Organic Anion Transporters (OAT) and Other SLC22 Transporters in Progression of Renal Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Statistical Analysis
2.2. TCGA RNA-Seq Analysis
2.3. Survival Analysis
2.4. Network Analysis
3. Results
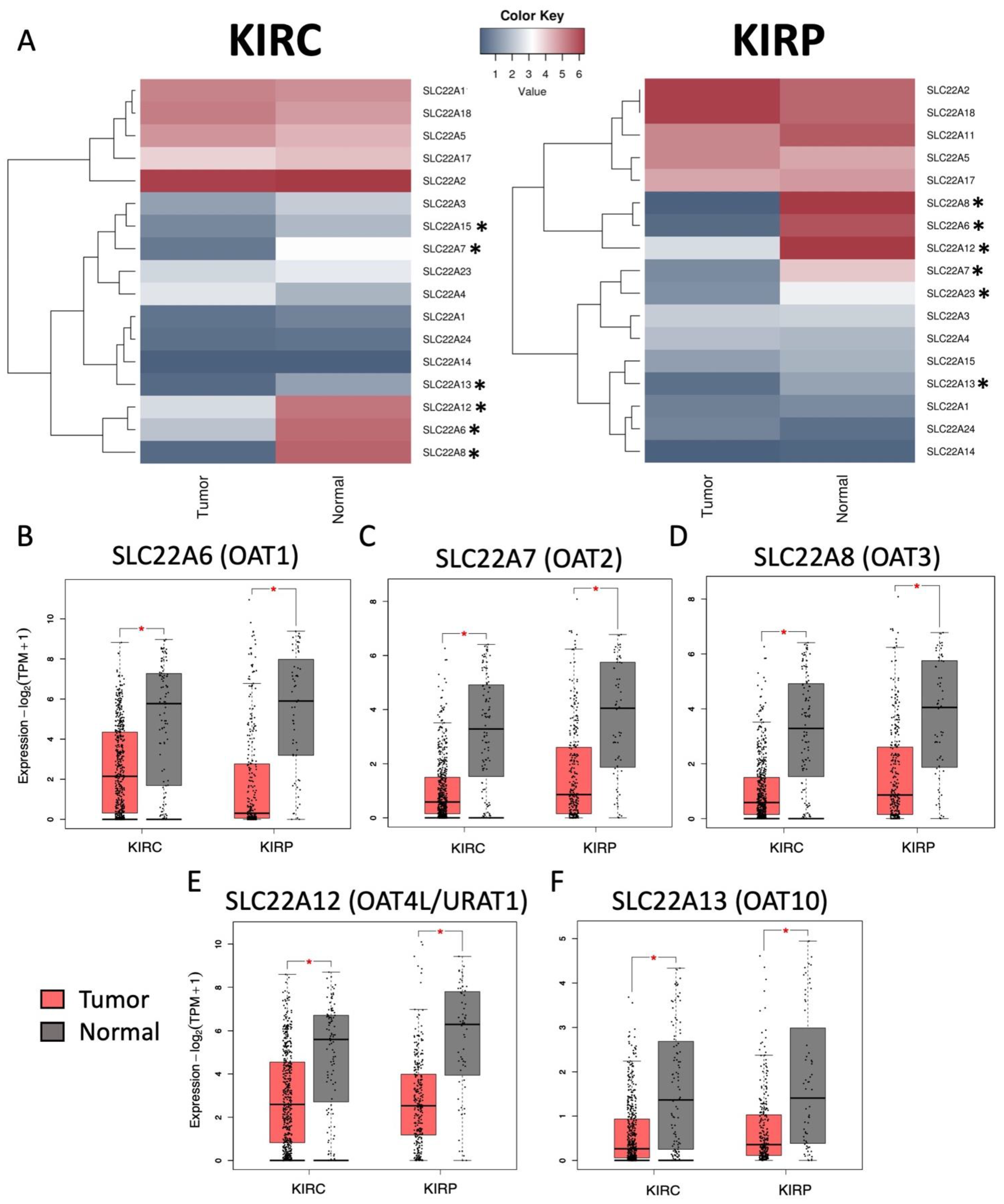
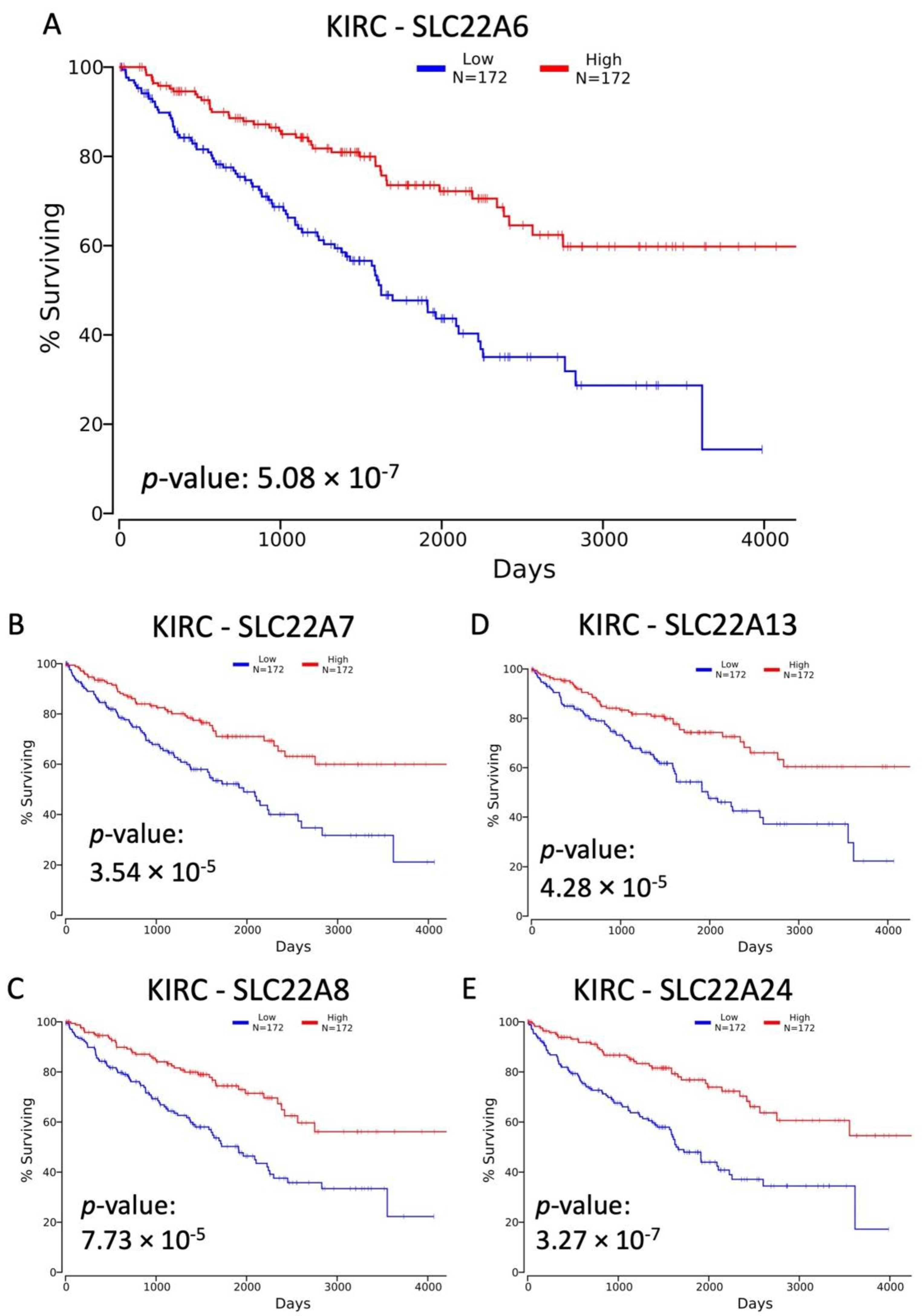
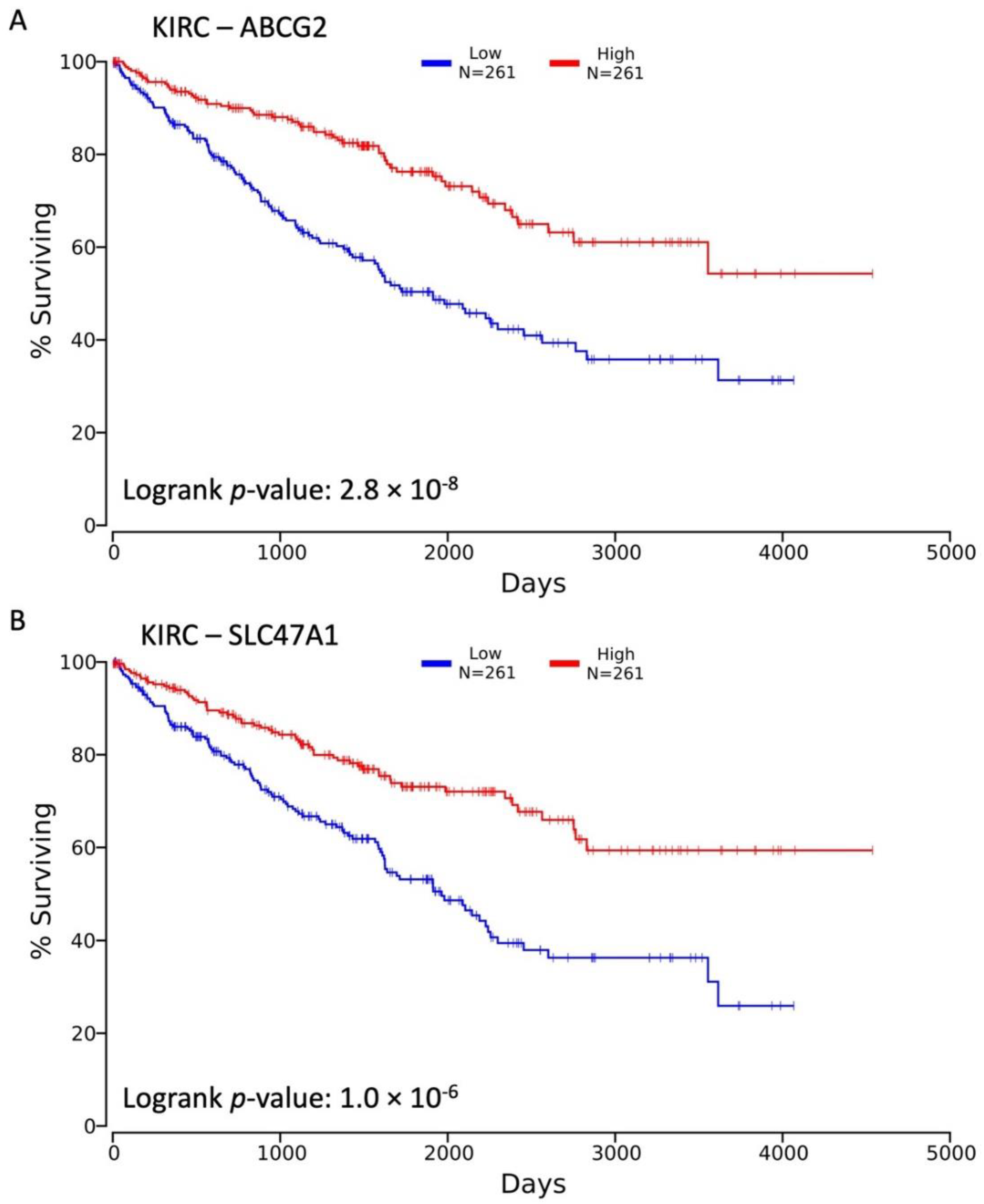
3.1. Decreased Expression of SLC22 Transporters in Kidney Cancers and Association with Poorer Outcomes
3.2. Variables Other Than OAT Family Expression Associated with Decreased Survival
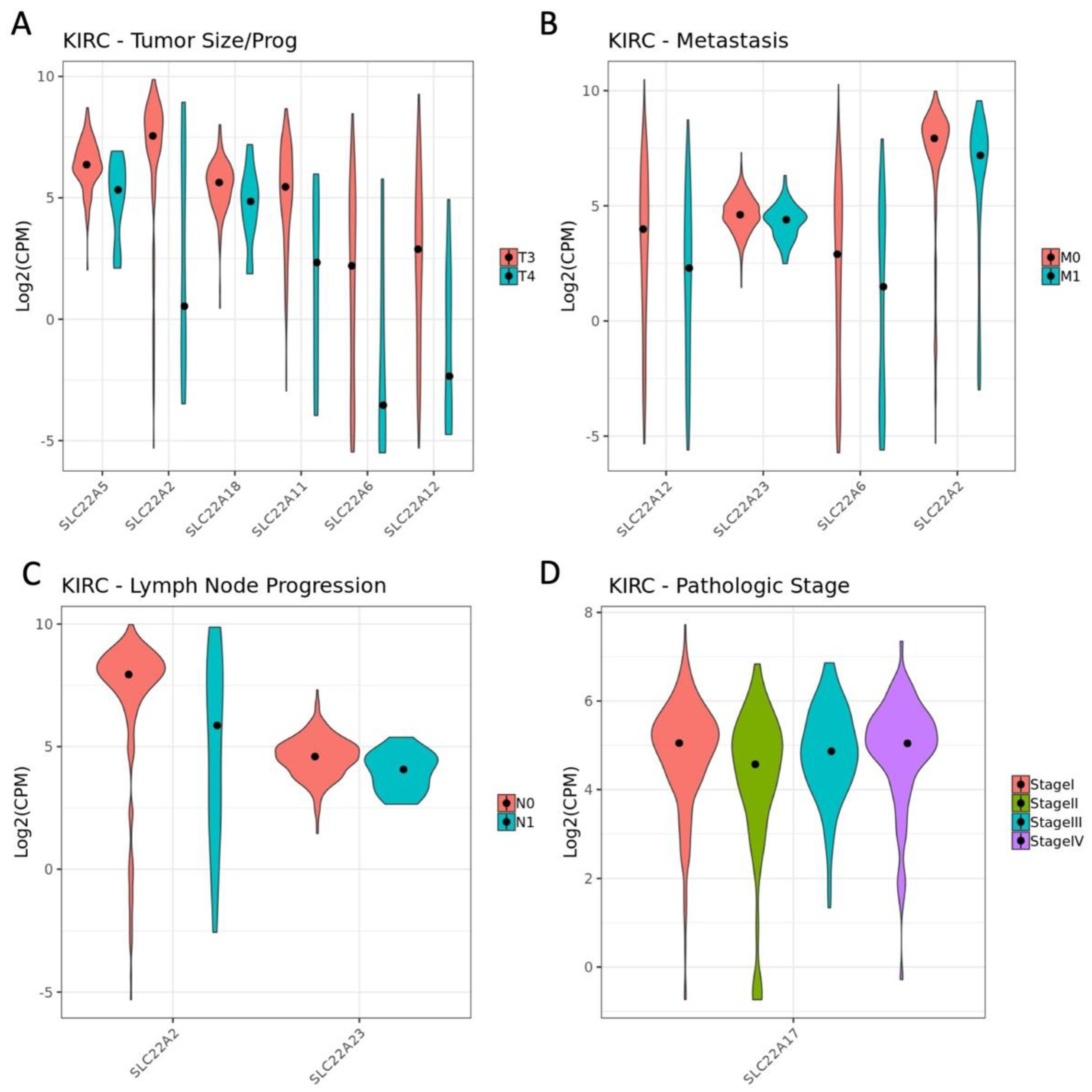
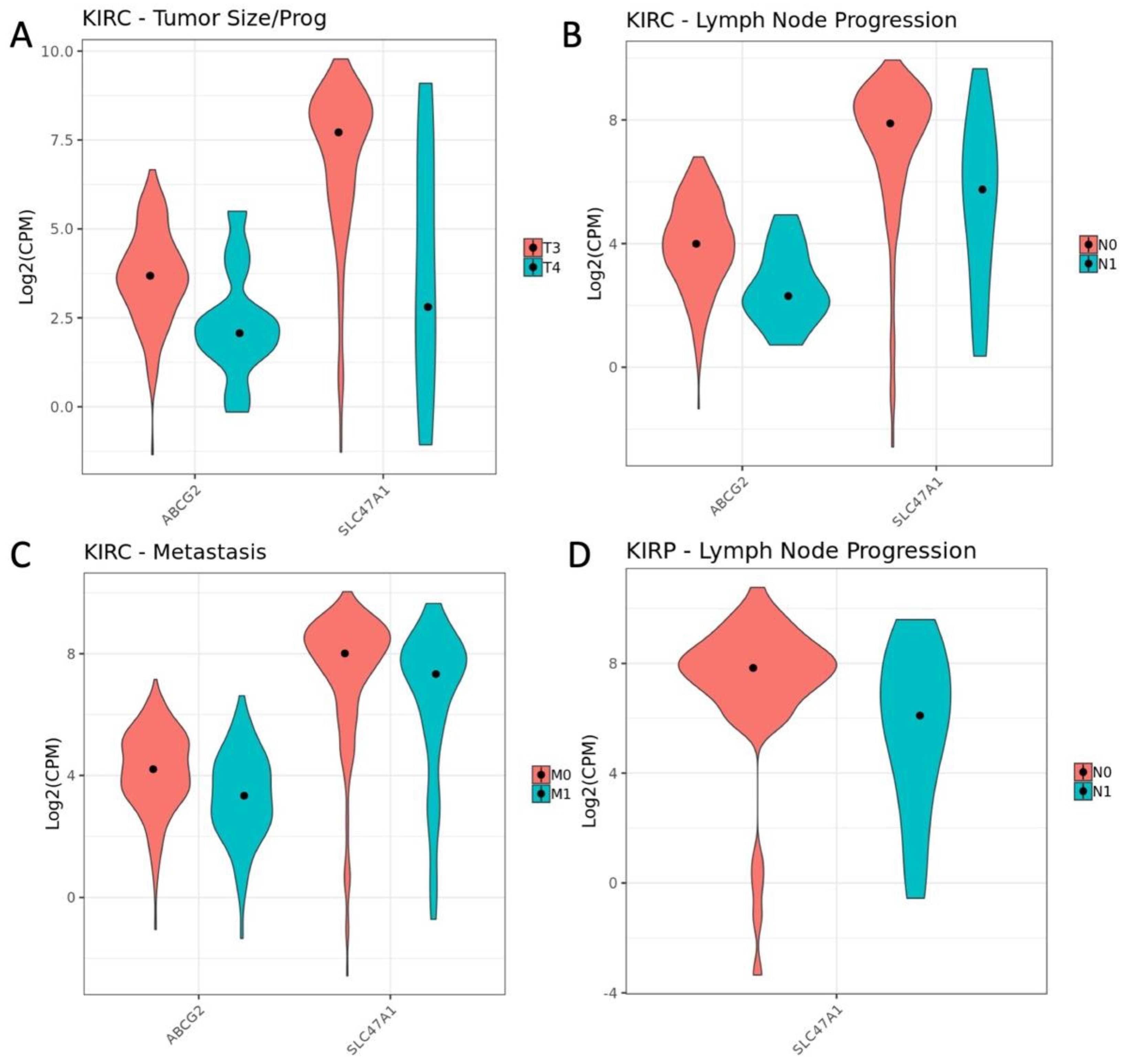
3.3. Influence of OAT (and Other SLC22 Transporter) Expression on Tumor Stage and Individual TNM Parameters in the Context of Overall Survival
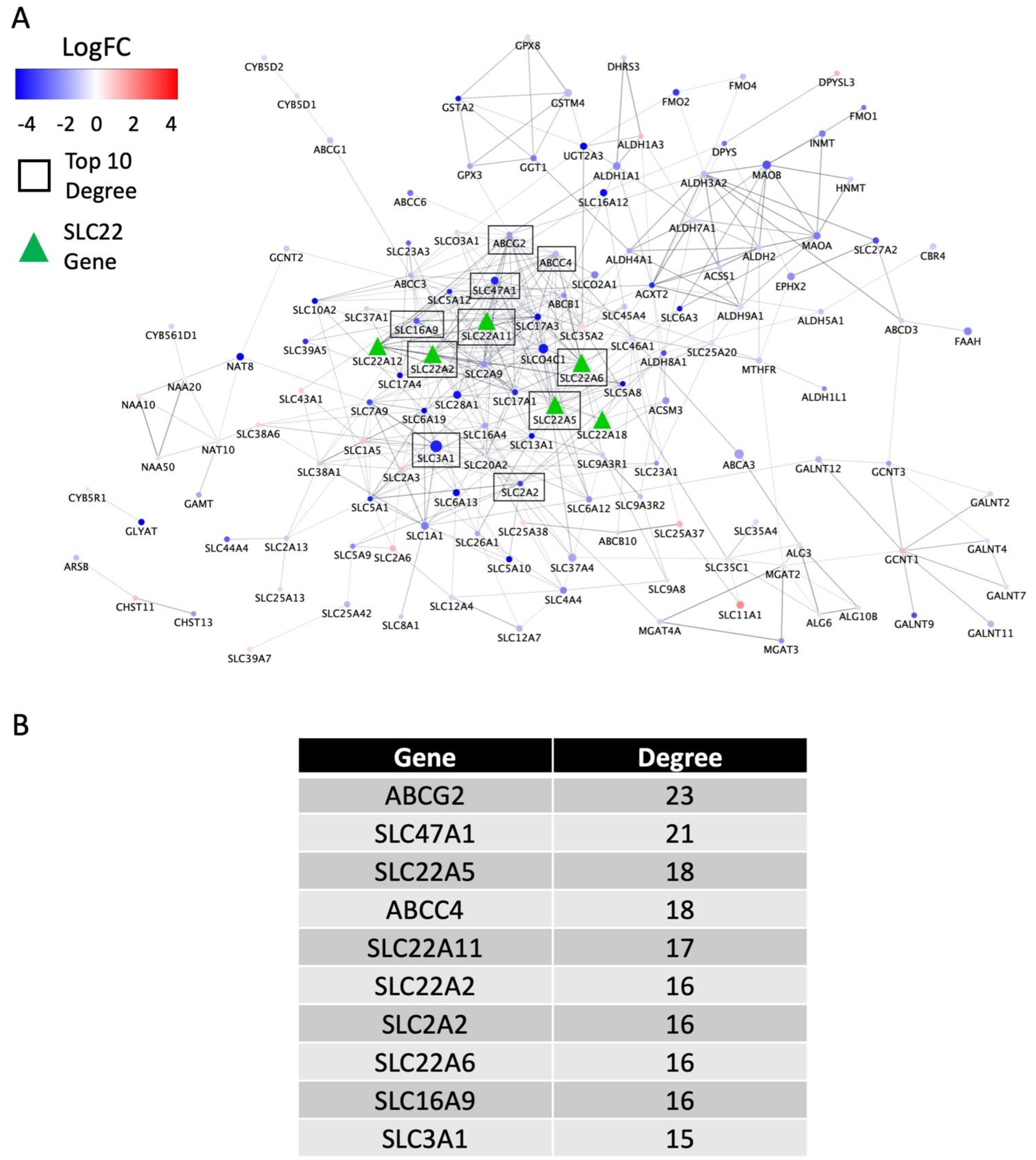
3.4. Biological Support and Network Interpretation of the Role of SLC22 Genes in KIRC and KIRP Disease Progression (in the Context of the Remote Sensing and Signaling Theory)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nigam, S.K. What do drug transporters really do? Nat. Rev. Drug Discov. 2014, 14, 29–44. [Google Scholar] [CrossRef]
- Zamek-Gliszczynski, M.J.; Taub, M.E.; Chothe, P.P.; Chu, X.; Giacomini, K.M.; Kim, R.B.; Ray, A.S.; Stocker, S.L.; Unadkat, J.D.; Wittwer, M.B.; et al. Transporters in Drug Development: 2018 ITC Recommendations for Transporters of Emerging Clinical Importance. Clin. Pharmacol. Ther. 2018, 104, 890–899. [Google Scholar] [CrossRef]
- Bush, K.T.; Singh, P.; Nigam, S.K. Gut-derived uremic toxin handling in vivo requires OAT-mediated tubular secretion in chronic kidney disease. JCI Insight 2020, 5, 133817. [Google Scholar] [CrossRef]
- Bush, K.T.; Wu, W.; Lun, C.; Nigam, S.K. The drug transporter OAT3 (SLC22A8) and endogenous metabolite communication via the gut–liver–kidney axis. J. Biol. Chem. 2017, 292, 15789–15803. [Google Scholar] [CrossRef]
- Estudante, M.; Soveral, G.; Morais, J.G.; Benet, L.Z. Insights into solute carriers: Physiological functions and implications in disease and pharmacokinetics. MedChemComm 2016, 7, 1462–1478. [Google Scholar] [CrossRef]
- Nigam, S.K. The SLC22 Transporter Family: A Paradigm for the Impact of Drug Transporters on Metabolic Pathways, Signaling, and Disease. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 663–687. [Google Scholar] [CrossRef]
- Yee, S.W.; Giacomini, K.M. Emerging Roles of the Human Solute Carrier 22 Family. Drug Metab. Dispos. 2022, 50, 1193–1210. [Google Scholar] [CrossRef]
- Lopez-Nieto, C.E.; You, G.; Bush, K.T.; Barros, E.J.G.; Beier, D.R.; Nigam, S.K. Molecular Cloning and Characterization of NKT, a Gene Product Related to the Organic Cation Transporter Family That Is Almost Exclusively Expressed in the Kidney. J. Biol. Chem. 1997, 272, 6471–6478. [Google Scholar] [CrossRef]
- Ahn, S.-Y.; Jamshidi, N.; Mo, M.L.; Wu, W.; Eraly, S.A.; Dnyanmote, A.; Bush, K.T.; Gallegos, T.F.; Sweet, D.H.; Palsson, B.; et al. Linkage of Organic Anion Transporter-1 to Metabolic Pathways through Integrated “Omics”-driven Network and Functional Analysis. J. Biol. Chem. 2011, 286, 31522–31531. [Google Scholar] [CrossRef]
- Liu, H.C.; Jamshidi, N.; Chen, Y.; Eraly, S.A.; Cho, S.Y.; Bhatnagar, V.; Wu, W.; Bush, K.T.; Abagyan, R.; Palsson, B.O.; et al. An Organic Anion Transporter 1 (OAT1)-centered Metabolic Network. J. Biol. Chem. 2016, 291, 19474–19486. [Google Scholar] [CrossRef]
- Wu, W.; Jamshidi, N.; Eraly, S.A.; Liu, H.C.; Bush, K.T.; Palsson, B.O.; Nigam, S.K. Multispecific Drug Transporter Slc22a8 (Oat3) Regulates Multiple Metabolic and Signaling Pathways. Drug Metab. Dispos. 2013, 41, 1825–1834. [Google Scholar] [CrossRef]
- Rosenthal, S.B.; Bush, K.T.; Nigam, S.K. A Network of SLC and ABC Transporter and DME Genes Involved in Remote Sensing and Signaling in the Gut-Liver-Kidney Axis. Sci. Rep. 2019, 9, 1–19. [Google Scholar] [CrossRef]
- Granados, J.C.; Bhatnagar, V.; Nigam, S.K. Blockade of Organic Anion Transport in Humans After Treatment With the Drug Probenecid Leads to Major Metabolic Alterations in Plasma and Urine. Clin. Pharmacol. Ther. 2022, 112, 653–664. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, H.; Fan, Y.; Yu, Z.; You, G. Regulation of organic anion transporters: Role in physiology, pathophysiology, and drug elimination. Pharmacol. Ther. 2020, 217, 107647. [Google Scholar] [CrossRef]
- Nigam, S.K.; Bush, K.T.; Bhatnagar, V.; Poloyac, S.M.; Momper, J.D. The Systems Biology of Drug Metabolizing Enzymes and Transporters: Relevance to Quantitative Systems Pharmacology. Clin. Pharmacol. Ther. 2020, 108, 40–53. [Google Scholar] [CrossRef]
- Zhu, C.; Nigam, K.B.; Date, R.C.; Bush, K.T.; Springer, S.A., Jr.; Wu, W.; Nigam, S.K. Evolutionary Analysis and Classification of OATs, OCTs, OCTNs, and Other SLC22 Transporters: Structure-Function Implications and Analysis of Sequence Motifs. PLoS ONE 2015, 10, e0140569. [Google Scholar] [CrossRef]
- Engelhart, D.C.; Granados, J.C.; Shi, D.; Milton, H., Jr.; Saier, S., Jr.; Baker, M.E.; Abagyan, R.; Nigam, S.K. Systems Biology Analysis Reveals Eight SLC22 Transporter Subgroups, Including OATs, OCTs, and OCTNs. Int. J. Mol. Sci. 2020, 21, 1791. [Google Scholar] [CrossRef]
- Nwafor, J.G.; Nowik, M.; Anzai, N.; Endou, H.; Wagner, C.A. Metabolic Acidosis Alters Expression of Slc22 Transporters in Mouse Kidney. Kidney Blood Press. Res. 2020, 45, 263–274. [Google Scholar] [CrossRef]
- Ganapathy, V.; Thangaraju, M.; Prasad, P.D. Nutrient transporters in cancer: Relevance to Warburg hypothesis and beyond. Pharmacol. Ther. 2009, 121, 29–40. [Google Scholar] [CrossRef]
- Yasui, Y.; Kudo, A.; Kurosaki, M.; Matsuda, S.; Muraoka, M.; Tamaki, N.; Suzuki, S.; Hosokawa, T.; Ueda, K.; Matsunaga, K.; et al. Reduced Organic Anion Transporter Expression Is a Risk Factor for Hepatocellular Carcinoma in Chronic Hepatitis C Patients: A Propensity Score Matching Study. Oncology 2014, 86, 53–62. [Google Scholar] [CrossRef]
- Lautem, A.; Heise, M.; Gräsel, A.; Hoppe-Lotichius, M.; Weiler, N.; Foltys, D.; Knapstein, J.; Schattenberg, J.M.; Schad, A.; Zimmermann, A.; et al. Downregulation of organic cation transporter 1 (SLC22A1) is associated with tumor progression and reduced patient survival in human cholangiocellular carcinoma. Int. J. Oncol. 2013, 42, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lovejoy, K.S.; Shima, J.E.; Lagpacan, L.L.; Shu, Y.; Lapuk, A.; Chen, Y.; Komori, T.; Gray, J.W.; Chen, X.; et al. Organic Cation Transporters Are Determinants of Oxaliplatin Cytotoxicity. Cancer Res. 2006, 66, 8847–8857. [Google Scholar] [CrossRef]
- Gu, J.; Wang, L.; Li, T.; Tang, S.; Wang, Y.; Zhang, W.; Jiang, X. Role and mechanism of organic cation transporter 3 in oxaliplatin treatment of colon cancer in vitro and in vivo. Oncol. Rep. 2019, 42, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Shnitsar, V.; Eckardt, R.; Gupta, S.; Grottker, J.; Müller, G.A.; Koepsell, H.; Burckhardt, G.; Hagos, Y. Expression of Human Organic Cation Transporter 3 in Kidney Carcinoma Cell Lines Increases Chemosensitivity to Melphalan, Irinotecan, and Vincristine. Cancer Res. 2009, 69, 1494–1501. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Gul, Z.G.; Liaw, C.W.; Mehrazin, R. Gender Differences in Incidence, Diagnosis, Treatments, and Outcomes in Clinically Localized Bladder and Renal Cancer. Urology 2020, 151, 176–181. [Google Scholar] [CrossRef]
- Sanchez, A.; Furberg, H.; Kuo, F.; Vuong, L.; Ged, Y.; Patil, S.; Ostrovnaya, I.; Petruzella, S.; Reising, A.; Patel, P.; et al. Transcriptomic signatures related to the obesity paradox in patients with clear cell renal cell carcinoma: A cohort study. Lancet Oncol. 2019, 21, 283–293. [Google Scholar] [CrossRef]
- Bukavina, L.; Bensalah, K.; Bray, F.; Carlo, M.; Challacombe, B.; Karam, J.; Kassouf, W.; Mitchell, T.; Montironi, R.; O’Brien, T.; et al. Epidemiology of Renal Cell Carcinoma: 2022 Update. Eur. Urol. 2022. [Google Scholar] [CrossRef]
- Lipworth, L.; Morgans, A.K.; Edwards, T.L.; Barocas, D.A.; Chang, S.S.; Herrell, S.D.; Penson, D.F.; Resnick, M.J.; Smith, J.A.; Clark, P.E. Renal cell cancer histological subtype distribution differs by race and sex. Br. J. Urol. 2015, 117, 260–265. [Google Scholar] [CrossRef]
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol. 2020, 11, 79–87. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Nigam, S.K.; Bush, K.T.; Martovetsky, G.; Ahn, S.-Y.; Liu, H.C.; Richard, E.; Bhatnagar, V.; Wu, W. The Organic Anion Transporter (OAT) Family: A Systems Biology Perspective. Physiol. Rev. 2015, 95, 83–123. [Google Scholar] [CrossRef]
- Nigam, S.K.; Bhatnagar, V. The systems biology of uric acid transporters. Curr. Opin. Nephrol. Hypertens. 2018, 27, 305–313. [Google Scholar] [CrossRef]
- Cancer Research UK. Kidney Cancer Survival Statistics. 2022. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/kidney-cancer/survival (accessed on 25 April 2022).
- Hu, D.G.; MacKenzie, P.I.; Nair, P.C.; McKinnon, R.A.; Meech, R. The Expression Profiles of ADME Genes in Human Cancers and Their Associations with Clinical Outcomes. Cancers 2020, 12, 3369. [Google Scholar] [CrossRef]
- Edemir, B. Identification of Prognostic Organic Cation and Anion Transporters in Different Cancer Entities by In Silico Analysis. Int. J. Mol. Sci. 2020, 21, 4491. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Y.; Liu, J.; Shou, Y.; Xiong, Z.; Xiong, H.; Xu, T.; Wang, Q.; Liu, D.; Liang, H.; et al. Low Expression Levels of SLC22A12 Indicates a Poor Prognosis and Progresses Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 659208. [Google Scholar] [CrossRef]
- Martovetsky, G.; Bush, K.T.; Nigam, S.K. Kidney versus Liver Specification of SLC and ABC Drug Transporters, Tight Junction Molecules, and Biomarkers. Drug Metab. Dispos. 2016, 44, 1050–1060. [Google Scholar] [CrossRef]
- Martovetsky, G.; Tee, J.B.; Nigam, S.K. Hepatocyte Nuclear Factors 4αand 1αRegulate Kidney Developmental Expression of Drug-Metabolizing Enzymes and Drug Transporters. Mol. Pharmacol. 2013, 84, 808–823. [Google Scholar] [CrossRef]
- Sweeney, D.E.; Vallon, V.; Rieg, T.; Wu, W.; Gallegos, T.F.; Nigam, S.K. Functional Maturation of Drug Transporters in the Developing, Neonatal, and Postnatal Kidney. Mol. Pharmacol. 2011, 80, 147–154. [Google Scholar] [CrossRef]
- Dai, X.; He, Q.; Jing, Z.; Yuan, J. Serum uric acid levels and risk of kidney cancer incidence and mortality: A prospective cohort study. Cancer Med. 2020, 9, 5655–5661. [Google Scholar] [CrossRef]
- Yim, K.; Bindayi, A.; McKay, R.; Mehrazin, R.; Raheem, O.A.; Field, C.; Bloch, A.; Wake, R.; Ryan, S.; Patterson, A.; et al. Rising Serum Uric Acid Level Is Negatively Associated with Survival in Renal Cell Carcinoma. Cancers 2019, 11, 536. [Google Scholar] [CrossRef]
- Li, X.; Li, Z.; Wu, X.; Zhang, M.; Xu, L.; Hao, X.; Li, H.; Qiao, P.; Wang, W. Serum uric acid variability increases the risk of postoperative chronic kidney disease in patients with renal cell carcinoma after radical nephrectomy. Urol. Oncol. Semin. Orig. Investig. 2021, 39, 500.e1–500.e7. [Google Scholar] [CrossRef]
- Wright, N.J.; Lee, S.-Y. Recent advances on the inhibition of human solute carriers: Therapeutic implications and mechanistic insights. Curr. Opin. Struct. Biol. 2022, 74, 102378. [Google Scholar] [CrossRef]
- Eraly, S.A.; Vallon, V.; Vaughn, D.A.; Gangoiti, J.A.; Richter, K.; Nagle, M.; Monte, J.C.; Rieg, T.; Truong, D.M.; Long, J.M.; et al. Decreased Renal Organic Anion Secretion and Plasma Accumulation of Endogenous Organic Anions in OAT1 Knock-out Mice. J. Biol. Chem. 2006, 281, 5072–5083. [Google Scholar] [CrossRef]
- Kaler, G.; Truong, D.M.; Sweeney, D.E.; Logan, D.W.; Nagle, M.; Wu, W.; Eraly, S.A.; Nigam, S.K. Olfactory mucosa-expressed organic anion transporter, Oat6, manifests high affinity interactions with odorant organic anions. Biochem. Biophys. Res. Commun. 2006, 351, 872–876. [Google Scholar] [CrossRef] [PubMed]
- Kaler, G.; Truong, D.; Khandelwal, A.; Nagle, M.; Eraly, S.A.; Swaan, P.; Nigam, S.K. Structural Variation Governs Substrate Specificity for Organic Anion Transporter (OAT) Homologs. J. Biol. Chem. 2007, 282, 23841–23853. [Google Scholar] [CrossRef] [PubMed]
- Vallon, V.; Rieg, T.; Ahn, S.Y.; Wu, W.; Eraly, S.A.; Nigam, S.K. Overlapping in vitro and in vivo specificities of the organic anion transporters OAT1 and OAT3 for loop and thiazide diuretics. Am. J. Physiol.-Ren. Physiol. 2008, 294, F867–F873. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Nagle, M.A.; Kouznetsova, V.L.; Tsigelny, I.F.; Nigam, S.K. Untargeted Metabolomics Identifies Enterobiome Metabolites and Putative Uremic Toxins as Substrates of Organic Anion Transporter 1 (Oat1). J. Proteome Res. 2011, 10, 2842–2851. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Bush, K.T.; Nigam, S.K. Key Role for the Organic Anion Transporters, OAT1 and OAT3, in the in vivo Handling of Uremic Toxins and Solutes. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Bhatnagar, V.; Richard, E.L.; Wu, W.; Nievergelt, C.M.; Lipkowitz, M.S.; Jeff, J.; Maihofer, A.X.; Nigam, S.K. Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: Potential role of remote sensing and signaling. Clin. Kidney J. 2016, 9, 444–453. [Google Scholar] [CrossRef]
- Eraly, S.A.; Vallon, V.; Rieg, T.; Gangoiti, J.A.; Wikoff, W.R.; Siuzdak, G.; Barshop, B.A.; Nigam, S.K. Multiple organic anion transporters contribute to net renal excretion of uric acid. Physiol. Genom. 2008, 33, 180–192. [Google Scholar] [CrossRef]
- Granados, J.C.; Richelle, A.; Gutierrez, J.M.; Zhang, P.; Zhang, X.; Bhatnagar, V.; Lewis, N.E.; Nigam, S.K. Coordinate regulation of systemic and kidney tryptophan metabolism by the drug transporters OAT1 and OAT3. J. Biol. Chem. 2021, 296, 100575. [Google Scholar] [CrossRef]
- Granados, J.C.; Nigam, A.K.; Bush, K.T.; Jamshidi, N.; Nigam, S.K. A key role for the transporter OAT1 in systemic lipid metabolism. J. Biol. Chem. 2021, 296, 100603. [Google Scholar] [CrossRef]
- Morrissey, K.M.; Wen, C.C.; Johns, S.J.; Zhang, L.; Huang, S.-M.; Giacomini, K.M. The UCSF-FDA TransPortal: A Public Drug Transporter Database. Clin. Pharmacol. Ther. 2012, 92, 545–546. [Google Scholar] [CrossRef]
- Torres, A.M.; Dnyanmote, A.V.; Granados, J.C.; Nigam, S.K. Renal and non-renal response of ABC and SLC transporters in chronic kidney disease. Expert Opin. Drug Metab. Toxicol. 2021, 17, 515–542. [Google Scholar] [CrossRef]
- Visentin, M.; Torozi, A.; Gai, Z.; Häusler, S.; Li, C.; Hiller, C.; Schraml, P.H.; Moch, H.; Kullak-Ublick, G.A. Fluorocholine Transport Mediated by the Organic Cation Transporter 2 (OCT2, SLC22A2): Implication for Imaging of Kidney Tumors. Drug Metab. Dispos. 2018, 46, 1129–1136. [Google Scholar] [CrossRef]
- Irshad, S.; Mahul-Mellier, A.-L.; Kassouf, N.; Lemarie, A.; Grimm, S. Isolation of ORCTL3 in a novel genetic screen for tumor-specific apoptosis inducers. Cell Death Differ. 2009, 16, 890–898. [Google Scholar] [CrossRef][Green Version]
- Ciarimboli, G.; Theil, G.; Bialek, J.; Edemir, B. Contribution and Expression of Organic Cation Transporters and Aquaporin Water Channels in Renal Cancer. In Reviews of Physiology, Biochemistry and Pharmacology; Springer: Cham, Switzerland, 2020; pp. 81–104. [Google Scholar] [CrossRef]
- National Cancer Institute. Renal Cell Cancer Treatment. 18 July 2022. Available online: https://www.cancer.gov/types/kidney/hp/kidney-treatment-pdq (accessed on 19 July 2022).
- Arcaniolo, D.; Manfredi, C.; Cindolo, L.; Marchioni, M.; Zukovski, E.P.; Mirone, V.; Anele, U.A.; Guruli, G.; Grob, B.M.; De Sio, M.; et al. Impact of Perioperative Blood Transfusions on the Outcomes of Patients Undergoing Kidney Cancer Surgery: A Systematic Review and Pooled Analysis. Clin. Genitourin. Cancer 2018, 17, e72–e79. [Google Scholar] [CrossRef]
- Woodward, O.M.; Köttgen, A.; Coresh, J.; Boerwinkle, E.; Guggino, W.B.; Köttgen, M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc. Natl. Acad. Sci. USA 2009, 106, 10338–10342. [Google Scholar] [CrossRef]
- Natarajan, K.; Xie, Y.; Baer, M.R.; Ross, D.D. Role of breast cancer resistance protein (BCRP/ABCG2) in cancer drug resistance. Biochem. Pharmacol. 2012, 83, 1084–1103. [Google Scholar] [CrossRef]
- Reustle, A.; Fisel, P.; Renner, O.; Büttner, F.; Winter, S.; Rausch, S.; Kruck, S.; Nies, A.T.; Hennenlotter, J.; Scharpf, M.; et al. Characterization of the breast cancer resistance protein (BCRP/ABCG2) in clear cell renal cell carcinoma. Int. J. Cancer 2018, 143, 3181–3193. [Google Scholar] [CrossRef]
- Wach, S.; Taubert, H.; Weigelt, K.; Hase, N.; Köhn, M.; Misiak, D.; Hüttelmaier, S.; Stöhr, C.G.; Kahlmeyer, A.; Haller, F.; et al. RNA Sequencing of Collecting Duct Renal Cell Carcinoma Suggests an Interaction between miRNA and Target Genes and a Predominance of Deregulated Solute Carrier Genes. Cancers 2019, 12, 64. [Google Scholar] [CrossRef]
| Symbol | EG ID | Cancer Type | Optimal Model * | KIRC Cox Coef. | Hazard Ratio |
|---|---|---|---|---|---|
| SLC22A2 | 6582 | KIRC | Base + race + treatment type | −0.433 | 0.649 |
| SLC22A4 | 6583 | KIRC | Base + race + treatment type | −0.332 | 0.717 |
| SLC22A5 | 6584 | KIRC | Base + stage | −0.354 | 0.702 |
| SLC22A6 | 9356 | KIRC | Base + race + treatment type | −0.413 | 0.661 |
| SLC22A7 | 10864 | KIRC | Base + race + prior malignancy | −0.319 | 0.727 |
| SLC22A8 | 9376 | KIRC | Base + race + treatment type | −0.465 | 0.628 |
| SLC22A11 | 55867 | KIRC | Base + race + treatment type | −0.384 | 0.681 |
| SLC22A12 | 116085 | KIRC | Base + race + treatment type | −0.389 | 0.678 |
| SLC22A24 | 283238 | KIRC | Base + race + treatment type | −0.442 | 0.643 |
| SLC22A2 | 6582 | KIRP | Base + prior malignancy | −0.911 | 0.631 |
| SLC22A13 | 9390 | KIRP | Base + race + lymph node involvement | −0.668 | 0.513 |
| SLC22A18 | 5002 | KIRP | Base + race + tumor size/prog | −0.644 | 0.525 |
| SLC22A24 | 283238 | KIRP | Base + race + stage | −0.757 | 0.469 |
| Sym | Alias | EG ID | Subclade * | Updated Grouping ** | Specificity | KIRC Cox Coef. | KIRC BH-adj p-val | KIRP Cox Coef. | KIRP BH-adj p-val |
|---|---|---|---|---|---|---|---|---|---|
| SLC22A1 | OCT1 | 6580 | OCT | OCT | Multi- | 0.2069 | &0.02783 | 0.0921 | 0.69794 |
| SLC22A2 | OCT2 | 6582 | OCT | OCT | Multi- | −0.3411 | +& 0.00033 | −0.9214 | 6.94× 10−5 |
| SLC22A3 | OCT3 | 6581 | OCT | OCT | Oligo- | 0.0074 | 0.94686 | −0.3733 | 0.08201 |
| SLC22A4 | OCTN1 | 6583 | OCTN | OCTN- related | Oligo- | −0.3159 | 0.00029 | −0.3289 | 0.16737 |
| SLC22A5 | OCTN2 | 6584 | OCTN | OCTN- related | Oligo- | −0.2901 | #0.00281 | −0.4169 | 0.07343 |
| SLC22A6 | OAT1 | 9356 | OAT | OATS1 | Multi- | −0.3473 | +0.00015 | −0.0640 | 0.82770 |
| SLC22A7 | OAT2 | 10864 | OAT | OATS2 | Oligo- | −0.2599 | 0.00814 | −0.3251 | 0.15414 |
| SLC22A8 | OAT3 | 9376 | OAT | OATS1 | Multi- | −0.3883 | 9.46× 10−5 | NA | NA |
| SLC22A11 | OAT4 | 55867 | OAT | OATS3 | Oligo- | −0.3384 | 0.00049 | −0.3131 | 0.12484 |
| SLC22A12 | URAT1 | 116085 | OAT | OATS3 | Mono- | −0.3211 | ^ 0.00135 | −0.3657 | 0.07921 |
| SLC22A13 | ORCTL3 | 9390 | OAT-like | OAT-like | Mono- | −0.2307 | 0.02281 | −0.4095 | 0.04680 |
| SLC22A14 | ORCTL4 | 9389 | OAT-like | OAT-like | N/A | 0.2288 | 0.01846 | −0.2331 | 0.32010 |
| SLC22A15 | FLIPT1 | 55356 | OCTN- related | OCTN-related | Mono- | 0.1598 | 0.08677 | 0.2117 | 0.33305 |
| SLC22A17 | BOCT | 51310 | OAT- related | OAT-related | Mono- | 0.0689 | 0.54035 | −0.2479 | 0.23082 |
| SLC22A18 | ORCTL2 | 5002 | OAT- related | OAT-related | N/A | −0.0784 | 0.47059 | −0.4283 | 0.04613 |
| SLC22A23 | N/A | 63027 | OAT- related | OAT-related | Oligo- | −0.0992 | 0.35122 | 0.4132 | 0.05213 |
| SLC22A24 | N/A | 283238 | OAT | OATS4 | Oligo- | −0.3146 | 0.00200 | −0.5923 | 0.00669 |
| Oncologic Variable | Comparison | KIRP-SLC22 (p adj. < 0.1) | KIRC-SLC22 (p adj. < 0.1) |
|---|---|---|---|
| Stage | Stage I v Stage II | 0 | 2 |
| Stage II v Stage III | 0 | 1 | |
| Stage III v Stage IV | 0 | 0 | |
| T | T1 v T2 | 0 | 0 |
| T2 v T3 | 0 | 0 | |
| T3. v T4 | 0 | 6 | |
| M | M0 v M1 | 0 | 4 |
| N | N0 v N1 | 5 | 2 |
| PriMalig | Yes v No | 0 | 0 |
| TxType | Pharm v Rad | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Whisenant, T.C.; Nigam, S.K. Organic Anion Transporters (OAT) and Other SLC22 Transporters in Progression of Renal Cell Carcinoma. Cancers 2022, 14, 4772. https://doi.org/10.3390/cancers14194772
Whisenant TC, Nigam SK. Organic Anion Transporters (OAT) and Other SLC22 Transporters in Progression of Renal Cell Carcinoma. Cancers. 2022; 14(19):4772. https://doi.org/10.3390/cancers14194772
Chicago/Turabian StyleWhisenant, Thomas C., and Sanjay K. Nigam. 2022. "Organic Anion Transporters (OAT) and Other SLC22 Transporters in Progression of Renal Cell Carcinoma" Cancers 14, no. 19: 4772. https://doi.org/10.3390/cancers14194772
APA StyleWhisenant, T. C., & Nigam, S. K. (2022). Organic Anion Transporters (OAT) and Other SLC22 Transporters in Progression of Renal Cell Carcinoma. Cancers, 14(19), 4772. https://doi.org/10.3390/cancers14194772






