---|---|---|---|---|---|---|---|
| CD8 Low | 359 | − | 1.00 | 333 | − | 1.00 | ||
| + | 0.33 (0.20–0.55) | + | 0.31 (0.19–0.53) | |||||
| p = 0.005 | p = 0.008 | |||||||
| CD8 High | 91 | − | 1.00 | 84 | − | 1.00 | ||
| + | 1.55 (0.60–4.02) | + | 1.31 (0.48–3.60) | |||||
| CD4 Low | 226 | − | 1.00 | 214 | − | 1.00 | ||
| + | 0.53 (0.30–0.92) | + | 0.50 (0.28–0.9) | |||||
| p = 0.37 | p = 0.42 | |||||||
| CD4 High | 224 | − | 1.00 | 84 | − | 1.00 | ||
| + | 0.35 (0.18–0.70) | + | 0.33 (0.16–0.71) | |||||
| FOXP3 Low | 227 | − | 1.00 | 209 | − | 1.00 | ||
| + | 0.34 (0.19–0.62) | + | 0.35 (0.18–0.65) | |||||
| p = 0.14 | p = 0.25 | |||||||
| FOXP3 High | 223 | − | 1.00 | 209 | − | 1.00 | ||
| + | 0.64 (0.35–1.18) | + | 0.56 (0.30–1.05) | |||||
| CD19 Low | 260 | − | 1.00 | 242 | − | 1.00 | ||
| + | 0.29 (0.17–0.50) | + | 0.28 (0.16–0.49) | |||||
| p = 0.003 | p = 0.003 | |||||||
| CD19 High | 190 | − | 1.00 | 176 | − | 1.00 | ||
| + | 1.22 (0.56–2.66) | + | 1.21 (0.53–2.76) | |||||
| PD1 Low | 257 | − | 1.00 | 242 | − | 1.00 | ||
| + | 0.46 (0.25–0.85) | + | 0.40 (0.21–0.76) | |||||
| p = 0.92 | p = 0.77 | |||||||
| PD1 High | 193 | − | 1.00 | 176 | − | 1.00 | ||
| + | 0.44 (0.24–0.81) | + | 0.42 (0.22–0.78) | |||||
| PD–L1 Low | 252 | − | 1.00 | 240 | − | 1.00 | ||
| + | 0.42 (0.25–0.71) | + | 0.37 (0.22–0.64) | |||||
| p = 0.61 | p = 0.45 | |||||||
| PD–L1 High | 198 | − | 1.00 | 178 | − | 1.00 | ||
| + | 0.53 (0.26–1.08) | + | 0.52 (0.24–1.13) | |||||
| IMMUNE1 Low | 246 | − | 1.00 | 228 | − | 1.00 | ||
| + | 0.35 (0.20–0.61) | + | 0.31 (0.18–0.56) | |||||
| p = 0.14 | p = 0.09 | |||||||
| IMMUNE1 High | 204 | − | 1.00 | 190 | − | 1.00 | ||
| + | 0.67 (0.34–1.33) | + | 0.68 (0.33–1.39) | |||||
| IMMUNE2 Low | 259 | − | 1.00 | 240 | − | 1.00 | ||
| + | 0.38 (0.21–0.68) | + | 0.32 (0.17–0.59) | |||||
| p = 0.31 | p = 0.14 | |||||||
| IMMUNE2 High | 191 | − | 1.00 | 178 | − | 1.00 | ||
| + | 0.59 (0.31–1.10) | + | 0.62 (0.33–1.18) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pousette, J.; Johansson, A.; Jönsson, C.; Fornander, T.; Lindström, L.S.; Olsson, H.; Perez-Tenorio, G. Prognostic and Predictive Significance of Stromal Tumor-Infiltrating Lymphocytes (sTILs) in ER-Positive/HER2−Negative Postmenopausal Breast Cancer Patients. Cancers 2022, 14, 4844. https://doi.org/10.3390/cancers14194844
Pousette J, Johansson A, Jönsson C, Fornander T, Lindström LS, Olsson H, Perez-Tenorio G. Prognostic and Predictive Significance of Stromal Tumor-Infiltrating Lymphocytes (sTILs) in ER-Positive/HER2−Negative Postmenopausal Breast Cancer Patients. Cancers. 2022; 14(19):4844. https://doi.org/10.3390/cancers14194844
Chicago/Turabian StylePousette, Jenny, Annelie Johansson, Carolin Jönsson, Tommy Fornander, Linda S. Lindström, Hans Olsson, and Gizeh Perez-Tenorio. 2022. "Prognostic and Predictive Significance of Stromal Tumor-Infiltrating Lymphocytes (sTILs) in ER-Positive/HER2−Negative Postmenopausal Breast Cancer Patients" Cancers 14, no. 19: 4844. https://doi.org/10.3390/cancers14194844
APA StylePousette, J., Johansson, A., Jönsson, C., Fornander, T., Lindström, L. S., Olsson, H., & Perez-Tenorio, G. (2022). Prognostic and Predictive Significance of Stromal Tumor-Infiltrating Lymphocytes (sTILs) in ER-Positive/HER2−Negative Postmenopausal Breast Cancer Patients. Cancers, 14(19), 4844. https://doi.org/10.3390/cancers14194844






